Abstract
We have conducted a comparative genomic analysis of several olfactory receptor (OR) genes that lie immediately 5′ to the V-α gene segments at the mouse and human T-cell receptor (TCR) α/δ loci. Five OR genes are identified in the human cluster. The murine cluster has at least six OR genes; the first five are orthologous to the human genes. The sixth mouse gene has arisen since mouse-human divergence by a duplication of a ∼10-kb block. One pair of OR paralogs found at the mouse and human loci are more similar to each other than to their corresponding orthologs. This paralogous “twinning” appears to be under selection, perhaps to increase sensitivity to particular odorants or to resolve structurally-similar odorants. The promoter regions of the mouse OR genes were identified by RACE-PCR. Orthologs share extensive 5′ UTR homology, but we find no significant similarity among paralogs. These findings extend previous observations that suggest that OR genes do not share local significant regulatory homology despite having a common regulatory agenda. We also identified a diverged TCR-α gene segment that uses a divergent recombination signal sequence (RSS) to initiate recombination in T-cells from within the OR region. We explored the hypothesis that OR genes may use DNA recombination in expressing neurons, e.g., to recombine ORs into a transcriptionally active locus. We searched the mouse sequence for OR-flanking RSS motifs, but did not find evidence to suggest that these OR genes use TCR-like recombination target sequences.
Chemosensory systems are among the oldest forms of communication between organisms and their environment. Throughout evolution, chemosensory receptor repertoires have undergone extensive diversification. Expansion and contraction of olfactory receptor (OR) gene families, recombination, gene conversion, translocation, and positive selection for functional change (Ben-Arie et al. 1993; Ngai et al. 1993; Glusman et al. 1996; Trask et al. 1998a; Sharon et al. 1999) are all hallmarks of a rapidly evolving olfactory subgenome. This propensity for change in OR repertoires may reflect the biological demands for adaptation to narrow, species-specific niches. The OR gene family is the largest gene family in mammalian genomes, with approximately 1000 genes arrayed in clusters at multiple chromosomal locations (Buck and Axel 1991; Trask et al. 1998b; Mombaerts 1999; Glusman et al. 2001).
In mammalian olfactory systems, the internal representation of the complex odorant world is accomplished largely by virtue of one fundamental organizing principle: Each neuron that binds odorants is dedicated to a single allele of a single receptor gene (Chess et al. 1994). Thus, odor quality is encoded by discrete patterns of neuronal activity that result from the specific subset of ORs stimulated by an odorant or odorant mixture (Vassar et al. 1993, 1994).
The transcriptional mechanisms responsible for ensuring that only a single OR gene is expressed per neuron are unknown. Transgenic experiments have shown that ∼3 kb surrounding an OR gene is sufficient to achieve normal expression patterns (Qasba and Reed 1998), yet comparative analyses of paralogous genes in three human and two mouse OR clusters have failed to reveal significant conservation in putative regulatory regions (Bulger et al. 2000; Sosinsky et al. 2000; Lane et al. 2001).
The striking similarities between the olfactory and immune systems have provoked speculation that the two systems might use a common regulatory strategy. Both systems achieve recognition of a vast array of ligands by dedicating each ligand-binding cell to a single receptor allele, which is selectively expressed from a large genomic repertoire. In the immune system, selective receptor expression is accomplished by DNA recombination at both the TCR and immunoglobulin loci. This strategy generates receptor diversity and permits adaptability and heritability in antigen-recognizing cells. Programmed DNA editing has emerged recurrently in evolution as a viable developmental strategy for gene control (e.g., Gierl et al. 1989; Klar 1990; Muller et al. 1991; Haselkorn 1992; Prescott 1992) and is an appealing model for regulation in the olfactory system. Recombination could ensure singular gene transcription in olfactory neurons and long-term commitment of basal cells responsible for regenerating the olfactory neuroepithelium. The apparent lack of extensive promoter homology among OR genes might be explained if OR transcription requires recombination into an active locus (or loci). The observation that recombination-activating genes (RAG), key components of the recombination mechanisms of the immune system, are expressed in the olfactory neurons of two different vertebrate species (Jessen et al. 1999, 2001) lends further credence to this model.
In this paper, we provide a comparative genomic analysis of the mouse and human OR clusters that are found 5′ to the TCR genes in both species. We identify orthologous relationships, characterize recent gene block duplication events, and describe paralogous ORs that appear to be subject to strong selection to be maintained as highly similar pairs. We have used 5′ RACE-PCR to identify transcription start sites and find that orthologs share extensive noncoding homology largely contained within the transcriptional unit. Our analysis reveals no strong sequence conservation, TATA-boxes, or conserved transcription initiator sites in OR promoter regions. We identify a functional TCR V-α gene segment (Vα1), which is significantly diverged from the other V-α segments and uses a recombination signal sequence (RSS) that differs markedly from consensus RSSs. Therefore, we were curious if the divergent RSS of Vα1 might be a relic of an ancestral recombination system, perhaps one used by surrounding OR genes. However, no Vα1-like RSSs are apparent near the OR genes. Thus, we find no evidence to support the hypothesis that RAG-mediated recombination plays a direct role in OR regulation.
