Abstract
KIAA0187 is a gene of unknown function that maps to 10q11 and has been subject to recent duplication events. Here we analyze 18 human paralogs of this gene and show that paralogs of exons 14–23 were formed through satellite-associated pericentromeric-directed duplication, whereas paralogs of exons 1–9 were created via chromosome-specific satellite-independent duplications. In silico, Northern, and RT-PCR analyses indicate that nine paralogs are transcribed, including four in which KIAA0187 exons are spliced onto novel sequences. Despite this, no new genes appear to have been created by these events. The chromosome 10 paralogs map to 10q11, 10q22, 10q23.1, and 10q23.3, forming part of a complex family of chromosome-specific repeats that includes GLUD1, Cathepsin L, and KIAA1099 pseudogenes. Phylogenetic analyses and comparative FISH indicates that the 10q23.1 and 10q23.3 repeats were created in 10q11 and relocated by a paracentric inversion 13 to 27 Myr ago. Furthermore, the most recent duplications, involving the KIAA1099 pseudogenes, have largely been confined to 10q11. These results indicate a simple model for the evolution of this repeat family, involving multiple rounds of centromere-proximal duplication and dispersal through intrachromosomal rearrangement. However, more complex events must be invoked to account for high sequence identity between some paralogs.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AJ298152 through AJ298168.]
It is now clear from analyses of specific sequence families (Trask et al. 1998, Eichler et al. 1997), chromosomal regions (Flint et al. 1997, Jackson et al. 1999, Horvath et al. 2000), and the draft human sequence (Bailey at al. 2001; Lander et al. 2001) that subtelomeric and pericentromeric regions of the human genome are significantly enriched in duplicated sequences. In pericentromeric regions, the tandem or local duplication of DNA can lead directly (through dosage effects) or indirectly (through subsequent microdeletion) to clinical phenotypes, including velocardiofacial syndrome (Edelmann et al. 1999) and Prader Willi/Angelman syndromes (Christian et al. 1999). In addition, large tracts of sequence are frequently transposed or translocated into pericentromeric locations and distributed between nonhomologous chromosomes in a centromere-specific manner (Eichler et al. 1999). Many sequences affected by the latter events are related to genes, including adrenoleukodystrophy (Eichler et al. 1997), keratinocyte growth factor (Zimonjic et al. 1997), and neurofibromatosis type I (Regnier et al. 1997) genes.
The cytogenetic co-localization of highly mutable genes of clinical importance and repeated rounds of sequence formation and rearrangement has led to speculation that these events may lead to the formation of new genes (Trask et al. 1998; Eichler 1999). Although tandem duplication per se is clearly central to the expansion and diversification of large gene families such as olfactory receptors and zinc finger genes, pericentromeric regions are known to be rich in heterochromatin, which is incompatible with transcription (Csink et al. 1997), making it unclear if the excess of duplicated material in these regions is of evolutionary significance. Detailed structural and transcriptional analyses of these regions is, therefore, important if we are to understand both the duplication mechanisms and their consequences.
We have previously analyzed ∼1 Mb of genomic sequence-linking pericentromeric satellites in 10q11 to the RET proto-oncogene (Guy et al. 2000) and found that interchromosomal duplication events had been confined to the satellite-rich proximal 250 kb of this sequence, which was devoid of transcripts. In contrast, the distal 850 kb contained evidence of multiple intrachromosomal duplication events in addition to the presence of three known genes and five novel transcripts. These results implied a model of pericentromeric sequence organization on this chromosome arm consisting of two distinct domains: (1) a proximal domain that is satellite rich, transcript poor, and prone to interchromosomal duplication; and (2) a distal domain that is satellite poor and prone to intrachromosomal rearrangement. The identification of similar interchromosomally duplicated, transcript-poor sequence tracts close to the centromere of chromosomes 22 and 21 (Ruault et al. 1999; Footz et al. 2001), which map proximal to well-characterized intrachromosomal duplications (Orti et al. 1998; Dunham et al. 1999; Edelmann et al. 1999), indicates that this basic organization may be typical of many pericentromeric regions within the human genome.
However, one gene in 10q11 is an apparent exception to this two-domain model of sequence organization. KIAA0187 is a gene of unknown function with a putative transmembrane domain that shares 40% identity to the Saccharomyces cerevisiae open reading frame (ORF) YPL217c at the protein level. It also shares significant identity along its entire length to mouse, rat, and nematode expressed sequence tags (ESTs; http://www.ncbi.nlm.nih.gov/UniGene/), consistent with functional conservation between diverse eukaryotes. The gene lies within the distal 10q11 sequence domain, telomeric of the intrachromosomally duplicated IFB12 and RSU1 pseudogenes, and centromeric of the intrachromosomally duplicated D10S141B locus (Guy et al. 2000). Despite its position, two paralogous fragments of KIAA0187 map to 22q11 (Dunham et al. 1999): one proximal to the satellite 3 array on this chromosome, the other linked to CAGGG and HSREP522 repeat sequences, which have been implicated in pericentromeric duplication events (Eichler et al. 1999). Furthermore, analysis of monochromosomal somatic cell hybrids indicate that additional paralogs of this gene are present on chromosomes 9, 12, 13, 14, 15, and 20 (Guy et al. 2000). Thus, unlike surrounding sequences, KIAA0187 appears to have been involved in interchromosomal duplication events, leading to a widely dispersed family of human paralogs.
To understand why this gene is an apparent exception to the physical separation of inter- and intrachromosomal duplications in 10q11 and to investigate the extent to which paralog formation has generated biological novelty, we have used the data within the human draft genomic sequence to establish the structure, transcriptional activity, and evolutionary history of human KIAA0187 paralogs. The results indicate that the paralogs have been generated by two distinct mechanisms, consistent with the two-domain model of pericentromeric organization in 10q11, and establish that a recent intrachromosomal rearrangement has been central to the dispersal of this gene family. However, no new genes appear to have been created by either dispersal mechanism, despite evidence for extensive transcription of KIAA0187 paralogs.
RESULTS
Physical Organization of the KIAA0187 Gene
Genomic sequence of the KIAA0187 gene and a cDNA containing the complete ORF were available (accession Nos. D80009 and AL02234; Nagese et al. 1996; Guy et al. 2000), making it straightforward to establish the intron/exon organization (see Methods). Analysis of publicly available transcripts identified two further ESTs (accession Nos. BE543163 and BE501103), which extended the cDNA by 26 bp and identified an additional 5′ exon (exon 1) within a CpG island ∼1.5 kb upstream of the putative initiation codon. The gene, therefore, contains a minimum of 23 exons, which extend over ∼50 kb of DNA in 10q11 (Fig. 1A).
Figure 1.
Structure of the KIAA0187 gene and its human paralogs. (A) Schematic of KIAA0187 intron/exon organization. The size of each intron is given in base pairs below the schematic. Nucleotide positions defining each exon are shown above the schematic, taking the A of the initiation codon as position 1 because the transcription start site has not been defined. All splice sites conform to the GT-AG rule. Untranslated regions are indicated in grey, and exon numbers are shown when resolution allows. (B) Physical structure of human paralogs. Solid horizontal lines indicate the extent of paralogy relative to the KIAA0187 gene. Solid vertical bars are shown when the terminus of paralogy can be defined; arrows are shown when it cannot be defined because of the incomplete nature of the sequence. Paralogous sequences that are contiguous within a single clone but are physically separated within the KIAA0187 gene are indicated by a dashed line joining the domains of paralogy. The positions of satellite arrays are shown if they are present within 1 kb of the terminus of paralogy. The chromosome 22 paralogs (ap000525/526 and d87003/d87018) have been characterized previously (Dunham et al. 1999; Eichler et al. 1999; Guy et al. 2000). Physical scale in kilobases is shown relative to Guy et al. (2000).
KIAA0187 Paralogs Fall Into Two Distinct Groups
Analysis of finished and high-throughput genomic sequence (see Methods) identified a total of 16 independent human paralogs (Fig. 1B). These vary in size from ∼4.5 kb (AL450334) to ∼32 kb (AP000525/526) and can be divided into two physically distinct groups: Five overlapping paralogs, defined here as proximal (AL135925, AL391137, AL136982, AC022400, AL450334), each contain sequence related to three or more proximal exons (1–9). None of these are linked to satellite arrays. In contrast, 10 out of the remaining 11 paralogs, defined here as distal, contain sequence related to two or more distal exons (14–23). Seven of these distal paralogs terminate within, or close to, a series of previously described satellite arrays, which include the CAGGG repeat (Eichler et al. 1999). In two cases, the second terminus is defined by a tract of α-satellite >15 kb in length (AC026273 and AC016424). The paralog within AP000525 and AP000526 is unique as it contains sequences 3′ of the gene in addition to distal exons (Fig. 1B).
