Abstract
Peroxisome proliferator-activated receptors (PPARs) are ligand-dependent transcription factors, and it is assumed that the biological effects of these receptors depend on interactions with recently identified coactivators, including steroid receptor coactivator-1 (SRC-1). We assessed the in vivo function of SRC-1 on the PPARα-regulated gene expression in liver by generating mice in which the SRC-1 gene was inactivated by gene targeting. The homozygous (SRC-1−/−) mice were viable and fertile and exhibited no detectable gross phenotypic defects. When challenged with a PPARα ligand, such as ciprofibrate or Wy-14,643, the SRC-1−/− mice displayed typical pleiotropic responses, including hepatomegaly, peroxisome proliferation in hepatocytes, and increased mRNA and protein levels of genes that are regulated by PPARα. These alterations were indistinguishable from those exhibited by SRC-1+/+ wild-type mice fed either ciprofibrate- or Wy-14,643-containing diets. These results indicate that SRC-1 is not essential for PPARα-mediated transcriptional activation in vivo and suggest redundancy in nuclear receptor coactivators.
Peroxisomes in hepatocytes can be induced to proliferate in response to structurally diverse nonmutagenic chemicals designated as peroxisome proliferators (1, 2). These agents form a broad group of compounds of industrial, pharmaceutical, and agricultural value and include certain phthalate ester plasticizers, leukotriene D4 antagonists, and hypolipidemic drugs, such as clofibrate, ciprofibrate and Wy-14,643, among others (2). The induction of peroxisome proliferation is mediated by peroxisome proliferator-activated receptor α (PPARα), a member of the family of ligand-dependent nuclear transcription factors that regulate the expression of genes associated with lipid metabolism and adipocyte differentiation (3–6). The PPAR subfamily has three isotypes (α, β or δ, and γ), which exhibit distinct patterns of tissue distribution and differ considerably in their ligand-binding domains and ligand specificities, suggesting that they possibly perform different functions in different cell types (5–8). Like other members of the nuclear receptor superfamily, PPARs possess a central DNA-binding domain that recognizes PPAR response elements (PPRE) in the promoter regions of target genes (2, 3, 9). PPARs form a heterodimer with the 9-cis-retinoic acid receptor (RXR), to bind PPRE in DNA and transcriptionally activate target genes (9).
Induction of peroxisome proliferation and the peroxisomal fatty acid β-oxidation system in rats and mice by sustained activation of PPARα either by exogenous or endogenous ligands leads to the development of liver tumors (10, 11). Recently, activation of PPARγ by its ligands, troglitazone and rosiglitazone, has been shown to induce a modest increase in the incidence of spontaneously-occurring colon tumors in Min+/− mice lacking one copy of the APC tumor suppressor gene (12, 13). Because these observations raise a potential concern of risk to humans, it becomes essential to explore the molecular mechanisms underlying tissue and species responses to PPAR ligands. Transcriptional stimulation of gene expression by nuclear receptors by ligands involves the participation of basal transcription factors, including TATA-binding protein and TFIIB, and nuclear receptor coactivator proteins to form a stable preinitiation macromolecular complex (14, 15). The coactivators identified during the past 3 years include: steroid receptor coactivator-1 (SRC-1) (16), SRC-2 [TIF-2/GRIP-1 (17, 18)], SRC-3 [ACTR (19)], AIB1 (20), p/CIP (21), and RAC3 (22), CREB-binding protein (CBP)/p300 (23, 24), PBP/TRAP220 (25, 26), PGC-1 (27), and ARA70 (28). We cloned and identified mouse SRC-1 (29), a homologue of human SRC-1 (16), and PBP (25) as coactivators of PPAR. To study the physiological role of SRC-1 in PPAR-mediated induction of pleiotropic responses, we inactivated the SRC-1 gene in mice by homologous recombination. We report that homozygous SRC-1 mutants are viable and fertile and respond to prototypical PPARα ligands in a manner similar to that of SRC-1+/+ wild-type mice, suggesting possible physiological redundancy in vivo of coactivators in PPARα-mediated transcriptional activation.
MATERIALS AND METHODS
Gene Targeting.