RESULTS AND DISCUSSION
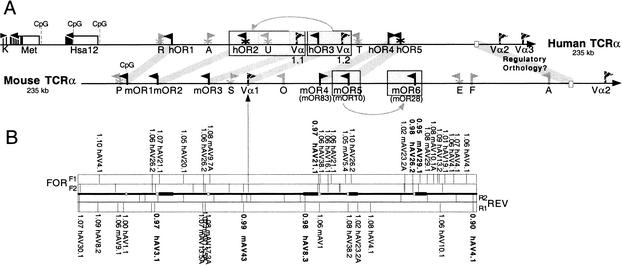
We have identified six OR genes in ∼200 kb of genomic sequence immediately 5′ to the mouse TCR-α/δ locus and five OR genes in the corresponding human region (Fig. 1). No further OR homology is found in the ∼65 kb of available sequence beyond hOR1. This ∼65-kb region contains three non-OR genes: the Hsa12 zinc finger gene, a gene encoding a methyl transferase, and the 3′ end of a gene (KIAA0737) whose function has not been characterized. CpG islands are associated with the upstream regions of Hsa12 and the methyltransferase gene. Available sequence in mouse extends 15.5 kb beyond mOR1, and no non-OR genes are detected in this region. Thus, it is possible that the mouse OR cluster extends further in this direction.
Figure 1.
Gene map and RSS profiling of the olfactory receptor regions 5′ to the mouse and human TCR-α/δ loci. (A) The ∼200-kb regions upstream of the TCR-α/δ loci are shown. Olfactory receptor (OR) genes are shown as black flags, and putative orthologs are indicated by thick gray lines that connect the mouse and human maps. The five human OR genes were named OR10G3, OR10G1P, OR10G2, OR4E2, and OR4E1P by Glusman et al. (2001). The mOR4, mOR5, and mOR6 genes are identical to previously identified mOR83, mOR10, and mOR28 OR genes, respectively (Tsuboi et al. 1999). V-gene segments are shown as striped flags (Vα). The position of a conserved 2-kb region of putative regulatory orthology is shown as open rectangles within the ∼80-kb region identified previously by Serizawa et al. (2000). Processed pseudogenes are shown as gray flags (R: retinoblastoma-binding protein-like; A: Arp3/actin2-like; U: ubiquitin-like; T: TBX2-like; P: proteosome component C8-like; S: signal peptidase-like; O: ODR4-like; E: enolase-like; F: FBRNP-like; AT: ATPase-like). Multiexon genes (K: Accession Code KIAA0737; M6A: methyl-transferase; ZNF: Hsa12 zinc finger genes) are indicated by black arrows, and individual exons are indicated by vertical lines (width according to exon size) beneath these arrows. In all cases, flags point in predicted transcriptional directions, and “X's” on the stems indicate pseudogenes. Block duplications are shaded and boxed. (B) RSS profiling for both strands (FOR, REV) of the mouse OR region. For each direction, the outer rectangles (F1, R1): A Hamming distance was computed between every known functional Vα and Vδ segment RSS for every position in the region. Positions (along with scores and the name of the most similar Vα-gene-segment RSS outside the rectangle) below cutoff threshold 1.1 are indicated in the figure (vertical lines within the rectangles for both strands). Stronger RSS signals below cutoff threshold 1.0 are bolded. The inner rectangles (F2, R2): the positions of conserved heptamer motifs (CACAGTG). Open boxes between forward and reverse plots indicate positions of OR coding sequences. Closed rectangles upstream of the mOR2, mOR4, mOR5, and mOR6 open boxes indicate the positions of 5′ UTRs (introns and exons).
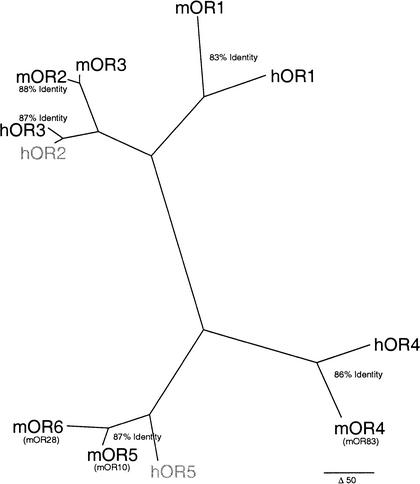
The first five OR genes in the mouse locus are orthologs to the five OR genes at the human locus. A molecular tree (Fig. 2) illustrates that the relative position and orientation of orthologs have been maintained within their respective clusters. Pairwise identities between orthologs range between 83% and 88%, consistent with levels of conservation observed between orthologs at the mouse and human P2- and β-globin-associated OR loci (Bulger et al. 2000; Lane et al. 2001).