Thirteen of the 16 paralogs have been integrated into the draft human sequence map (Lander et al. 2001). However, because of the difficulty of mapping clones that contain recently duplicated DNA (Bailey et al. 2001), we independently analyzed the map position of these clones using sequence analysis and FISH (Table 1). With two exceptions (AC026273 and AC013411), all clones containing distal paralogs have been integrated into pericentromeric contigs within the draft human sequence (see legend to Table 1). Consistent with this, they hybridize to multiple pericentromeric locations in FISH analyses, and all hybridize to their draft map positions (Table 1, in bold). Furthermore, with the exception of AP000525/6, all contain tracts of two or more tandem repeat sequences with a known pericentromeric distribution (α-satellite, satellite II, and the CAGGG repeat). These repeats account for between 3.8% and 31.4% of each clone. In contrast, four of the proximal paralogs can be mapped to 10q11, 10q22, 10q22.2-q23, and 10q23, as they are present within clones that contain genes previously mapped to these locations (Deloukas et al. 1998). The FISH results are consistent with these positions (Table 1, underlined and in bold), although several clones give two hybridization signals on 10q, whereas one (AL391137) hybridized to multiple locations within the human genome. The clone containing the fifth proximal paralog (AL450334) is not within the University of California, Santa Cruz, draft sequence but is currently integrated into a Sanger Centre bacterial artificial chromosome contig that maps to 10q11 (Bentley et al. 2001). The only tandemly repetitive sequences within these clones are microsatellites, which account for <2% of each clone (Table 1). It is clear, therefore, that the proximal and distal paralogs have both distinct physical distributions and sequence contexts.
Table 1.
Sequence Features Within Clones Containing KIAA0187 Paralog
| Clone (accession no.) | Paralog classification | Long tandem repeats in clones | % Tandem repeats | Known genes in clone(s) | Draft map position | FISH mapping |
|---|---|---|---|---|---|---|
| AC026273 | distal | Alpha Sat, CATTT, CAGGG | 31.4 | — | Not placed | 1cen, 2cen, 9p12, 9q12, 13cen, 16cen, 22cen |
| AC068279 | distal | GCTG, AluSx, CATTT, CAGGG | 21.2 | — | 2q13 | 1qcen, 2cen, 2q13, 9cen, 15q11, 16p11 |
| AC018696 | distal | AluSx, CAGC, Sat II, CATTT, CAGGG | 18.4 | — | 2p13 | n.d. |
| AC024573 | distal | CATTT, CAGGG, CAGC, AluSx | 13.2 | — | 2p11 | n.d. |
| AC016424 | distal | CAGGG, CATTT, Alpha Sat | 12.9 | — | 2p11 | 1cen, 2cen, 7cen, 9p12, 9q12, 13nor, 14cen, 16p11, 22q11+nor cen |
| AC010098 | distal | CAGC, CATATT, AT complex, CAGGG | 10.7 | — | 2p11 | 1cen, 2cen, 16cen |
| D87003/18 | distal | CAGGG, CATTT | 9.7 | — | 22q11 | n.d. |
| AC013411 | distal | CAGGG, CATTT | 6.2 | — | 1 unplaced | 1cen, 2cen, 13cen, 15cen, 16cen |
| AC018963 | distal | AluSx, CATTT, CAGGG | 4.7 | — | 15q11.2 | 1q11, 2cen, 9p12, 9q12, 16q11, 15q11 |
| AC036220 | distal | CAGGG, CATTT | 3.8 | — | 16p12.3 | 1cen, 2cen, 14cen, 16cen |
| AP000525/6 | distal/proximal | — | 0.8 | — | 22q11 | n.d. |
| AL450334 | proximal | — | 0.0 | — | Not present | 10q11, 10q23 |
| AL391137 | proximal | — | 1.97 | Annexin A8 (Chr10: 69–72cM) | 10q11.2 | 2q23, 6q11, 8pter, 8q23, 10q11, 12q22, 16pter |
| AC022400 | proximal | — | 0.44 | Heparan N-deacetylase (97–98cM) | 10q22.2 | 10q11, 10q22 |
| AL135925 | proximal | — | 1.17 | Lung surfactant protein D Chr10: 98–107cM) | 10q22.3 | 10q11, 10q23.1 |
| AL136982 | proximal | — | 0.57 | GLUDI (Chr10: 114–199cM) | 10q23.2 | 10q11, 10q23.3 |
Paralog classification is based on exons present (see text for details). AluSx refers to a tandem array of up to 43 Alu elements, mostly of the AluSg/x subfamily. The only tandem repeats in the proximal clones are di-, tri-, and tetranucleotide repeats. Genetic intervals defining gene positions were taken from Deloukas et al. (1998). Current map positions for each clone are derived from the UCSC sequence (December 2000 build, http://genome.ucsc.edu/).
Proximal and Distal Paralogs Are Expressed
Two paralogs of KIAA0187 in 22q11 (AP000526.1 and D87003.2) have associated ESTs, indicating that they are transcribed (Dunham et al. 1999). To investigate expression further, we performed an in silico analysis (see Methods), which established that KIAA0187-related ESTs are derived from a minimum of nine loci, two of which are not currently represented within genomic sequence (Fig. 2A). Although a large number of transcripts from these loci contain intronic sequence, six loci have associated ESTs that are spliced, and four include sequences unrelated to KIAA0187. One of these (Fig. 2A, unassigned cluster 2) contains KIAA1087 exons spliced to aquaporin-related sequence (data not shown). Northern analyses (Fig. 2B) confirm that KIAA0187 is widely expressed, producing an ∼4-kb transcript in all adult tissues tested. Furthermore, probes from exons 10 and 23 identify a muscle-specific transcript ∼1 kb in size, indicating that this gene is alternatively spliced. An intron 19 probe identifies a weak transcript of ∼2.5 kb in size (presumed to be derived from the D87003/018 paralog in 22q11 because of the large number of ESTs from this locus), but no other transcripts could be detected by this method. We therefore investigated transcription by designing PCR primers specific for individual paralogs. This approach is complicated by the high sequence identity between loci, but we were able to confirm that three distinct transcripts (from 10q11, 10q22, and unassigned cluster 2) are expressed at low levels in a wide variety of adult tissues (Fig. 2C). This analysis also confirmed the heterogeneity of transcript structure implied by the EST data. For instance, the AA677615 primers give products consistent with exon 7 being spliced out in some transcripts and retained in others, whereas the AI62619 primers give products consistent with the 83-bp intron 16 being correctly spliced in some transcripts and retained in others (multiple products indicated in Fig. 2C). Despite this evidence of widespread transcription, analysis of the coding potential of all paralogs identified frameshift mutations or stop codons in all loci relative to the functional gene. For example, AC026273 possesses a stop codon in exons 16 and 20 (positions 2618 and 3155 of d80009), AC022400 has a frameshift in exon 3 (position 303 of d80009), and the ESTs within unassigned cluster 2 (Fig. 2) have a frameshift in exon 20 (position 3253 of d80009). This strongly indicates that all paralogs of KIAA0187, including AP000526.1, which has been previously defined as a putative gene (Dunham et al. 1999), are pseudogene fragments.
Figure 2.