P1 Genomic clone (#11189) containing the SRC-1 gene was obtained by screening a mouse 129/Sv P1 bacteriophage library (Genome Systems, St. Louis) by using PCR with primers 5′-TGACAGTAATTCTGGAATGTCAAT-3′ and 5′-GGGATTGCTGCTCTGGGAAC-3′. P1 clone DNA was subjected to restriction analysis, and a 16-kb PstI fragment was isolated for use in the construction of targeting vector (Fig. 1A). This fragment covered two exons: exon A, containing residues 317Met-366Arg and exon B, containing residues 367Glu-873Arg (29). The targeting construct was assembled in the pPNT targeting vector by using a 4.5-kb NcoI/HindIII fragment and a 4-kb PstI/HindIII fragment of homologous mouse SRC-1 genomic sequences 5′ and 3′, respectively, of the phosphoglycerate kinase promoter/neomycin-resistance complementary DNA (phosphoglycerate kinase-neomycin) (Fig. 1A). The targeting vector also contained the herpes simplex thymidine kinase (hsv-tk) gene, which allowed the use of a positive–negative selection scheme. The final construct, designated pPNT-SRC-1, is illustrated in Fig. 1A.
Figure 1.
Generation of SRC-1-deficient mice. (A) Schematic representation of the mouse SRC-1 gene, targeting vector, and the structure of the locus after gene targeting. Two exons, A and B, in the SRC-1 gene are shown as closed boxes. Restriction sites are indicated. Location of hybridization probes used for Southern blot analysis and of PCR primers used for genotyping are shown. (B) Southern blot analysis of genomic DNA. Genomic DNA (5 μg) isolated from the tail tips of 2- to 3-week-old pups was digested with PstI, transferred to membranes, and hybridized with the 5′ probe (probe 1 shown in A). Lanes: SRC-1+/+ wild type, SRC-1+/− heterozygous, SRC-1−/− homozygous mice. The 16-kb band corresponds to the wild-type allele and the 7.5-kb band corresponds to the rearranged allele. (C) RT-PCR analysis of liver (L), kidney (K), and colon (C) of SRC-1+/+ and SRC-1−/− mice by using primers from different regions (see Methods). The expected sizes of the PCR products (465, 240, and 1,533 bp) are indicated. Lanes +/+, wild type and −/−, homozygote. The SRC-1 transcript is not detected in tissues of SRC-1−/− mice. (D) Northern blot analysis using Poly(A)+ RNA (5 μg/lane) isolated from liver probed with SRC-1 or β-actin cDNA reveals absence of SRC-1 transcript in SRC-1 null mouse liver. Note the presence of β-actin mRNA both. (E) Western blot analysis for SRC-1 expression in wild-type and SRC-1 mutant mice. Liver extracts (100 μg) were immunoblotted by using a monoclonal antibody; no immunodetectable SRC-1 protein is seen in SRC-1−/− mouse.
Generation of SRC-1 Mutant Mice.
NotI-linearized targeting vector (30 μg) was electroporated into BK4 embryonic stem (ES) cells that were selected in 200 μg/ml G418 and 2 μM ganciclovir (30, 31). One hundred G418-ganciclovir-resistant ES colonies were picked up and subjected to Southern analysis; seven were identified as having one normal and one targeted disrupted allele. Two positive ES clones were injected into 3.5-day-old C57/BL6J blastocysts and transferred into pseudopregnant CBAF1 foster female recipients. The resulting chimeras were mated with C57/BL6J mice and germline transmission was ascertained by coat color and confirmed by Southern analysis with probes 1 and 2. F1 heterozygous siblings for the disrupted SRC-1 gene were then mated to obtain homozygous SRC-1 null mice.
Genotype of SRC-1 Mutant Mice.
The offspring from subsequent breeding were genotyped by PCR amplification and confirmed by Southern analysis as needed. Two primers, primers P1 (5′-CCACCATCCAACAACAACATGG-3′) and P2 (5′-AGCACTGTTGTCGCTGTTGTC-3′), derived from exon B of SRC-1 shown in Fig. 1A, were designed to a detect wild-type allele and two primers, primers P3 (5′-TGAATGAACTGCAGGACGAGG-3′) and P4 (5′-CCACAGTCGATGAATCCAGAA-3′) from the neomycin cassette were used to detect the SRC-1 targeted allele.
Reverse Transcription–PCR (RT–PCR).
To confirm the absence of SRC-1 mRNA in SRC-1−/− mice, RT-PCR was performed by using primers 5′-TTTCAAGAAGTGATGACTCGTGG-3′ in exon A either with 5′-GGGATTGCTGCTCTGGGAAC-3′or with 5′-CCAGGATTGACTGAGGGATT-3′, designed from the deleted region and the region 5′- of the deleted region in exon B, with resulting products of 465 bp and 240 bp, respectively. Another pair of primers, 5′-TGACAGTAATTCTGGATG-3′ in exon B undeleted region and 5′-AACTGGTTATCGATCGCTT-3′ from the exon following exon B amplified the 1,533-bp fragment. Total RNA extracted from liver, kidney, and colon from wild-type and SRC-1−/− mice was used in the one-step RT-PCR system (GIBCO/BRL) for cDNA synthesis.