Figure 2.
Phylogenetic reconstruction of mouse and human OR genes at the TCR-α/δ locus. Paup (Sinauer Associates) parsimony tree for the six mouse (prefix “m”) and five human (prefix “h”) OR genes at the TCR-α/δ locus is shown. Previously published names for mOR4, mOR5, and mOR6 are shown in parentheses (25). The two human pseudogenes are shown in light gray. Percent nucleotide identities for orthologs are indicated within orthologous clades. Scale bar for branch length (50 nt changes) is shown.
Overall, pairwise paralogous nucleotide identities range from 55% to 98%, indicative of both ancient paralogous relationships and very recent duplications within the clusters. The mOR6 gene, for example, is the result of a mouse-specific duplication of a ∼10-kb block containing mOR5. The mOR5 and mOR6 coding sequences are 93% identical, and the entire duplicated blocks are ∼85% identical at the nucleotide level.
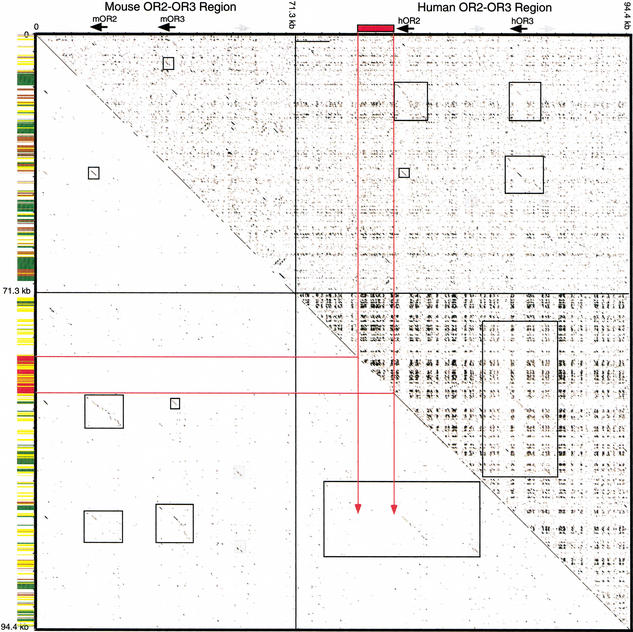
Dot-plot analysis of the human sequence also provides evidence for a duplication of a ∼ 14-kb block, which produced the OR2-OR3 gene pair and a second Vα1 gene (Fig. 3). Several observations suggest that this duplication predated the divergence of mouse and humans. First, in human, noncoding regions in the two blocks are ∼26% diverged (>30% substitution level), consistent with the duplication having occurred around the time of or before the mouse-human split (Li et al. 1996). Second, the mouse and human orthologous loci for OR2 and OR3 align throughout both duplication units; the 5′ mouse unit is most similar to the 5′ human unit, and the 3′ mouse unit is most similar to the 3′ human unit (Fig. 3). Third, a LINE1 repeat of the L1MA4 subfamily is present in one, yet cleanly absent in the other duplication unit in human (position 93432–104265 bp in the human sequence; also see Fig. 3), indicating that it inserted after the tandem duplication. L1MA4 copies were fixed in our genome during the time of the eutherian radiation (Smit et al. 1995). Therefore, this duplication likely took place during or before the radiation of placental mammals. We note that the second human Vα1 segment (Vα1.1) does not have an ortholog in mouse. Assuming that the OR2-OR3/Vα1.1-Vα1.2 duplication occurred before mouse and human diverged, we postulate that the mouse Vα1.1 ortholog has since been deleted.
Figure 3.
Dotplot analysis of the mOR2-mOR3 and hOR2-hOR3 region. A concatenated sequence file containing 71.3-kb mouse genomic sequence surrounding the mOR2-mOR3 region and 94.4-kb human genomic sequence surrounding the hOR2-hOR3 region was plotted against itself. Above the diagonal is the plot of unmasked sequence; below the diagonal is a plot of RepeatMasker sequence. Breaks in the diagonal line indicate positions of masked repeats. Along the vertical axis, repeat content is summarized by color-coded bars: low complexity (light gray), simple repeats (dark gray), LTRs (brown), SINEs (yellow), LINEs (green), and the L1MA4 noted in the text (red). Along the top axis, black arrows indicate the positions of the mOR2, mOR3, hOR2, and hOR3 coding regions; gray arrows indicate the positions of Vα1 gene segments; red rectangles indicate the position of the L1MA4 repeat inserted within the hOR2 block. Within the plot, red lines indicate the regions of the L1MA4 repeat. The duplication unit extends beyond the L1MA4 repeat, although this is not obvious in the dotplot because the sequences preceding the insertion are short and disrupted by numerous Alu repeat insertions. Black boxes in the plot surround homologous portions of the OR gene blocks. The Vα1 gene homology is noted with light gray shaded boxes in the plot. Note that the mOR2 block has more extensive homology with the hOR2 block than the hOR3 block, and the mOR3 block has more extensive homology with the hOR3 block than the hOR2 block (upper right/lower left quadrants). Also note the extensive hOR2 and hOR3 paralogous block homology (lower right quadrant) as compared to the mOR2 and mOR3 paralogous blocks, which are more significantly diverged (upper left quadrant).