Transcriptional analysis of KIAA0187. (A) Structure of expressed sequence tags (ESTs) related to KIAA0187. The structure of KIAA0187-related ESTs is shown relative to the exon/intron organization of the functional gene with ESTs from this locus shown above the schematic. Two clusters of ESTs could not be unambiguously assigned to any genomic sequence (see Methods). The four ESTs in unassigned cluster 1 share seven nucleotides that are not present in any genomic paralog, whereas the six ESTs in unassigned cluster 2 share six unique nucleotides (data not shown), indicating that they are not derived from any of the known genomic loci. ESTs labeled with an asterisk (aa047147 from the functional gene and aa994753 from d87003/018 in 22q11) represent ∼20 ESTs with very similar structures that have been omitted for clarity. Four ESTs contain sequence unrelated to the 10q11 gene: ESTs AA677615 (10q11) and AA725634 (22q11) are both spliced to anonymous sequence; EST W90094 (not placed) contains exons 16 and 17 spliced onto an Alu element; and ESTs AI653509, AA759017, and AA707452 (unassigned cluster 2) contain exons 20 and 21 spliced onto Aquaporin-related sequences. (B) Northern blots of polyA+ RNA probed with KIAA0187 exons. Panels labeled E3, E10, E21, and E23 and intron 19 were probed with fragments specific for exons 3, 10, 21, and 23 and intron 19, respectively. The three transcripts are indicated with arrows. (C) RT-PCR analyses of KIAA0187 paralogs. The cDNAs shown are as follows: H, heart; B, brain, Pl, placenta; Lu, lung; Li, liver; K, kidney; and Pa, pancreas. The accession number of the ESTs used to design primers specific for the three clusters is shown to the left of each panel, as is the intronic/exonic origin of the two primers. (Middle) The reverse primer is specific for the novel sequence within AA677615 (see A). The first positive control in the top and middle panels is 10 ng of the parent EST. In the bottom panel, it is 50 ng of genomic DNA. The second positive control in all panels is a G3PDH cDNA template amplified using the G3PDH primers (see Methods). The marker is a 100-bp ladder (Promega).
Proximal and Distal Paralogs Have Been Created at Different Times
The structure, position, and sequence context of the proximal and distal paralogs indicate that they may have been duplicated independently. To confirm this, we performed pairwise and multiple alignments between paralogs (Fig. 3; Table 2). The functional KIAA0187 gene shares 94.43% to 96.02% similarity to the distal paralogs (Table 2). However, their diverse structure means that only eight can be aligned over a distance >1 kb. The maximum likelihood tree generated from this alignment is shown in Figure 3A. The branch leading to KIAA0187 is at least twice as long as any other terminal branch. This is most compatible with the functional gene being the ancestral locus, especially as the intronic nature of the aligned sequences makes selection unlikely. The distal gene fragments therefore appear to have been derived from one initial duplication event, whereas their structure (Fig. 1B) indicates that this event involved the duplication of ∼60 kb of DNA extending from exon 14 to ∼40 kb distal of KIAA0187. The topology of the tree further indicates that this sequence was originally duplicated into the pericentromeric region of chromosomes 16 or 22, was duplicated intrachromosomally on chromosome 2, and underwent further duplication to chromosomes 1 and 15. If we assume a neutral substitution rate of 1.5 × 10−9 to 2.0 × 10−9 per site per year (Miyamoto et al. 1987; Sakoyama et al. 1987), the pairwise distances (Table 2) indicates that the initial duplication of distal exons occurred 13 to 17 Myr ago (0.052 substitutions/site), whereas the most recent duplication (involving paralogs AC018696 and AC068279) occurred ∼1.7 to 2.3 Myr ago (0.007 substitutions/site).
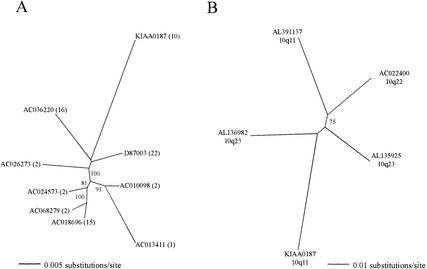
Figure 3.
Phylogenetic analysis of KIAA0187 paralogs. The scale for branch lengths is shown, and nodes with >50% bootstrap support are indicated. (A) Maximum-likelihood tree of distal paralogs. Derived from an 1892-bp alignment spanning positions 694294–696183 of the 10q11 sequence (Guy et al. 2000), which contains no exonic sequence. The chromosomal assignment for each clone is shown in brackets. (B) Maximum-likelihood tree of proximal paralogs. Derived from a 1909-bp alignment spanning positions 666815–668710 of the 10q11 sequence (Guy et al. 2000), which contains 235 bp of exonic sequence. The map position on chromosome 10 is shown for each sequence.
Table 2.
Pairwise Distances Betweeen KIAA0187 paralog
| A. KIAA0187—Distal paralog (1892 bp) | ||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | % sim. to KIAA0187 | Alignment length (bp) | |
| AC068279 | — | 95.42 | 6300 | |||||||
| AC026273 | 0.022 | — | 95.95 | 18061 | ||||||
| AC024573 | 0.012 | 0.019 | — | 95.48 | 6308 | |||||
| D87003/18 | 0.020 | 0.019 | 0.016 | — | 96.02 | 16610 | ||||
| AC036220 | 0.026 | 0.025 | 0.023 | 0.022 | — | 95.90 | 26239 | |||
| AC010098 | 0.015 | 0.021 | 0.013 | 0.019 | 0.025 | — | 94.98 | 10801 | ||
| AC018696 | 0.007 | 0.023 | 0.011 | 0.021 | 0.028 | 0.015 | — | — | 95.30 | 11140 |
| AC013411 | 0.026 | 0.032 | 0.024 | 0.030 | 0.036 | 0.018 | 0.026 | 94.43 | 3609 | |
| KIAA0187 | 0.042 | 0.041 | 0.040 | 0.038 | 0.043 | 0.040 | 0.041 | 0.052 | — | — |
| B. KIAA0187—Proximal paralog (1909 bp) | ||||||||||
| 1 | 2 | 3 | 4 | % sim. to KIAA0187 | Alignment length (bp) | |||||
| AL136982 | — | 92.80 | 18254 | |||||||
| AL135925 | 0.058 | — | 91.19 | 5787 | ||||||
| AC022400 | 0.063 | 0.055 | — | 92.54 | 14876 | |||||
| AL391137 | 0.069 | 0.059 | 0.058 | — | 92.81 | 9930 | ||||
| KIAA0187 | 0.077 | 0.070 | 0.073 | 0.082 | — | |||||
Kimura 2 parameter distances between the functional KIAA0187 gene and 11 paralogs are shown, calculated over the sequence ranges employed in the phylogenetic analyses (Fig. 4). The % similarity of each paralog to KIAA0187 is also shown. These are calculated over the full alignment length with the exception of AC013411 where unfinished sequence prevents a complete alignment. The % similarity shared between KIAA0187 and the paralogs with a physical structure that precluded inclusion in the multiple alignments (with alignment lengths) are as follows: AL450334, 92.68% (4451 bp); AC018963, 95.42% (14891 bp); AC016424, 95.60% (12106 bp); and AP000525/6, 91.25% (31819 bp).
In contrast to the distal paralogs, the proximal paralogs only share 91.19% to 92.8% similarity to the functional gene (Table 2). Furthermore, the topology of the phylogenetic tree derived from proximal sequences is distinct from the tree of distal paralogs (Fig. 3B), as all internal branches are short and all terminal branches are of similar lengths. This makes it impossible to infer the temporal order of paralog creation and indicates that they were created in a relatively rapid burst of duplication. Making the same assumptions concerning neutral substitution rates, we estimate that this process began ∼21 to 28 Myr ago (0.083 substitutions/site) and ended 14 to 18 Myr ago (0.055 substitutions/site; Table 2). Although accurate estimates for the timing of these duplication events will require independent calibration of the neutral mutation rate within this sequence family, the results presented here clearly indicate that in addition to having distinct sequence contexts and genomic distributions, the proximal paralogs were created before the distal paralogs with little or no temporal overlap.