Treatment with Peroxisome Proliferators.
Wild-type and SRC-1−/− mice were fed powdered diet with or without PPARα ligands, ciprofibrate (0.0125% wt/wt) or Wy-14,643 (0.1% wt/wt) for 4 to 14 days. For cell proliferation analysis, mice were given bromodeoxyuridine (0.5 mg/ml) in drinking water for 4 days and their livers processed for immunohistochemistry. To assess the dose response, groups of three wild-type and SRC-1−/− mice were given a single intragastric dose of ciprofibrate (150, 75, 37.5, 18.75, or 0 mg/kg body weight) and killed 24 h later.
Western Blot Analysis.
Liver extracts were subjected to 10% SDS/PAGE and transferred to nitrocellulose membranes. Immunoblotting was performed by using rabbit polyclonal antibodies against rat peroxisomal acyl-CoA oxidase (AOX), peroxisomal L- and D-type bifunctional protein (enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase) (L-PBE, and D-PBE), peroxisomal 3-ketoacyl-CoA thiolase (THL), catalase (CAT), urate oxidase, sterol carrier protein x, short-chain acyl-CoA dehydrogenase (SCAD), medium-chain acyl-CoA dehydrogenase (MCAD), and very long-chain acyl-CoA dehydrogenase (VLCAD), as described (32). Antibodies against SRC-1 were from a commercial source, or were a generous gift of Bert O’Malley, Baylor College of Medicine, Waco, TX. The membranes were incubated with the primary antibody followed by alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma).
Northern Hybridization.
For northern analysis, total RNA (20 μg) extracted from wild-type and SRC-1−/− mice by Trizol reagent (GIBCO/BRL) was glyoxylated, separated on 0.8% agarose gel, and transferred to nylon membrane. cDNA probes used for Northern blotting included AOX, L-PBE, THL, cytochrome P450 (CYP)4A1, CYP4A3, β-actin, and ribosomal RNA (28 S). Poly(A)+ RNA extracted from liver was used for SRC-1 analysis. Changes in mRNA levels were estimated by densitometric scanning of autoradiograms.
Histology and Electron Microscopy.
For light microscopy, tissues were fixed in 10% neutral buffered formalin and embedded in paraffin by using standard procedures. Sections (4-μM thick) were cut and stained with hematoxylin and eosin. For cytochemical localization of catalase, tissues were fixed in 1.5% glutaraldeyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 4 h at 4°C and processed as described (10, 30). Semithin sections, without counterstain, were examined by light microscopy. Ultrathin sections for electron microscopy were contrasted with uranyl acetate and lead citrate.
RESULTS
Disruption of PPAR-Binding Site of SRC-1 Coactivator.
The mouse SRC-1 cDNA that we and others have cloned (23, 24, 29) is the homologue of human SRC-1(16). We have shown that it functions as a coactivator for PPARs (29). We identified two PPAR binding regions in SRC-1 (29); one region located between residues 620–789 displayed stronger interaction with PPAR than the second one, located at the carboxy terminus (residues 1231–1447). The region between amino acids 620 and 789 contains three highly conserved LXXLL motifs, demonstrated recently to be sufficient and necessary for the binding of several coactivators to nuclear receptors (21, 33). The carboxy-terminal region of mouse SRC-1 contains one LXXLL motif (29). Accordingly, we decided to disrupt the region containing the first PPAR interacting domain with three LXXLL motifs. We isolated a 16-kb SRC-1 genomic fragment that contained two exons, one of which, designated exon B, includes amino acid residues 620–789. The gene targeting vector, pPNT-SRC-1, was designed to delete this region (exon B shown in Fig. 1A) and to replace it with phosphoglycerate kinase-neomycin gene (Fig. 1A). A correct gene targeting event would result in a protein, if any, that lacks the three critical LXXLL motifs functional in this region.
Generation of Mice with a Modified SRC-1 Gene.
pPNT-SRC-1 was introduced into BK4 ES cells by electroporation. Of the 100 G418-ganciclovir-resistant colonies analyzed by Southern blotting, seven displayed one normal and one disrupted SRC-1 allele. ES cells from two clones were injected into C57BL/6J blastocysts to generate chimeras. Chimeras from both lines were able to transmit the mutant allele to the offspring after outbreeding with C57/B6J.