The identity of the coding regions of OR2 and OR3 paralogs is anomalously high given the age of the duplication that gave rise to this gene pair. The coding sequences of the OR2 and OR3 paralogs are ∼98% identical in both species, as compared to 74% and <60% similarity in the surrounding noncoding regions of these genes in human and mouse, respectively. This coding sequence similarity is remarkable given the twofold difference in estimated molecular clock rates for these species (Li et al. 1996). Gene conversion is possible between neighboring coding regions. However, these conversion events would have had to involve the same pairs of genes in both species and be timed such that the resulting paralogs are 2% diverged in both species. Rather, it is likely that there has been selection to maintain OR2 and OR3 as a pair, perhaps to permit resolution of structurally similar odorants. Another example of paralogous twins is evident at the mouse and human P2 OR loci (Lane et al. 2001).
Numerous processed pseudogenes have inserted into the OR subregions of the mouse and human TCR loci since mouse-human divergence. At least four independent insertions have occurred in each species, and none of these eight genes is present in both species (Fig. 1). Intriguingly, a 1341-bp open reading frame of an ODR4-like sequence is present in the mouse cluster. In Caenorhabditis elegans, the ODR4 gene chaperones olfactory receptors to the neuronal cell surface (Dwyer et al. 1998). However, the mouse ODR4 homolog in this cluster is most likely a processed pseudogene, because it lacks introns, has remnants of a poly(A) tail, and is missing from the human locus. Furthermore, two putative human orthologous cDNAs (GenBank AK000171 and AK000512) exist that are 84% identical to the mouse ODR4-like gene. These human cDNAs are encoded by a 14-exon gene on BAC 173P17 (GenBank AF172081), which maps to human chromosome 1q25 (Carpten et al. 2000). Thus, the functional mouse ODR4-like gene is likely to be multiexon and reside at a location syntenic to human 1q25 (i.e., not near the TCR locus at 14D1-D2). Moreover, a candidate functional mouse ODR4 cDNA (GenBank BC003331) has been identified that is 0.8% diverged from the ODR4-like homolog at the TCR locus. The functional form of this gene could play a role in OR targeting in neurons and be a potentially important cofactor in the effort to express OR genes in heterologous cell types.
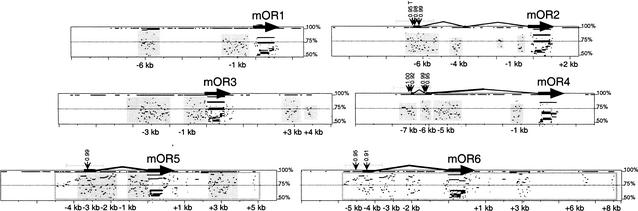
All six mouse OR genes have complete open-reading frames and are, therefore, presumably functional. So far, we have identified cDNAs for four of the mouse genes. The 5′ RACE-PCR products for these four mouse OR transcripts indicate that each has at least one upstream intron (Fig. 4). Transcription start sites range from 4–7 kb upstream of the coding sequence. In no case do we find introns that span exons of other genes.
Figure 4.
PIPmaker analysis illustrating genomic sequence similarity in the vicinity of the mouse OR genes. Gene homology in the vicinity of the six mouse OR genes is plotted by PipMaker (Schwartz et al. 2000). Coding sequences are shown as thick black arrows. Upstream exons (horizontal) and introns (bent) as determined by 5′ RACE-PCR are shown as thin black lines. Positions relative to translation start are shown in kilobases (kb). Gene structures for mOR5 (= mOR10), mOR6 (= mOR28), and mOR4 (= mOR83) isoforms are identical to those reported in Tsuboi et al. (1999). Each gene was compared to all other mouse and human OR genes at the TCR-α/δ loci. Detected sequence homology (>50% identity) is plotted according to position in the test gene and level of sequence similarity. Light grey boxes surround homology detected exclusively between a mouse OR gene and its human ortholog. The only paralogous homology detected is between mOR5 and mOR6 (because of a block duplication), and this block of homology for both genes is surrounded by grey-framed, open rectangles. Regions ± 1 kb from transcription start sites (grey horizontal bars) were analyzed for promoter signals. Positions of NNPP promoter hits with scores >0.90 are shown (suffix “T” indicates TATA, no suffix indicates non-TATA). Positions of RepeatMasker sequences are indicated as breaks in horizontal black lines at the top (100% identity level) of each box.