Proximal Paralogs Are Linked to Other Independently Duplicated Pseudogenes
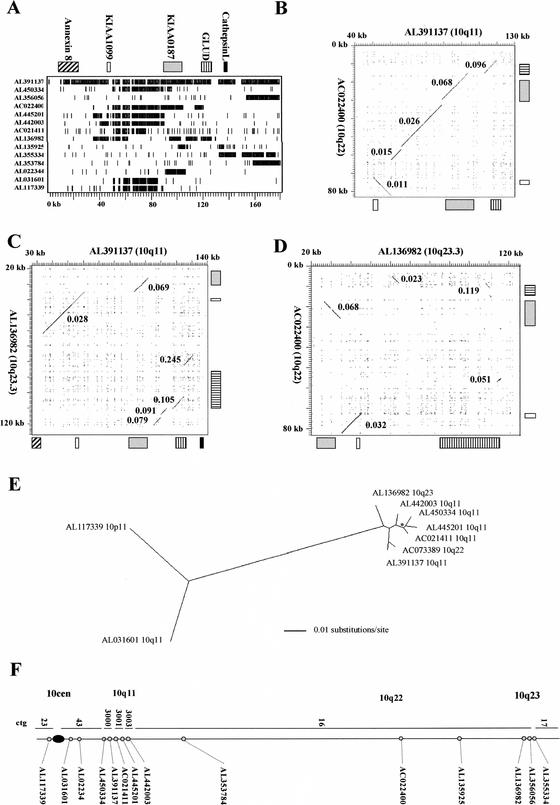
To obtain more information on the formation of the proximal paralogs, we investigated the sequence context of one paralog present within finished sequence in detail (AC0391137). A BLASTN analysis identified nonprocessed GLUD1 and Cathepsin L pseudogenes and processed KIAA1099 pseudogenes within this clone and within clones containing other KIAA0187 paralogs on chromosome 10 (Fig. 4A). This indicated that the KIAA0187 paralogs map within previously uncharacterized chromosome-specific pseudogene clusters. To investigate the structure of these and to establish if each cluster was formed by a single duplication event, sequence relationships between three clones in which the linear order of pseudogenes has been established (al391137, ac022400, and al136982) were analyzed further (Fig. 4B–D). The pseudogenes, which span 50 to 70 kb in all clones, are in the same order in AL391137 (10q11) and AC022400 (10q22), although a 10-kb region containing the KIAA1099 pseudogene is in different orientations in the two clones (Fig. 4B). Strikingly, the sequence divergence between the two pseudogene clusters decreases in a linear fashion from 0.096 at one end of the cluster (GLUD1 pseudogenes) to 0.011 at the other (KIAA1099 pseudogenes). In contrast, although the orientation of sequences within the 10q23.3 cluster is the same as that in the 10q11 cluster, the linear order is different, with the position of KIAA0187 and KIAA1099 sequences being reversed (Fig. 4C). The GLUD1 sequences are again more highly diverged than the KIAA1099 sequences, a pattern that is also apparent within the comparison of the 10q22 and 10q23.3 clusters (Fig. 4D), in which structural similarities are least pronounced. Collectively, these comparisons indicate that the different pseudogenes within the clusters have shared their most recent common ancestors at different times. If we make the same assumptions concerning neutral mutation rates as before, the data indicate that the GLUD1 sequences diverged ∼30 to 40 Myr ago (pairwise distances of 0.096, 0.105, and 0.119); the KIAA0187 sequences, ∼17 to 23 Myr ago (pairwise distances of 0.068, 0.069, and 0.068); and the KIAA1099 sequences, ∼2.8 to 10 Myr ago (pairwise distances of 0.011, 0.028, and 0.032). This provides strong evidence that the linked pseudogenes in each cluster were formed by independent duplication events.
Figure 4.
Analysis of pseudogene clusters containing KIAA0187 paralogs. (A) Human genomic blast hits to AL391137. The position of all independent human clones with >90% sequence identity to AL391137 over >2 kb are shown relative to RepeatMasked AL391137 sequence (hit 1 is the self-comparison). The positions of gene-related sequences within AL391137 are indicated above, with the scale in kilobases indicated below. Numbers refer to the clones shown in F. The GLUD1-related sequences identified include the functional GLUD1 gene (within hit 8) and two nonprocessed pseudogene fragments, one of which (GLUDP3 within hit 4) has been previously mapped to 10q22 (Deloukas et al. 1993). The Cathepsin L–related sequences include two pseudogenes, CTSLL1 and CTSLL1-2 (within hits 10 and 1, respectively), which have been mapped previously to 10q (Bryce et al. 1994). (B–D) Dot matrix analyses of 10q pseudogene clusters. The positions of gene-related sequences are shown for each clone. Kimura 2 parameter distances for each individual region of high identity are indicated. The highly diverged match in C (K2P = 0.245) is owing to a cluster of Alu elements and is assumed to be coincidental. (E) Maximum-likelihood tree of KIAA1099 paralogs generated from a 10952-bp alignment spanning nucleotides 55178–59761 of AL391137. The scale (in substitutions/site) for the branch lengths is shown. All branchpoints have >95% bootstrap support with a single exception (asterisk), which has 81% support. (F) Distribution of sequences related to AL391137 on chromosome 10. The position of each clone identified in the BLAST analysis within Sanger Centre contigs (Bentley et al. 2001) is shown. Cytogenetic locations (established by FISH; Table 1) are also shown.
Most KIAA1099 Duplications Have Occurred in 10q11
The most recent duplications in these pseudogene clusters have involved sequences flanking the KIAA1099 paralogs. To investigate the dynamics of these, a phylogenetic tree of these sequences was constructed (Fig. 4E). The tree contains two principle clades. The two sequences within the smaller clade (AL117339 and AL031601) lie within a previously identified duplication of ∼250 kb between 10p11 and 10q11 (Jackson et al. 1999), and the branch lengths are consistent with previous estimates for the timing of this event (25 to 30 Myr ago; Hearn 2000). In contrast, both the terminal and internal branches within the large clade are short (<0.012 substitutions/site for terminal branches, <0.007 for internal branches; data not shown), indicating that these paralogs shared a single common ancestor 6.3 to 8.3 Myr ago (data not shown). Finally, all the clones in this analysis map within ordered contigs, allowing their gross distribution to be established (Fig. 4F). Most map to a single cluster that spans three small contigs (3001–3003) within a 4-Mb region of 10q11 (Bentley et al. 2001), indicating that most KIAA1099 duplications have been tandem or local events.
Dispersal of 10q Paralogs Has Involved a 10q11:10q23 Inversion
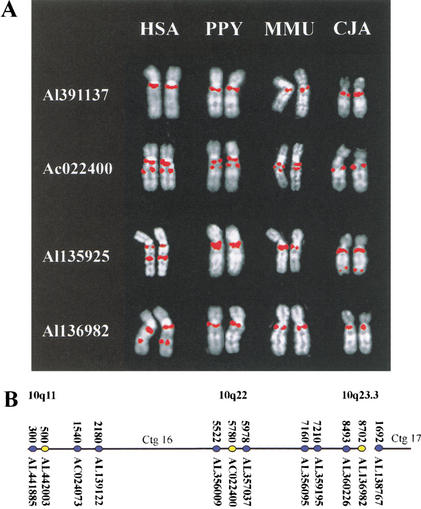
The high sequence identities between pseudogenes in 10q11, 10q22, and 10q23.3 indicate either that chromosomal rearrangement has disrupted locally duplicated sequences or that more complex processes such as chromosome-specific transpositional duplication or gene conversion have occurred. In an effort to distinguish between these possibilities, clones containing four of the KIAA0187 paralogs (from 10q11, 10q22, 10q23.1, and 10q23.3; Table 2) were used for comparative FISH analyses (Fig. 5A). The clone containing the 10q11 paralog (AL391137) hybridizes to 10q11 or Xq11 in all primate species analyzed (X is the ortholog of human chromosome 10 in great apes). The clone containing the 10q22 paralog (AC022400) hybridizes to both 10q11 and 10q22 in human and to Xq11 and Xq13 in PPY and gives two centromere proximal signals in MMU. The clones containing the 10q23 paralogs (AL135925 and AL136982) hybridize to 10q23.1 and 10q23.3, respectively, in human in addition to giving a 10q11 signal. However, the 10q23 signals are not observed in the PPY, MMU, and CJA hybridizations. This indicates that the ancestral position of the sequences within these clones (including the GLUD1 and Lung Surfactant protein D genes) was 10q11-q13 and that the current position of the paralogs in 10q22 and 10q23 has involved intrachromosomal sequence movement.
Figure 5.