Mice heterozygous for the disrupted SRC-1 gene were phenotypically normal. These heterozygous F1 mice were intercrossed to obtain homozygous mutant offspring (Fig. 1B). The F1 progeny exhibited the predicted Mendelian frequency of 25% homozygous mutant offspring and the SRC-1−/− mice exhibited no apparent morphological abnormalities. Both male and female homozygous mice grew normally, survived longer than 10 months, and were fertile. No significant differences in body weight and liver weight were found between age-matched SRC-1+/+ and SRC-1−/− mice. The absence of expression of the SRC-1 transcript in liver, kidney, and colon of SRC-1 null mice was confirmed by RT-PCR (Fig. 1C). Northern blot analysis using Poly(A)+ RNA isolated from the liver also showed the absence of SRC-1 mRNA in SRC-1−/− mouse (Fig. 1D).Western blotting revealed no immunodetectable SRC-1 protein in the liver of SRC-1 mutant mice (Fig. 1E).
Induction of Peroxisome Proliferation in SRC-1 Mutant Mice.
Wild-type mice treated with a peroxisome proliferator, such as ciprofibrate or Wy-14,643, exhibit profound increases in the number and volume density of peroxisomes in their liver cells (1). To assess the impact of SRC-1 gene disruption, if any, on PPARα-regulated pleiotropic responses, including liver cell proliferation and hepatic peroxisome proliferation, we studied the inductive response after dietary feeding of ciprofibrate or Wy-14,643. No significant differences in hepatomegaly and bromodeoxyuridine labeling were noted between wild-type and SRC-1 null mice treated with a peroxisome proliferator for 4 days (Fig. 2). Light microscopic evaluation of sections of liver that were processed to visualize peroxisomal catalase revealed marked increases in the number and size of peroxisomes in hepatic parenchymal cells of SRC-1+/+ wild-type and SRC-1 null mice treated with a peroxisome proliferator (Fig. 3). The magnitude of peroxisome proliferation, as assessed by light (Fig. 3) and electron microscopy (not illustrated), was similar in both groups. Thus, SRC-1 null mice exhibited no resistance to either ciprofibrate or Wy-14,643 at the dose level administered, as evidenced by hepatomegaly and peroxisome proliferation.
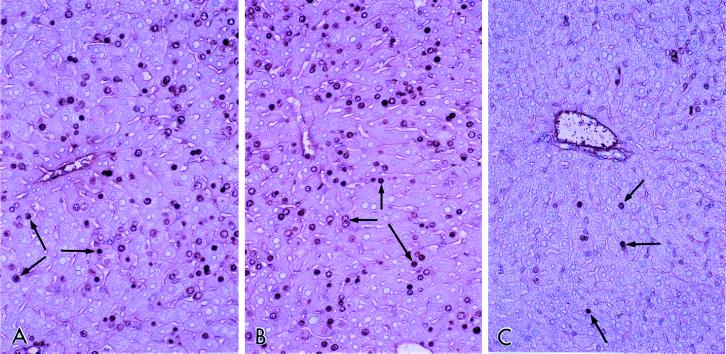
Figure 2.
Cell proliferation in the liver. SRC-1+/+ wild type and SRC-1−/− mice were fed Wy-14,643 (0.1% wt/wt) in the diet for 4 days and were also given bromodeoxyuridine in drinking water (0.5 mg/ml). Liver sections were processed for immunohistochemical demonstration of bromodeoxyuridine uptake. Nuclear labeling (arrows) is seen in hepatocytes. (A) SRC-1+/+ and (B) SRC-1−/− mice fed Wy-14,643. (C) SRC−/− mice fed a control diet.
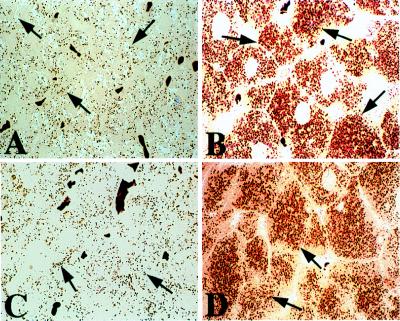
Figure 3.
Peroxisome proliferation in liver parenchymal cells of wild-type and SRC-1 mutant mice treated with ciprofibrate. Light microscopic appearance of liver as revealed in semithin (0.5 μM thick) sections of tissue that was processed for the cytochemical localization of peroxisomal catalase by using the alkaline 3′,3′-diaminobenzidine substrate. A and C represent SRC-1+/+ wild-type and SRC-1−/− mice, respectively, maintained on control chow. Peroxisomes appear as brown dots (arrows) randomly distributed in the cytoplasm. B and D represent SRC-1+/+ and SRC-1−/− mice, respectively, which were fed a diet containing ciprofibrate for 2 weeks. Both wild-type and SRC-1 mutant mice display a robust degree of peroxisome proliferation, as evidenced by numerous brown granules (arrows).