We have examined noncoding sequences in the OR clusters for conserved motifs that might be involved in the regulation of these genes. PipMaker analyses (Schwartz et al. 2000) show that, with the exception of recent duplications, noncoding sequence has been conserved only between orthologs, and this homology typically extends only a few hundred base pairs upstream of transcription start sites (TSSs). We find strong non-TATA promoter signals upstream of some but not all OR TSSs (Fig. 4). Regions upstream of the TSSs lack homology with other OR clusters and other gene families represented in GenBank. Because OR transgenes with as little as 3 kb of upstream genomic sequence transcribe in the appropriate cell types and within the native zones of the olfactory epithelium (Qasba and Reed 1998), it is likely that cis motifs play a role in OR transcriptional regulation. Our results suggest that putative cis regulatory sequences may be small and/or scattered, thus requiring more refined techniques to identify.
The expression of the mOR6 transgene is dependent on sequences that reside well within the TCR locus, 45–125 kb upstream of the mOR6 coding sequence (Serizawa et al. 2000, in which the mOR6 gene was named mOR28). The ∼80-kb region required for mOR6 expression contains three Vα gene segments of the TCR cluster, a small region of similarity to vacuolar proton ATPase, and a 250-bp region of homology with mouse type IIB intracisternal A-particle (IAP). Within this 80-kb putative regulatory region, we find a 2-kb region 68 kb upstream of the mOR6 gene that is homologous to a region 33 kb upstream of the hOR5 gene at the human locus (Fig. 1). Within this 2-kb noncoding region are four patches of especially high-sequence homology between mouse and human: an 84-bp sequence with 82% identity, a 38-bp sequence with 89% identity, a 20-bp sequence with 100% identity, and a 28-bp sequence with 93% identity. This cross-species homology may be the consequence of selective pressure. Therefore, these specific sequences are candidate regulatory motifs that could account for the mOR6 transgene result. If this region is also required for the transcription of the other OR genes at this locus, it could be a locus-control region (LCR) or an insulator to partition the TCR and olfactory regulatory domains. This orthology resides at the boundary between the olfactory and TCR clusters, an appropriate position for a genomic insulator.
One model able to account for singular expression of OR genes and consistent with apparent lack of paralogous homology and strong promoters invokes recombination of OR sequences into an active OR locus in the genome. This model predicts that OR genes share signal sequences near the transcriptional unit that would direct recombination into an active locus. Because OR transgenes can be expressed from constructs that lack 3′ noncoding sequences (Qasba and Reed 1998), RSSs in regions upstream of the 5′ UTR would, therefore, be sufficient to direct these putative recombination events. We explored this hypothesis by screening OR regions of the mouse TCR locus for RSS-like motifs using a profile derived from multiple alignments of the known functional V-gene segment RSSs. We identified orphan RSSs (RSSs not associated with V-α segments) in the region, but no pattern of RSSs common to multiple OR genes emerges (Fig. 1). For example, we do not identify RSS motifs immediately 5′ to transcription start sites, which would be expected if these regions were recombined adjacent to an active promoter.
Interestingly, there are few RSS-like sequences other than the functional downstream RSS in the ∼40-kb region surrounding the Vα1 gene, a functional recombination target (cDNA GenBank accession codes: AF012171, X55824, D12895, Z49903, U51446), and the only known functional non-OR gene so far identified within an OR cluster. This apparent RSS void around Vα1 suggests that orphan RSSs are tolerated only if they are not a distraction to functional RSSs.
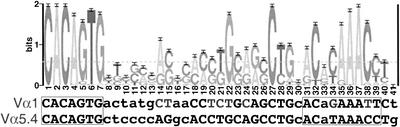
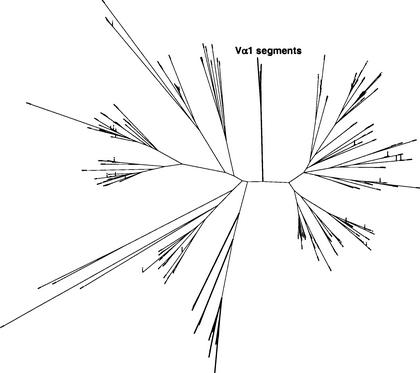
During these analyses, we discovered that the Vα1 gene segment has a lower-scoring RSS than orphan RSSs in the region. The Vα1 RSS is significantly diverged from the RSS consensus identified for the other functional Vα gene segments (Fig. 5). In addition, the Vα1 gene-coding sequence is significantly diverged from other V-α gene segments (Fig. 6). With the thought that the Vα1 RSS may be more representative of sequence motifs that might be involved with recombination within the olfactory region, we performed two additional searches aimed at identifying more divergent RSSs surrounding OR genes. First, we searched using the CACAGTG heptamer motif conserved in every known functional RSS, including the Vα1 RSS. Second, we computed Hamming distances (for a definition, see http://www.its.bldrdoc.gov/projects/t1glossary2000/_hamming_distance.html) between every known RSS and every subsequence in the cluster. We recorded significant similarity to the Vα1-like RSS or any other functional RSS variant regardless of similarity to an average RSS. Although we identified several candidate RSS motifs by this analysis (Fig. 1), we found none with a highest homology with Vα1. This result argues against the hypothesis that the divergent Vα1-like RSS resembles putative olfactory-specific signals. We also find no RSS motifs at a relative position common to more than one OR gene. These results argue against the hypothesis that RAG-mediated recombination involving TCR-like RSSs occurs to achieve selective expression of OR genes.