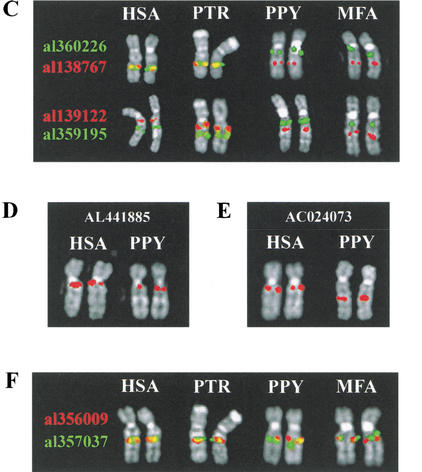
Comparative analysis of primates. Species used are as follows: hsa, human; PTR, chimpanzee; PPY, orangutan; MMU, rhesus monkey; MFA, crab-eating macaque; and CJA, common marmoset. Individual chromosomes (HSA 10 or syntenic equivalent) rather than complete metaphase spreads are shown for clarity. The species origin of each pair of chromosomes is indicated at the top of each panel. The identity of the probes used in C and F is indicated by the color of the accession number to the left of each panel. For the relative position of probes used in A, see Table 1. For the relative position of probes used in C–F, see B. (A) Comparative mapping of clones containing proximal KIAA0187 paralogs. A clone containing the functional KIAA0187 gene hybridizes specifically to 10q11 in all four species. A signal is observed close to the telomere of the long arm in CJA using AL135925, indicating further lineage-specific dispersal of sequences in this clone. (B) Relative position of clones used in analyses of rearrangement. The position of each clone within Sanger Centre contigs 16 and 17 are shown in blue. The position of 10q11, 10q22, and 10q23.3 paralog clusters are shown in yellow. Contigs 16 and 17 extend from 10q11 to 10q25, with a single gap in 10q23.3. (Bentley et al. 2001). (C) Comparative analysis of clones flanking the 10q23.3 paralog cluster. (D) Delineation of proximal inversion breakpoint. (E) Comparative analysis of clones flanking the 10q22 paralog cluster.
To establish if this movement has been associated with intrachromosomal rearrangement, clones flanking the 10q22 and 10q23 paralogs in AC022400 and AL136982 were used in a series of cohybridization experiments. The physical location of these clones relative to the paralog clusters is shown in Figure 5B, and the hybridization results are shown in Figure 5, C through F. In human (HSA) and chimpanzee (PTR) clones flanking the 10q23.3 paralog cluster (AL360226 and AL138767), each give hybridization signals in 10q23 that overlap to produce a yellow signal, consistent with their map positions. However, in orangutan (PPY) and macaque (MFA), AL138767 produces a signal in 10q23 (phylogenetically Xq23 in PPY) but AL360226 hybridizes to 10q11 (Xq11 in PPY), consistent with paracentric inversion on the lineage leading to human (Fig. 5C). To corroborate this conclusion, we analyzed the relative order of clones AL356009 and AL356095, which map between the putative breakpoints of the inversion. Both produce discrete hybridization signals in human (Fig. 5C), consistent with their position on chromosome 10 (Fig. 5B). However, in MFA they map closer to the centromere and their order is reversed, consistent with a 10q11:10q23 paracentric inversion. The distal breakpoint of this inversion is localized between AL136982, which is within the inversion, and AL138767 (Fig. 5A–C). Further mapping of bacterial artificial chromosomes from the 10q11-q21 region localized the proximal breakpoint of this inversion to between AL441885 (which lies within the 10q11 cluster of KIAA1099 paralogs) and ac024073 (Fig. 5D).
An equivalent analysis using clones flanking the 10q22 pseudogene cluster (AL356009 and AL357037) failed to find evidence of significant intrachromosomal rearrangements (Fig. 5F), as flanking probes map to 10q13-10q22 in all primates analyzed. However, the fact that the signals clearly overlap in HSA and PTR but are discrete in MFA does indicate that some local rearrangement may also have affected this region of 10q.
Duplication of KIAA0187 Paralogs Predated the 10q11:10q23 Inversion
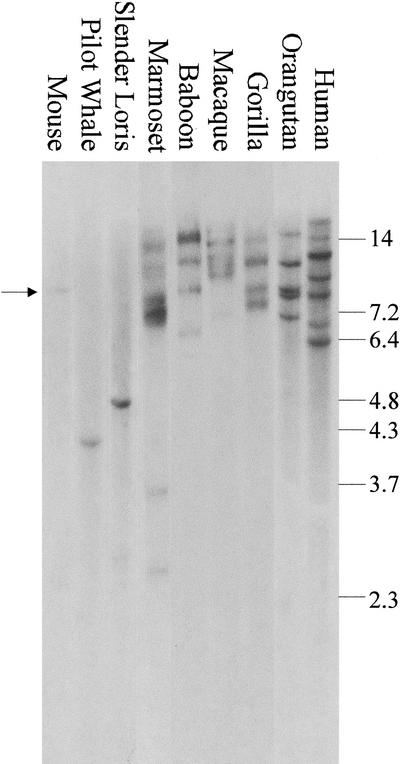
Based on current estimates of the neutral substitution rate in primates (Miyamoto et al. 1987, Sakoyama et al. 1987), our phylogenetic analysis indicates that duplication of KIAA0187 paralogs on 10q began 21 to 28 Myr ago. If this is so, it would mean that they were created in 10q11, before the 10q11:10q23 inversion that occurred after the divergence of orangutan from other great apes (Fig. 5C). Because this conclusion is central to our understanding of how these sequences have dispersed, we probed a Southern blot of EcoRI-digested mammalian DNAs with exon 3 of KIAA0187 (Fig. 6). A single weak hybridizing band is observed in mouse (arrowed), pilot whale, and slender loris (a pro-simian), whereas four to seven discrete hybridizing fragments are observed in one New World monkey (marmoset), two Old World monkeys (macaque and baboon), orangutan, gorilla, and human. This supports the conclusion that the duplication of proximal KIAA0187 exons began before the divergence of Old World primate and apes and indicates that it may have occurred before the divergence of Old and New World primates ∼40 Myr ago (Goodman 1999). These results therefore provide direct evidence that the KIAA0187 paralogs in 10q23.1 and 10q23.3 were created in 10q11 before the 10q11:10q23 inversion.
Figure 6.
Zoo blot of mammalian and primate species. DNAs were digested with EcoRI and probed with a PCR product specific for exon 3 of KIAA0187 (see Methods) The weak band of hybridization in mouse is indicated with an arrow. Size of marker fragments are given in kilobase pairs.
DISCUSSION
We have analyzed the structure, transcription, and evolution of KIAA0187-related loci within the human draft sequence to establish why this gene is an apparent exception to the two-domain model of pericentromeric organization previously established for the 10q11 region (Guy et al. 2000). Using criteria for inclusion in the analyses that are likely to underestimate the true number of loci within draft sequence data (see Methods), we have identified a minimum of 18 human loci related to the KIAA0187 gene. Of these, 16 can be classified into one of two distinct groups (which we define as proximal or distal) based on structure, chromosomal position, and proximity to tandemly repeated DNA. The remaining two are only represented by ESTs, indicating that further human KIAA0187 paralogs may remain to be identified and cannot be classified accurately, although their structure implies that they belong to the distal group.
KIAA0187: A Structurally Heterogeneous Expressed Pseudogene Family
Numerous human genes have been duplicated into and between pericentromeric locations, but there have been few cases in which transcription of derivative loci has been confirmed and characterized in detail. A notable exception is the creatine transporter gene in 16q11 (Eichler et al. 1996), although transcription of this gene is confined to testis (Iyer et al. 1996). As a result, the widespread expression of the distal KIAA0187 paralogs, which are tightly linked to satellite sequences, is unusual. Transcripts from AC026273 are of particular interest as they are derived from a ∼10-kb paralog sandwiched between satellite 3 sequences and >35 kb of α-satellite. Although surprising, this is consistent with the observations that a transgene placed between centromeric and telomeric satellites can be efficiently expressed, albeit from a heterologous promoter (Bayne et al. 1994), and provides evidence that satellite-rich regions are not totally devoid of transcriptional activity. Furthermore, the identification of hybrid transcripts, including one containing aquaporin-related sequences, provided further evidence that the juxtaposition of duplications from different genes has the potential to contribute to exon shuffling, a process that has occurred extensively during eukaryotic evolution (Patthy 1999). However, the only distal KIAA0187 paralog in which expression has been confirmed by Northern hybridization is >5 Mb telomeric of pericentromeric satellite on chromosome 22 (Dunham et al. 1999). The high expression of this locus relative to other paralogs can therefore be rationalized in terms of a more open chromatin environment.