PPARα-Regulated Fatty Acid-Metabolizing Enzymes in the Liver of SRC-1 Mutant Mice.
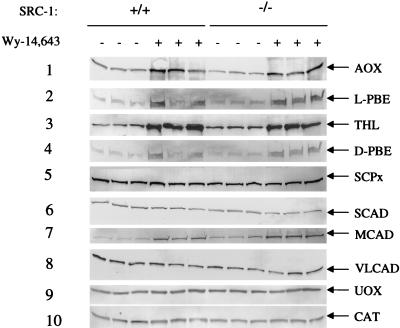
To assess the influence of SRC-1 on PPARα function, we evaluated fatty acid-metabolizing enzymes in SRC-1 null mutants by immunoblotting. We examined constitutive and inducible levels of fatty acid-metabolizing enzymes in liver. Constitutive levels of hepatic peroxisomal fatty acid β-oxidation enzyme expression of AOX, L-PBE, and THL were not significantly different between wild-type and SRC-1 null mice (Fig. 4). These three enzymes were induced >25-fold in both wild-type and SRC-1 null mice fed a diet containing Wy-14,643, a PPARα ligand, for two weeks (Fig. 4). The constitutive levels of expression in liver of peroxisomal D-PBE, sterol carrier protein x, CAT, and urate oxidase in wild-type and SRC-1-deficient mice were essentially similar and only a ≈3-fold increase in the levels of these proteins occurred after Wy-14,643 administration in both SRC-1+/+ and SRC-1−/− mice (Fig. 4). We also determined the constitutive expression of hepatic mitochondrial enzymes SCAD, MCAD, and VLCAD that are involved in lipid metabolism and found no significant differences between wild-type and SRC-1 null mice. There was a significant increase in the expression of MCAD in both SRC-1+/+ and SRC-1−/− mouse liver following Wy-14,643 treatment. We also examined the effect of administration of ciprofibrate on fatty acid-metabolizing enzymes and found no significant differences in responsiveness between SRC-1+/+ and SRC-1−/− mice (data not shown).
Figure 4.
Western blot analysis of selected fatty acid-metabolizing and other enzymes in liver. Liver homogenates from SRC-1+/+ and SRC-1−/− mice fed either control or Wy-14,643-containing diet were subjected to SDS/PAGE and immunoblotting (three mice for each group). AOX (lane 1; 5 μg), L-PBE (lane 2; 20 μg), THL (lane 3; 5 μg), D-PBE (lane 4; 20 μg), sterol carrier protein x (lane 5; 10 μg), SCAD (lane 6; 20 μg), MCAD (lane 7; 20 μg), VLCAD (lane 8; 2 μg), urate oxidase (lane 9; 2 μg) and CAT (lane 10; 2 μg).
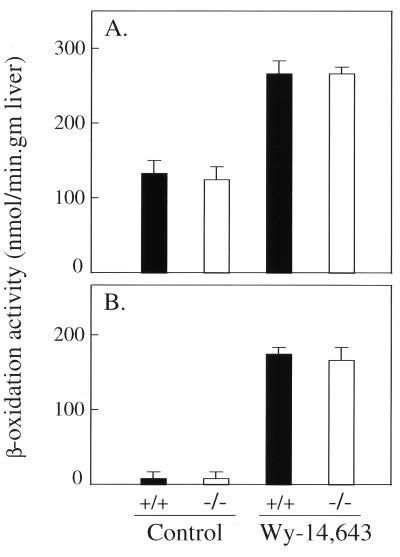
To extend these observations further, we measured total hepatic β-oxidation using palmitic acid (C-16). The basal level of total fatty acid β-oxidation in wild-type and SRC-1 null mice was similar and, as expected, administration of Wy-14,643, a PPARα ligand, caused a significant increase in fatty acid metabolism in wild-type and SRC-1−/− mice (Fig. 5A). The constitutive level of peroxisomal β-oxidation in the liver of SRC-1 null mice was similar to that found in the wild-type mice (Fig. 5B). Both SRC-1+/+ and SRC-1−/− mice exhibited an identical increase in β-oxidation activity after Wy-14,643 treatment (Fig. 5B).
Figure 5.
Hepatic fatty acid β-oxidation in SRC-1+/+ wild type and SRC-1−/− mice. Fatty acid β-oxidation of palmitic acid was measured and the values expressed as nmol/min per gm liver. (A) Mitochondrial plus peroxisomal and (B) peroxisomal. Solid bars and open bars are from wild-type and SRC-1 null mice, respectively, fed a control or Wy-14,643-containing (0.1% wt/wt) diet for 7 days. A significant difference was not found between wild-type and SRC-1 null mutants fed a peroxisome proliferator, but the induction is significant in both when compared with their respective untreated controls.