Figure 5.
Sequence LOGO of RSS based on a profile of V-gene segments in the TCR-α/δ loci. The divergent RSS for the mouse Vα1 segment is compared to V-gene consensus RSS motifs. Within the Vα1 RSS motif, capital letters indicate residues in which there is strong consensus, and gray letters indicate residues that deviate from consensus.
Figure 6.
A molecular tree of the V-gene segments. The phylogenetic isolation of the mouse and human Vα1 gene segments (bold) is illustrated by a molecular tree of vertebrate V-gene segments.
However, a DNA recombination model in the olfactory system cannot be excluded. There are at least 45 human transposase-like genes that, like RAG1 and RAG2, are derived from transposons (Smit 1999; Lander et al. 2001). Each presumably has its own target sequence and potential function. In addition, our computational analyses were confined to relatively simple comparisons of primary sequences. Subtle recombination signals, perhaps related to three-dimensional structure or accompanying cofactor binding sites, might be missed by our analyses. A definitive test of this model awaits examination of the genomic context of an expressed OR gene in a homogeneous population of neurons.
The analyses presented here add to the paradox of OR gene regulation. Although functional studies suggest the existence of many common levels of transcriptional control, which together achieve the expression of a single allele of a single gene in each neuron and zone-specific expression within the confines of the olfactory epithelium, available genomic sequences have provided few clues to this regulatory puzzle. The fact that the TCR and OR gene families have a similar transcriptional agenda (e.g., allelic exclusion and restricted expression of only one of a number of similar clustered genes) and are colocalized in the genome could be because of overlapping regulatory features. Indeed, the diverged Vα1 TCR gene segment is expressed from within an OR genomic region, and mOR6 transcription is dependent on sequences within the TCR genomic region. However, we find no additional evidence to support the hypothesis that these two gene families are interdependent or use common regulatory mechanisms (e.g., recombination) that might account for their overlapping genomic relationships.
METHODS
Sequence Data
The sequences considered in this paper were generated previously in our laboratory (Boysen et al. 1997; Glusman et al. 2001) and are available in the GenBank database (accession codes: mouse TCR α/δ locus NT_002581; human TCR α/δ U85199, U85198, U85197, U85196, and U85195). Before the availability of the genome sequence of the mouse TCR α locus and the subsequent revision of the nomenclature, mouse Vα1 was known as Vα19. Olfactory receptor genes have been named in accordance with genomic position (5′ OR1, OR2…3′) for convenience, using the prefix “m” for mouse and “h” for human genes. The five human OR genes were named OR10G3, OR10G1P, OR10G2, OR4E2, and OR4E1P by Glusman et al. (2001). The mOR4, mOR5, and mOR6 mouse OR genes were named mOR83, mOR10, and mOR28 by Tsuboi et al. (1999).
Isolation of 5′ OR Exons by RACE
The olfactory epithelium from seven B6CBAF1/J adult mice was dissected, and 1.3 μg of poly(A)+ mRNA was isolated using oligo(dT) cellulose (Stratagene). Preparation of cDNA and RACE protocols were essentially as described in the Marathon cDNA Amplification and Advantage cDNA PCR kits (Clontech), using antisense PCR primers within the coding region of the mouse OR genes.
Genomic Analysis Tools
Repeat content was determined by RepeatMasker (Smit and Green, version of June 6, 2000; A.F. Smit and P. Green, unpubl.) with RepBase 5.03 as a reference repeat library. Mapping of noncoding sequence homology was aided by PipMaker (Schwartz et al. 2000). The following genomic analysis tools available at the Baylor College of Medicine Search Launcher (http://www.hgsc.bcm.tmc.edu) were used: Genie (Kulp et al. 1996; Reese et al. 1997), TSSG (Solovyev et al., in prep.), TSSW (Solovyev et al., in prep.), NNPP (Reese and Eeckman 1995; Reese et al. 1996), and MatInspector/ TRANSFAC (Quandt et al. 1995).