Expression of the proximal paralogs is less surprising, given their interstitial positions and the fact that several of the Cathepsin L pseudogenes on 10q (CSTLL2 and CSTLL3) are also expressed (Bryce et al. 1994). Despite this, we can find no clear evidence to indicate that any of these transcripts are expressed at a high level or have the potential to encode novel proteins. However, in the absence of appropriate model systems, it will be difficult to categorically rule out a function for these transcripts. Numerous expressed pseudogene families created during primate evolution have now been identified on other chromosomes, including the GGT1 pseudogenes in 22q11 (Dunham et al. 1999), the HERC-2 pseudogenes in 15q11 (Ji et al. 2000b), and p47-phox and GTF21 pseudogenes in 7q11 (DeSilva et al. 1999), indicating that these are a common feature of our genome. Recent analysis of the fate of gene duplications in eukaryotes indicates a surprisingly high gene duplication rate of ∼0.02 duplications/locus per Myr, but a relatively short half-life for duplicated genes of only ∼7.3 Myr (Lynch and Conery 2000). With a mammalian gene number of ∼30,000 (Lander et al. 2001), the identification of a large number of gene-related transcripts in a transient state of decay is therefore not surprising.
Distal and Proximal Paralogs: Subfamilies Created by Two Distinct Processes
It is clear from our analyses that the KIAA0187 paralogs have been created by two mechanisms during primate evolution. The phylogenetic analysis, FISH data, and linkage to pericentromeric repeats are all consistent with the distal paralogs being created via pericentromeric-directed duplication (Eichler et al. 1997). The termination of these paralogs at both CAGGG repeats and α-satellite provided further evidence that these repeats are involved in the duplication mechanism (Regnier et al. 1997; Eichler et al. 1999). Furthermore, the evolutionary relationship between the distal paralogs conforms to the two-step model of pericentromeric-directed duplication (Eichler et al. 1997; Horvath et al. 2000), as there is no evidence of recurrent duplication of the functional gene. Similar dynamics have been documented for adrenoleukodystrophy (Xq28; Eichler et al. 1997) and neurofibromatosis type I (17q11; Luitjen et al. 2000). Critically, the identification of only one interchromosomal duplication event involving the functional KIAA0187 gene does not seriously undermine the validity of the two-domain model of pericentromeric organization for 10q11, which would predict, based on the KIAA0187 gene being ∼500 kb distal of pericentromeric satellites in 10q11, that it should be prone to intrachromosomal duplication as opposed to interchromosomal duplication (Guy et al. 2000).
In contrast, and consistent with the two-domain model, the proximal paralogs have been created by exclusively intrachromosomal events and are closely linked to other pseudogenes, including GLUD1, KIAA1099, and Cathepsin L–related sequences. The functional GLUD1 gene was assigned to 10q23 in 1989 (Jung et al. 1989), and although at least five human paralogs exist, the only nonprocessed paralogs identified to date map to 10q (Deloukas et al. 1993; present study). The active Cathepsin L gene maps to 9q21-22 (Fan et al. 1989), but again, all pseudogenes identified to date map to 10q (Bryce et al. 1994; present study), and sequence analysis indicates that these have been created following a single duplication of the active gene from chromosome 9 to 10 ∼40 to 50 Myr ago (Bryce et al. 1994). Furthermore, although the functional KIAA1099 gene maps to 2q24 (Unigene cluster Hs159377), all chromosome 10 paralogs of this gene are processed, indicating that the chromosome 10 paralogs have been formed subsequent to an RNA-mediated transposition event from chromosome 2 to 10. Collectively, therefore, these pseudogenes represent a previously uncharacterized chromosome-specific repeat family.
Chromosome-specific low-copy-number repeats have now been identified on a large number of chromosomes, including chromosomes 15 (Ji et al. 2000b; Pujana et al. 2001), 16 (Loftus et al. 1999), and 22 (Dunham et al. 1999). All have multiple copies within centromere-proximal cytogenetic locations. Our analysis of the KIAA1099 pseudogenes is noteworthy in that it identifies a focus of recent local duplication within an ∼4-Mb centromere-proximal region (Bentley et al. 2001) <2.5 Mb telomeric of centromeric satellites (Guy et al. 2000), similar to the organizational pattern observed on other chromosomes. Given the large number of clinical phenotypes associated with microdeletion or duplication mediated by low-copy repeats on other chromosomes (for review, see Ji et al. 2000a), it follows that the repeats described here must be considered good candidates for involvement in the rare deletions that have been reported for the 10q11-q23 region (for review, see Deloukas et al. 2000).
The 10q23 Pseudogenes Were Created in 10q11 and Dispersed by Intrachromosomal Rearrangement
Several chromosome-specific repeat families have a complex organization, including the chromosome 16 low copy repeats (LCRs) (Loftus et al. 1999) and the LCR15 repeats, which are distributed over at least four major cytogenetic locations between 15q11 and 15q26 (Pujana et al. 2001). However, the 10q clusters characterized here are unusual because although each pseudogene family appears to have been created in a relatively short evolutionary time period, different pseudogenes within an individual cluster were created independently. Specifically, although sequence duplications appear to have occurred as recently as 3 Myr ago (KIAA1099 events), adjacent sequences have been duplicated ∼25 to 30 Myr and 30 to 40 Myr ago (KIAA0187 and GLUD1 events, respectively), indicating that the same physical regions have been subject to repeated bursts of duplication. Given the distributed nature of these pseudogene clusters, the obvious question is how has this occurred? Our Southern analyses indicate that the KIAA0187 paralogs were duplicated before the 10q11:10q23 inversion identified by our FISH analyses. We can therefore conclude that the KIAA0187 sequences now present in 10q23.1 and 10q23.3 in human were created in 10q11. Furthermore, because the active GLUD1 gene was also moved from 10q11 to 10q23.3 by this inversion, and our alignments indicate that the GLUD1 duplications predate the KIAA0187 duplications, we can also conclude that the GLUD1 pseudogene present in 10q11 was the result of a local duplication event that occurred when the functional gene also mapped to 10q11. This, together with the more recent burst of KIAA1099 duplication in 10q11 identified by our phylogenetic analysis, indicates that 10q11 has been a focus for continual local duplication events for at least the last 30 to 40 Myr.
A Model for the Spread of Chromosome-Specific Repeats on 10q
Collectively, these results allow us to consider a simple model for the evolution of dispersed chromosome-specific repeats on 10q involving local centromere-proximal duplication followed by dispersal through intrachromosomal rearrangement. This model is attractive for a number of reasons. First, it is generally consistent with the distribution of foci of recent intrachromosomal duplication within the human genome. In addition to the centromere-proximal duplications associated with clinical phenotypes (cited above), there are numerous examples of large centromere-proximal tandem duplications identified as de novo variants within asymptomatic individuals (Barber et al. 1998, 1999; Ritchie et al. 1998). This centromere bias has recently been confirmed within the human draft sequence, in which there is a 2.7-fold enrichment of intrachromosomally duplicated sequence within 2 Mb of human centromeres that is not observed within subtelomeric regions (Bailey et al. 2001). The misalignment between centromeric satellite arrays, which are approximately two orders of magnitude larger than telomeric arrays and are known to show extensive length polymorphism (Warburton and Willard 1996, Guy et al. 2000), also provides a plausible explanation for much higher levels of duplication close to centromeres. Reduced meiotic recombination in these regions (Jackson et al. 1996, Lander et al. 2001) may also contribute to a higher retention frequency of duplications once they are formed. Finally, although large syntenic blocks exist between widely diverged species, a low-resolution genetic map of baboon has identified changes in marker order relative to human on 15 out of 22 autosomes, including one change consistent with the inversion described here (Rogers et al. 2000), whereas comparative analyses between S. cerevisiae and Candida abicans (Seoighe et al. 2000) and between Fugu and man (McLysaght et al. 2000) uncovered unexpectedly high frequencies of intrachromosomal rearrangement. This implies that cryptic intrachromosomal rearrangements are sufficiently common in diverse eukaryotic lineages to rapidly disperse locally duplicated sequences, irrespective of whether the duplicated DNA is mechanistically involved in the rearrangement process as indicated by recent analyses of man/mouse syntenic breaks on human chromosome 19 (Dehal et al. 2001).