Expression of mRNAs.
To further affirm PPARα activation, the structural findings are extended by analysis of the hepatic mRNA levels by Northern blotting. Constitutive levels of AOX, L-PBE, THL, and CYP4A1 mRNA were similar in the livers of both wild-type and SRC-1 null mice (Fig. 6). The AOX, L-PBE, THL, and CYP4A1 hepatic mRNA levels increased markedly and identically in both wild-type and SRC-1-deficient mice fed a peroxisome proliferator (Fig. 6).
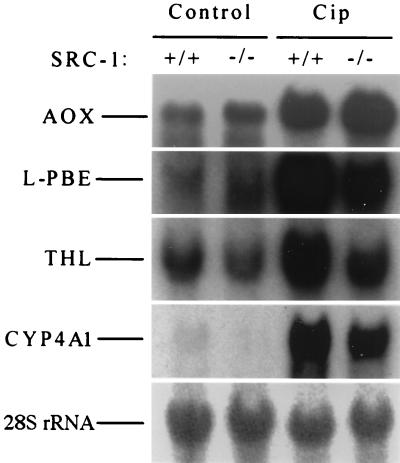
Figure 6.
Northern blot analysis of total RNA extracted from liver of SRC-1+/+ wild type and SRC-1−/− mice. RNA (20 μg) from mice fed a control or ciprofibrate- (0.0125% wt/wt) containing diet (Cip) for 2 weeks was probed with different random-primed 32P-labeled cDNA probes as shown. AOX, L-PBE, THL, CYP4A1, and CYP4A3 (not shown) are genes that contain PPRE in their promoters and are regulated by PPARα. 28S rRNA is used as loading indicator.
Dose Response to Ciprofibrate.
To determine whether SRC-1 null mice are possibly less sensitive to a PPARα ligand at lower dose levels than wild-type mice, we administered a single dose of ciprofibrate (ranging from 0 to 150 mg/kg body weight) by gavage and analyzed changes in hepatic CYP4A1 and L-PBE mRNA levels 24 hr after dosing by Northern blotting. Both wild-type and SRC-l null mice responded in a similar manner to different dose levels of ciprofibrate administered, indicating that response to PPARα ligand is not subdued in SRC-1-deficient mice (data not shown).
DISCUSSION
Ligand-inducible transcriptional activation by nuclear receptors is now known to involve the participation of proteins termed coactivators that link the receptors with the basal transcriptional apparatus (14–16, 23, 34). The interaction of coactivators with nuclear receptors is ligand dependent and occurs through the carboxy-terminal helical region of the receptor, referred to as activation function-2 domain (15, 35). This region serves as the ligand-binding pocket and undergoes a conformational change that facilitates protein–protein interactions, enabling the recruitment of coactivator proteins for efficient gene transcription (23). Some of these coactivators, for example CBP/p300 (36), SRC-1 (37), and ACTR (19), possess intrinsic histone acetyltransferase activities capable of modifying the chromatin organization of the target gene promoter regions.
Of the various nuclear receptor coactivators identified to date, SRC-1 (29), CBP/p300 (38, 39), PBP (25), and PGC-1 (27) have been shown to mediate transcriptional activation of PPARs. Mammalian PPARs are members of the nuclear receptor superfamily of ligand-activated transcription factors that bind to PPRE as heterodimers with RXR and regulate expression of genes involved in lipid homeostasis and differentiation (3, 7, 10, 40). PPRE is a degenerate direct repeat of the canonical AGGTCA sequence separated by a single nucleotide (so-called DR1) (2, 9, 35, 41) that is located in the promoter region of PPAR-regulated genes. Several structurally diverse synthetic peroxisome proliferators have long been known to induce predictable pleiotropic responses by a receptor-mediated mechanism (1, 2), and the recent identification of additional synthetic and natural agonists of PPARα and PPARγ raises issues regarding the cell- and species-specific nature of their biological effects in extrapolating potential risk to humans from long-term exposure (11–13, 42). Responses to PPAR agonists may well be influenced by pharmacokinetics, relative abundance of PPAR isotypes in specific cell types and their affinity for agonists, the nature of PPRE in the upstream regions of target genes, the extent of competition or crosstalk among nuclear transcription factors for PPAR heterodimerization partner RXR, and the modulating role of coactivators and corepressors (29, 30, 43). Given the plethora of pleiotropic responses elicited by natural and synthetic PPAR agonists and their extensive clinical use and importance, it is of great interest and relevance to dissect the molecular basis for each of the above variables. Our laboratory has characterized mouse SRC-1 (29) and PBP (25) genes and identified them as PPAR coactivators. Studies from other laboratories have shown that CBP/p300 (38, 39) and PGC-1 (27) also enhance transactivation by PPARs. As a first step in exploring the physiological role in vivo of various coactivators in nuclear receptor activity in general and in PPAR-regulated gene expression in particular, we have used homologous recombination to disrupt the mouse SRC-1 gene and have investigated the response of SRC-1 null mice to PPARα agonists.