RSS Analysis
An RSS profile was generated from a multiple alignment of the RSSs of all known functional V-α genes, in which predictions of functionality were based on the presence of the V-gene segment in expressed mRNAs. The inclusion or exclusion of RSSs of V-gene segments not definitely known to have function did not significantly impact our results. A profile is a tabulation of the frequency of each residue at each position in an alignment. The V-α profile was used to screen the OR region for orphan RSS-like sequences not associated with V-α gene segments. Each position in the OR region was assigned a score equivalent to the probability that it was generated from the RSS profile. If a position in the OR region achieves a high-profile score, this indicates that the identified position is the start of a sequence with high similarity to the consensus RSS sequence. Additionally, Hamming distances were computed between every known functional RSS and every position in the OR region. Each character comparison was weighted by the information content of the corresponding position in the RSS profile. For each position, the shortest distance was reported with cutoff threshold 1.1, which was chosen empirically to limit the number of reported scores. A 100-kb random control sequence with 50% GC content produced six hits with score <1.1 and none <1.0. If a position in the OR region produces a low Hamming score, this indicates that this position is the start of a sequence that is very similar to one of the known functional RSSs, which may or may not be highly similar to the consensus RSS sequence. Sequences were also screened for the conserved CACAGTG heptamer motif found in many RSSs. All analyses were performed in both the forward and reverse directions.
Acknowledgments
We thank the Richard Axel laboratory at Columbia University, in particular Tyler Cutforth, for providing mouse OR 5′ RACE data. The authors also thank numerous individuals in the Department of Molecular Biotechnology Core Sequencing facility for their ongoing efforts in this project. This work was supported by the NIH grants R01 DC04209 and R01 HG01475.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rlane@fhcrc.org; FAX (206) 667-6524.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.197901.
REFERENCES
- Ben-Arie N, Lancet D, Taylor C, Khen M, Walker N, Ledbetter DH, Carrozzo R, Patel K, Sheer D, Lehrach H, et al. Olfactory receptor gene cluster on human chromosome 17, possible duplication of an ancestral receptor repertoire. Hum Mol Genet. 1993;3:229–235. doi: 10.1093/hmg/3.2.229. [DOI] [PubMed] [Google Scholar]
- Boysen C, Simon MI, Hood L. Analysis of the 1.1-Mb human alpha/delta T-cell receptor locus with bacterial artificial chromosome clones. Genome Res. 1997;7:330–338. doi: 10.1101/gr.7.4.330. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors, a molecular basis for odor recognition. Cell. 1991;65:165–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Bulger M, Bender MA, van Doorninck JH, Wertman B, Farrell CM, Felsenfeld G, Groudine M, Hardison R. Comparative structural and functional analysis of the olfactory receptor genes flanking the human and mouse beta-globin gene clusters. Proc Natl Acad Sci. 2000;97:14545–14560. doi: 10.1073/pnas.97.26.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpten JD, Makalowska I, Robbins CM, Scott N, Sood R, Connors TD, Bonner TI, Smith JR, Faruque MU, Stephan DA, et al. A 6-Mb high-resolution physical and transcription map encompassing the hereditary prostate cancer 1 (HPC1) region. Genomics. 2000;64:1–14. doi: 10.1006/geno.1999.6051. [DOI] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Dwyer ND, Troemel ER, Sengupta P, Bargmann CI. Odorant receptor localization to olfactory cilia is mediated by ODR-4, a novel membrane-associated protein. Cell. 1998;93:455–466. doi: 10.1016/s0092-8674(00)81173-3. [DOI] [PubMed] [Google Scholar]
- Gierl A, Saldler H, Peterson PA. Maize transposable elements. Annu Rev Genet. 1989;23:71–85. doi: 10.1146/annurev.ge.23.120189.000443. [DOI] [PubMed] [Google Scholar]
- Glusman G, Clifton S, Roe B, Lancet D. Sequence analysis in the olfactory receptor gene cluster on human chromosome 17, recombinatorial events affecting receptor diversity. Genomics. 1996;37:147–160. doi: 10.1006/geno.1996.0536. [DOI] [PubMed] [Google Scholar]
- Glusman G, Rowen L, Lee I, Boysen C, Roach JC, Smit AF, Wang K, Koop BF, Hood L. Comparative genomics of the human and mouse T cell receptor loci. Immunity. 2001;15:337–349. doi: 10.1016/s1074-7613(01)00200-x. [DOI] [PubMed] [Google Scholar]
- Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- Haselkorn R. Developmentally regulated gene rearrangements in prokaryotes. Annu Rev Genet. 1992;26:113–130. doi: 10.1146/annurev.ge.26.120192.000553. [DOI] [PubMed] [Google Scholar]