Despite the appeal of this model, it would predict a 10q11:10q22 inversion within the last 3 Myr of human evolution to account for the very high sequence identity between the KIAA1099 paralogs in AL391137 (10q11) and AC022400 (10q22). We can find no evidence for such events using probes flanking the 10q22 paralog cluster. We are therefore forced to hypothesize either a larger number of cytogenetically cryptic paracentric inversions in the 10q11-10q22 region to explain the relocation of the 10q22 cluster, or the action of more complex processes such as gene conversion or chromosome-specific duplicative transposition. The latter, although unusual, appears to be the more plausible explanation for several reasons. First, sequences related to the proximal exons of KIAA0187 are present in marmoset (a New World monkey) in addition to Old World monkeys and apes, indicating that the proximal duplications may have occurred >40 Myr ago, considerably earlier than predicted by the sequence data under an assumption of neutrality (21 to 28 Myr ago). This could be caused by an unusually low neutral substitution rate in these paralogs, independent amplification of KIAA0187 exons in marmoset, or selection acting on the sequence, although the latter appears unlikely given the degenerate nature of the loci and the fact that the sequence used for the proximal alignment was 88% noncoding. However, it is noteworthy that similar discrepancies between the estimated timing of duplications obtained by low-resolution comparative FISH and those obtained from sequence data have been observed in analyses of the LCR22 repeats (Shaikh et al. 2001), the repeats responsible for Williams syndrome (DeSilva et al. 1999), and the large pericentromeric duplication on chromosome 21 (Orti et al. 1998). This raises the further possibility that sequence exchange (or conversion) over megabase distances between existing domains of paralogy may be common within chromosome-specific repeats. These complications make it clear that although we can conclude that intrachromosomal rearrangement has been central to the dispersal of the 10q pseudogene clusters characterized here, it will be necessary to perform detailed comparative mapping and sequencing in other primate species if we are to fully understand the evolutionary dynamics of the KIAA0187 sequence family in particular and chromosome-specific repeats in general.
METHODS
In Silico Analysis of KIAA0187 Paralogs
Human paralogs of KIAA0187 were identified by querying the nonreduncant and high-throughput genomic divisions of EMBL using BLASTN (Altschul et al. 1990). This identified 24 genomic entries that shared >80% sequence identity to the query sequence over a region of >3.0 kb. Graphical overviews of the extent of sequence identity between clones were obtained using NIX (Williams et al. 1998). To prevent the inclusion of overlapping sequences, any clones sharing >99.0% identity over >1 kb were excluded from subsequent analyses (AC025039, AC025268, AC037447, AP001214, AC022934, AP001229, AC024972, and AC023099). It is possible that these represent further paralogs of the gene. The 16 remaining paralogs were aligned to the functional gene (accession No. AL022344) using GenomeDotter, an in-house Dot matrix program that plots the output of RepeatMasker (A.F.A. Smit and P. Green, unpubl.), and Blast_2 sequences (Tatusova and Madden 1999). Eleven paralogs were within unfinished sequence. Eight of these were each contained within individual assembly fragments, allowing their structure to be defined, whereas the remaining three (ac013411, ac010098, and ac036220) were sufficiently complete to allow partial characterization (see Fig.1B). Long tandem repeats were identified using Tandem Repeat Finder (Benson 1999) The percent identity of each paralog to the KIAA0187 genomic sequence was determined using BESTFIT (Genetics Computer Group 1991). When appropriate, Kimura 2 parameter distances between paralogs were established using Alignscorer (Horvath et al. 2000), following alignment using Align (http://genome.cs.mtu.edu/align/align.html).
In Silico Analysis of KIAA0187-Related ESTs
To determine the intron/exon organization of KIAA0187, the cDNA (accession No. D80009) and overlapping ESTs (BE543163 and BE501103) were aligned to nucleotides 650001–750000 of the sequence presented by Guy et al. (2000) using est_genome (available through the UK HGMP Resource Centre). Transcripts related to KIAA0187 are present in three Unigene clusters: Hs.10848, Hs.231614, and Hs.288876. Additional ESTs were identified by using the cDNA to query the EST division of EMBL using BLASTN. The basic internal structure of each was established using NIX. Additional sequence data was derived from some clones to provide further details of their internal structure (accession Nos. AJ298152 through AJ298168). Because most ESTs only span 1–4 exons, they were binned into groups based on their structure for subalignment. Each group was then aligned with the cDNA and KIAA0187-related genomic sequence using Megalign (DNAstar). In-house software (Alignsearch) was then used to extract and display nucleotide positions that differed between all sequences within these alignments, allowing ESTs to be assigned to specific genomic loci based on diagnostic nucleotide positions.
EST Sequencing
Sequence data was generated from individual ESTs by amplifying plasmids in appropriate selective media using standard techniques (Sambrook et al. 1989) before isolating DNA using Qiagen purification kits (Qiagen) according to manufacturer instructions. Approximately 100 ng of template was used for each sequencing reaction, and all sequencing reactions were performed using an ABI PRISM BigDye cycle sequencing kit according to manufacturer instructions (PE Applied Biosystems) and were analyzed using an ABI377 (PE Applied Biosystems).
Northern Hybridization
Probes were generated by PCR and purified using a QiaQuick PCR purification kit (Qiagen). Primer pairs used are as follows (5′-3′): exon 3F/3R, gcatcatattccagtggttg and atctcagtcaacttct gccgg; exon 10F/10R, gatgccaaaggaggaaaaacaaa and actctggc aattagctggtgaca; exon 21F/21R, tgtcttcatgcgaacttggtat and gagtccttgttcgyctttagtc; exon 23F/23R, agcggcacctgcacaataaaga and tgaggcaggcagaggaaagtaaga; and intron 19F/19R, gtttgag cagcatttattga and ccctacaggtacagcaagat.
Probes were labelled with α32P-dCTP via random hexamer priming. Northern blots (Clontech) were prehybridized for 1 h at 65°C in ExpressHyb solution (Clontech) containing 0.1 mg/mL denatured sheared salmon sperm DNA and were hybridized for 2 h at 65°C in the same solution. Filters were washed according to manufacturer instructions and exposed to Kodak XAR X-ray film for 1 to 7 d at −70°C with an intensifying screen. Radioactivity was allowed to decay naturally between successive hybridizations.
RT-PCR Analyses
Panels of cDNAs derived from adult tissues (Clontech) were analyzed according to manufacturer recommendations. In addition to the standard controls, primers specific for exon 10 of the KIAA0187 gene were analyzed and found to amplify cDNA from all tissues. The following primer pairs specific for ESTs from paralogous loci were used: exon3F/intron4R (from W23168), gcatcatattccagtggttg and cggggtaaaataacctat cac; exon7F/AA677R (from AA677615), gatggaagatttgacaaacc and tccatggaaaagtcagaggtg; and exon16F/intron17R (from AI627619), gcrttgagattgaaaatgttcc and actccaaaccacctccctgct.
Phylogenetic Analyses
PAUP version 4.0b8 (Sinauer Associates) was used to construct maximum-likelihood trees using an exhaustive search method under an HKY85 model of molecular evolution (Hasegawa et al. 1985). Estimates of the γ-distribution of among-site rate variation and the proportion of invariant sites were then obtained for each maximum-likelihood tree and one round of Tree Bisection and Reconnection branch swapping was performed. For each tree, 1000 replicates of a neighbour joining bootstrap using the maximum-likelihood settings obtained by the above procedure were also performed. Insertions and deletions were considered missing data and excluded from all analyses. Trees were also constructed using maximum parsimony, and comparable topologies were obtained in all cases (data not shown). Because many of the alignments included sequences that are currently in finishing, the nucleotide positions used for each alignment are only defined relative to one finished European Molecular Biology Laboratory entry (see figure legends).
Fluorescence In Situ Hybridization
Metaphase spreads were obtained from human and primate cell lines and hybridized in situ with probes labeled with biotin by nick translation as described by Lichter et al. (1990), with minor modifications described by Antonacci et al. (1995). Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princetown Instruments). Flourescence signals were recorded separately as grey scale images, and pseudocolouring and merging of images was performed using Adobe PhotoShop software.