Disruption of the SRC-1 gene by deletion of an exon that contains three conserved helical motifs of the consensus sequence LXXLL necessary for interaction with nuclear receptors led to the generation of SRC-1 null mutants. These mutant animals grow and reproduce normally, suggesting that their somatic and sexual development is essentially normal. The major finding of this study is that mice lacking the SRC-1 gene display no discernible differences when compared with wild-type littermates. These observations suggest that the absence of this coactivator does not adversely affect the developmental and physiological profiles, implying possible redundancy of coactivators in nuclear receptor function. In this study, mice lacking SRC-1 did not exhibit any appreciable differences, as compared with wild-type animals, in liver weight, hepatocellular proliferation, increases in peroxisome population in liver cells, or changes in the levels of expression of enzymes involved in fatty acid metabolism when challenged with PPARα ligands. The absence of significant differences in response between wild-type and SRC-1 mutant mice after exposure to peroxisome proliferators suggests that SRC-1 coactivator does not play an essential role in PPARα-regulated transcriptional activation in vivo. Alternatively, the SRC-1 is so pivotal that the loss of its function is compensated for by other, possibly redundant, coactivators, such as CBP, PBP, PGC-1, or other yet-to-be-discovered proteins.
In this study, we examined the immediate or early responses to PPARα agonists in the liver of SRC-1 mutant mice and, judging from the robust peroxisome proliferative response, one can speculate that these animals would most likely develop liver tumors after chronic exposure to peroxisome proliferators. Additional studies are nonetheless needed to test the response of SRC-1 null mice to PPARγ ligands and other stresses before one can conclude with certainty that SRC-1 does not play an appreciable role in PPAR-regulated transcriptional activation in vivo. Because SRC-1 and other coactivators identified to date interact in vitro with other nuclear receptors, the SRC-1 null mice would serve as valuable animal models to test the functional implications of this coactivator in vivo on the functioning of other nuclear receptors. In a recent report, SRC-1 gene-disrupted mice were found to exhibit somewhat subdued responses to sex hormonal stimuli after orchiectomy or ovarietcomy, although the intact animals were fertile and appeared hormonally indistinguishable from SRC-1+/+ mice (44). The partial hormone resistance of target tissues in castrated SRC-1 null mice suggests that coactivators such as SRC-1 may function more efficiently in certain cell types and may possibly play a more specific role in transcriptional activity in vivo of a subset of nuclear receptors (40, 43, 45). Additional data are needed to explore the role of various coactivators by generating mutant mice with defects in one or more coactivator functions.
Acknowledgments
This work was supported in part by National Institutes of Health Grants R37GM23750 (to J.K.R.) and HL42630 (to N.M.), Veterans Administration merit awards (to A.V.Y. and M.S.R.), and the Joseph L. Mayberry, Sr., Endowment Research Fund.