- Jessen JR, Willett CE, Lin S. Artificial chromosome transgenesis reveals long-distance negative regulation of rag1 in zebrafish. Nat Genet. 1999;23:15–16. doi: 10.1038/12609. [DOI] [PubMed] [Google Scholar]
- Jessen JR, Jessen TN, Vogel SS, Lin S. Concurrent expression of recombination activating genes 1 and 2 in zebrafish olfactory sensory neurons. Genesis. 2001;29:156–162. doi: 10.1002/gene.1019. [DOI] [PubMed] [Google Scholar]
- Klar AJ. Regulation of fission yeast mating-type interconversion by chromosome imprinting. Dev Suppl. 1990;3:3–8. [PubMed] [Google Scholar]
- Kulp D, Haussler D, Reese MG, Eeckman FH. ISMB-96 St. Louis, MO. 1996. A generalized Hidden Markov Model for the recognition of human genes in DNA. [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaun C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;15:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lane RP, Cutforth T, Young J, Athanasiou M, Friedman C, Rowen L, Evans G, Axel R, Hood L, Trask BJ. Genomic analysis of orthologous mouse and human olfactory receptor loci. Proc Natl Acad Sci. 2001;98:7390–7395. doi: 10.1073/pnas.131215398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Ellsworth DL, Krushkal J, Chang BH, Hewett-Emmett D. Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol Phylogenet Evol. 1996;5:182–187. doi: 10.1006/mpev.1996.0012. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- Muller F, Wicky C, Spicher A, Tobler H. New telomere formation after developmentally regulated chromosomal breakage during the process of chromatin diminution in Ascaris lumbricoides. Cell. 1991;97:815–822. doi: 10.1016/0092-8674(91)90076-b. [DOI] [PubMed] [Google Scholar]
- Ngai J, Chess A, Dowling MM, Necles N, Macagno ER, Axel R. Coding of olfactory information, topography of odorant receptor expression in the catfish olfactory epithelium. Cell. 1993;72:667–680. doi: 10.1016/0092-8674(93)90396-8. [DOI] [PubMed] [Google Scholar]
- Prescott DM. Cutting, splicing, reordering and elimination of DNA sequences in hypotrichous ciliates. Bioessays. 1992;14:317–324. doi: 10.1002/bies.950140505. [DOI] [PubMed] [Google Scholar]
- Qasba P, Reed RR. Tissue and zonal-specific expression of an olfactory receptor transgene. J Neurosci. 1998;18:227–236. doi: 10.1523/JNEUROSCI.18-01-00227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector—New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH. The Seventh Internation Genome Sequencing and Analysis Conference. Hilton Head Island, SC. 1995. Novel neural network algorithms for improved eukaryotic promoter site recognition. [Google Scholar]
- Reese MG, Harris NL, Eeckman FH. Large scale sequencing specific neural networks for promoter and splice site recognition. In: Hunter L, Klein TE, editors. Biocomputing: Proceedings of the 1996 Pacific Symposium. Singapore: World Scientific Publishing; 1996. pp. 2–7. [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Proceedings of the First Annual International Conference on Computational Molecular Biology. (RECOMB) Santa Fe, NM. 1997. Improved splice site detection in Genie. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawawa F, Asano M, Sudo K, Sakagami J, Sakano H, Ijiri T, et al. Mutually exclusive expression of odorant receptor transgenes. Nat Neurosci. 2000;3:687–693. doi: 10.1038/76641. [DOI] [PubMed] [Google Scholar]
- Sharon D, Glusman G, Pilpel Y, Khen M, Gruetzner F, Haaf T, Lancet D. Primate evolution of an olfactory receptor cluster, diversification by gene conversion and recent emergence of pseudogenes. Genomics. 1999;63:227–245. doi: 10.1006/geno.1999.5900. [DOI] [PubMed] [Google Scholar]
- Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Smit AF, Toth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Sosinsky A, Glusman G, Lancet D. The genomic structure of human olfactory receptor genes. Genomics. 2000;70:49–61. doi: 10.1006/geno.2000.6363. [DOI] [PubMed] [Google Scholar]
- Trask BJ, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, et al. Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum Mol Genet. 1998a;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- Trask BJ, Massa H, Brand-Arpon V, Chan K, Friedman C, Nguyen OT, Eichler E, van den Engh G, Rouquier S, Shizuya H, et al. Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum Mol Genet. 1998b;7:2007–2020. doi: 10.1093/hmg/7.13.2007. [DOI] [PubMed] [Google Scholar]
- Tsuboi A, Yoshihara S, Yamazaki N, Kasai H, Asai-Tsuboi H, Komatsu M, Serizawa S, Ishii T, Matsuda Y, Nagawa F, et al. Olfactory neurons expressing closely linked and homologous odorant receptor genes tend to project their axons to neighboring glomeruli on the olfactory bulb. J Neurosci. 1999;19:8409–8418. doi: 10.1523/JNEUROSCI.19-19-08409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]