ABBREVIATIONS
- PPAR
peroxisome proliferator-activated receptor
- SRC-1
steroid receptor coactivator-1
- PPRE
peroxisome proliferator response element
- RXR
9-cis-retinioc acid receptor
- CBP
CREB-binding protein
- AOX
fatty acyl-CoA oxidase
- CYP
cytochrome P450
- L-PBE and D-PBE
L- and D-type peroxisomal bifunctional protein (enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase)
- THL
peroxisomal 3-ketoacyl-CoA thiolase
- CAT
catalase
- SCAD
short-chain acyl-CoA dehydrogenase
- MCAD
medium chain acyl-CoA dehydrogenase
- VLCAD
very long-chain acyl-CoA dehydrogenase
- ES
embryonic stem
- RT-PCR
reverse transcription–PCR
References
- 1.Reddy J K, Krishnakantha T P. Science. 1975;190:787–789. doi: 10.1126/science.1198095. [DOI] [PubMed] [Google Scholar]
- 2.Reddy J K, Chu R. Ann N Y Acad Sci. 1996;804:176–201. doi: 10.1111/j.1749-6632.1996.tb18616.x. [DOI] [PubMed] [Google Scholar]
- 3.Issemann I, Green S. Nature (London) 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 4.Lee S S-T, Pineau T, Drago J, Lee E J, Owens J W, Kroetz D L, Fernandez-Salguero P M, Westphal H, Gonzalez F J. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y, Alvares K, Huang Q, Rao M S, Reddy J K. J Biol Chem. 1993;268:26817–26820. [PubMed] [Google Scholar]
- 6.Tontonoz P, Hu E, Graves R A, Budavari A I, Spiegelman B M. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 7.Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 8.Jain S, Pulikuri S, Zhu Y, Qi C, Kanwar Y S, Yeldandi A Y, Rao M S, Reddy J K. Am J Pathol. 1998;153:349–354. doi: 10.1016/s0002-9440(10)65577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kliewer S A, Umesono K, Noonan D J, Heyman R A, Evans R M. Nature (London) 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy J K, Azarnoff D L, Hignite C F. Nature (London) 1980;283:397–398. doi: 10.1038/283397a0. [DOI] [PubMed] [Google Scholar]
- 11.Fan C-Y, Pan J, Usuda N, Yeldandi A V, Rao M S, Reddy J K. J Biol Chem. 1998;273:15639–15645. doi: 10.1074/jbc.273.25.15639. [DOI] [PubMed] [Google Scholar]
- 12.Lefebvre A-M, Chen I, Desreumaux P, Najib J, Fruchart J-C, Geboes K, Briggs M, Heyman R, Auwerx J. Nat Med. 1998;4:1053–1057. doi: 10.1038/2036. [DOI] [PubMed] [Google Scholar]
- 13.Saez E, Tontonoz P, Nelson M C, Alvarez J G A, U, T M, Baird S M, Thomazy V A, Evans R M. Nat Med. 1998;4:1058–1061. doi: 10.1038/2042. [DOI] [PubMed] [Google Scholar]
- 14.Janknecht R, Hunter T. Nature (London) 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 15.Glass C K, Rose D W, Rosenfeld M G. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 16.Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 17.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 18.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 20.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 21.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Gomes P J, Chen J D. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, et al. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 24.Yao T-P, Ku G, Zhou N, Scully R, Livingston D M. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y, Qi C, Jain S, Rao M S, Reddy J K. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 26.Yuan C-X, Ito M, Fondell J D, Fu Z-Y, Roeder R G. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 28.Yeh S, Chang C. Proc Natl Acad Sci USA. 1997;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y, Qi C, Calandra C, Rao M S, Reddy J K. Gene Expression. 1996;6:185–195. [PMC free article] [PubMed] [Google Scholar]
- 30.Fan C-Y, Pan J, Chu R, Lee D, Kluckman K D, Usuda N, Singh I, Yeldandi A V, Rao M S, Maeda N, et al. J Biol Chem. 1996;271:24698–24710. doi: 10.1074/jbc.271.40.24698. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt J V, Su G H-T, Reddy J K, Simon M C, Bradfield C A. Proc Natl Acad Sci USA. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aoyama T, Peters J M, Iritani N, Nakajima T, Furihata K, Hashimoto T, Gonzalez F J. J Biol Chem. 1998;273:5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 33.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 34.Gerritsen M E, Williams A J, Neish A, Moore S, Shi Y, Collins T. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schultz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogryzko V, Schiltz R A, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 37.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, et al. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 38.Dowell P, Ishmael J E, Avram D, Peterson V J, Nevrivy D J, Leid M. J Biol Chem. 1997;272:33435–33443. doi: 10.1074/jbc.272.52.33435. [DOI] [PubMed] [Google Scholar]
- 39.Mizukami J, Taniguchi T. Biochem Biophys Res Commun. 1997;240:61–64. doi: 10.1006/bbrc.1997.7602. [DOI] [PubMed] [Google Scholar]
- 40.Nolte R T, Wisely G B, Westin S, Cobbs J E, Lambert M H, Kurokawa R, Rosenfeld M G, Wilson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 41.Palmer C N A, Hsu M-H, Groffin K J, Johnson E F. J Biol Chem. 1995;270:16114–1621. doi: 10.1074/jbc.270.27.16114. [DOI] [PubMed] [Google Scholar]
- 42.Seed B. Nat Med. 1998;4:1004–1005. doi: 10.1038/1990. [DOI] [PubMed] [Google Scholar]
- 43.Qi C, Zhu Y, Reddy J K. Cell Biochem. Biophys. in press; 1999. [DOI] [PubMed] [Google Scholar]
- 44.Xu J, Qiu Y, DeMayo F J, Tsai S Y, Tsai M J, O’Malley B W. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 45.Westin S, Kurokawa R, Nolte R T, Wisley G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Nature (London) 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]