Acknowledgments
The financial support of the Associazione Italiana Ricerca sul Cancro, Telethon, and Wellcome Trust (Grants 049859 and 059369) are gratefully acknowledged, as are a short term fellowship from European Molecular Biology Organization (M.V.) and a studentship from the Medical Research Council (UK) (T.H). Primate DNAs were obtained from the Institute of Zoology, London. Dr. E.C. Holmes provided useful advice on the phylogenetic analyses.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mjackson@hgmp.mrc.ac.uk; FAX 44 191 241 8666.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.213702.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antonacci R, Marzella R, Finelli P, Lonoce A, Forabosco A, Archiadiacono N, Rocchi M. A panel of subchromosomal painting libraries representing over 300 regions of the human genome. Cytogenet Cell Genet. 1995;68:25–32. doi: 10.1159/000133882. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne RA, Broccoli D, Taggart MH, Thomson EJ, Farr CJ, Cooke HJ. Sandwiching of a gene within 12 kb of a functional telomere and alpha satellite does not result in silencing. Hum Mol Genet. 1994;3:539–546. doi: 10.1093/hmg/3.4.539. [DOI] [PubMed] [Google Scholar]
- Barber JC, Cross IE, Douglas F, Nicholson JC, Moore KJ, Browne CE. Neurofibromatosis pseudogene amplification underlies euchromatic cytogenetic duplications and triplications of proximal 15q. Hum Genet. 1998;103:600–607. doi: 10.1007/s004390050875. [DOI] [PubMed] [Google Scholar]
- Barber JC, Reed CJ, Dahoun SP, Joyce CA. Amplification of a pseudogene cassette underlies euchromatic variation of 16p at the cytogenetic level. Hum Genet. 1999;104:211–218. doi: 10.1007/s004390050938. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Deloukas P, Dunham A, French L, Gregory SG, Humphray SJ, Mungall AJ, Ross MT, Carter NP, Dunham I, et al. The physical maps for sequencing human chromosomes 1, 6, 9, 10, 13, 20 and X. Nature. 2001;409:942–943. doi: 10.1038/35057165. [DOI] [PubMed] [Google Scholar]
- Bryce SD, Lindsay S, Gladstone AJ, Braithwaite K, Chapman C, Spurr NK, Lunec J. A novel family of cathepsin L–like (CTSLL) sequences on human chromosome 10q and related transcripts. Genomics. 1994;24:568–576. doi: 10.1006/geno.1994.1667. [DOI] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region 15q11-q13. Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- Csink AK, Sass GL, Henikoff S. Drosophila heterochromatin: retreats for repeats. In: vanDriel R, Otte A, editors. Nuclear organization, chromatin structure, and gene expression. Oxford, UK: Oxford University Press; 1997. pp. 223–35. [Google Scholar]
- Dehal P, Predki P, Olsen AS, Kobayashi A, Folta P, Lucas S, Land M, Terry A, Ecale Zhou CL, Rash S, et al. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science. 2001;293:104–111. doi: 10.1126/science.1060310. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Dauwerse JG, Moschonas NK, van Ommen GJ, van Loon AP. Three human glutamate dehydrogenase genes (GLUD1, GLUDP2, and GLUDP3) are located on chromosome 10q, but are not closely physically linked. Genomics. 1993;17:676–681. doi: 10.1006/geno.1993.1389. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, McKusick KB, Beckmann JS, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Deloukas P, French L, Meitinger T, Moschonas NK. Report of the third international workshop on human chromosome 10 mapping and sequencing Cytogenet. Cell Genet. 2000;90:1–12. doi: 10.1159/000015653. [DOI] [PubMed] [Google Scholar]
- DeSilva U, Massa H, Trask BJ, Green ED. Comparative mapping of the region of human chromosome 7 deleted in Williams syndrome. Genome Res. 1999;9:428–436. [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Dunham I, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Lu F, Shen Y, Antonacci R, Jurecic V, Doggett NA, Moyzis RK, Baldini A, Gibbs RA, Nelson DL. Duplication of a gene-rich cluster between 16p11.1 and Xq28: A novel pericentromeric-directed mechanism for paralogous genome evolution. Hum Mol Genet. 1996;5:899–912. doi: 10.1093/hmg/5.7.899. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Budarf ML, Rocchi M, Deaven LL, Doggett NA, Baldini A, Nelson DL, Mohrenweiser HW. Interchromosomal duplications of the adrenoleukodystrophy locus: A phenomenon of pericentromeric plasticity. Hum Mol Genet. 1997;6:991–1002. doi: 10.1093/hmg/6.7.991. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Archidiacono N, Rocchi M. CAGGG repeats and the pericentromeric duplication of the hominoid genome. Genome Res. 1999;9:1048–1058. doi: 10.1101/gr.9.11.1048. [DOI] [PubMed] [Google Scholar]
- Fan Y-S, Bayers MG, Eddy RL, Joseph LJ, Sukhatme VP, Chan SJ, Shows TB. Cathepsin L (CTSL) is located in the chromosome 9q21-22 region: A related sequence is on chromosome 10. Cytogenet Cell Genet. 1989;51:996. [Google Scholar]
- Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett NA, King A, Higgs DR. The relationship between chromosome structure and function at a human telomeric region. Nature Genet. 1997;15:252–257. doi: 10.1038/ng0397-252. [DOI] [PubMed] [Google Scholar]
- Footz TK, Brinkman-Mills P, Banting GS, Maier SA, Riazi MA, Bridgland L, Hu S, Birren B, Minoshima S, Shimizu N, et al. Analysis of the cat eye syndrome critical region in humans and the region of conserved synteny in mice: A search for candidate genes at or near the human chromosome 22 pericentromere. Genome Res. 2001;11:1053–1070. doi: 10.1101/gr.154901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetics Computer Group. Program manual for the GCG package. Madison, WI: GCG; 1991. [Google Scholar]
- Goodman M. The genomic record of humankind's evolutionary roots Am. J Hum Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Spalluto C, McMurray A, Hearn T, Crosier M, Viggiano L, Miolla V, Archidiacono N, Rocchi M, Scott C, et al. Genomic sequence and transcriptional profile of the boundary between pericentromeric satellites and genes on human chromosome arm 10q. Hum Mol Genet. 2000;9:2029–2042. doi: 10.1093/hmg/9.13.2029. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating the human-ape split by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hearn T. “Organisation, expression and evolution of Kruppel-type zinc finger genes in human chromosomal region 10p11.2-q11.2.” Ph.D. thesis. UK: University of Newcastle Upon Tyne; 2000. [Google Scholar]
- Horvath JE, Viggiano L, Loftus BJ, Adams MD, Archidiacono N, Rocchi M, Eichler EE. Molecular structure and evolution of an α-satellite non–α-satellite junction at 16p11. Hum Mol Genet. 2000;9:113–123. doi: 10.1093/hmg/9.1.113. [DOI] [PubMed] [Google Scholar]
- Iyer GS, Krahe R, Goodwin LA, Doggett NA, Siciliano MJ, Funanage VL, Proujansky R. Identification of a testis-expressed creatine transporter gene at 16p11.2 and confirmation of the X-linked locus to Xq28. Genomics. 1996;34:143–146. doi: 10.1006/geno.1996.0254. [DOI] [PubMed] [Google Scholar]
- Jackson MS, See CG, Mulligan LM, Lauffart BF. A 9.75-Mb map across the centromere of human chromosome 10. Genomics. 1996;33:258–270. doi: 10.1006/geno.1996.0190. [DOI] [PubMed] [Google Scholar]
- Jackson MS, Rocchi M, Thompson G, Hearn T, Crosier M, Guy J, Kirk D, Mulligan L, Ricco A, Piccininni S, et al. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications, and unstable sequences with homologies to telomeric and other centromeric locations. Hum Mol Genet. 1999;8:205–215. doi: 10.1093/hmg/8.2.205. [DOI] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genomic Res. 2000a;10:596–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- Ji Y, Rebert NA, Joslin JM, Higgins MJ, Schultz RA, Nicholls RD. Structure of the highly conserved HERC2 gene and of multiple partially duplicated paralogs in human. Genome Res. 2000b;10:319–329. doi: 10.1101/gr.10.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KY, Warter S, Rumpler Y. Assignment of the GDH loci to human chromosomes 10q23 and Xq24 by in situ hybridization Ann. Genet. 1989;32:109–110. [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lichter P, Tang C-JC, Call K, Hermanson G, Evans GA, Housman D, Ward D. High resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, Kim UJ, Sneddon VP, Kalush F, Brandon R, Fuhrmann J, Mason T, Crosby ML, Barnstead M, Cronin L, et al. Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics. 1999;60:295–308. doi: 10.1006/geno.1999.5927. [DOI] [PubMed] [Google Scholar]
- Luijten M, Wang Y, Smith BT, Westerveld A, Smink LJ, Dunham I, Roe BS, Hulsebos TJ. Mechanism of spreading of the highly related neurofibromatosis type 1 (NF1) pseudogenes on chromosomes 2, 14 and 22. Eur J Hum Genet. 2000;8:209–214. doi: 10.1038/sj.ejhg.5200434. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- McLysaght A, Enright AJ, Skrabanek L, Wolfe KH. Estimation of synteny conservation and genome compaction between pufferfish (Fugu) and human. Yeast. 2000;17:22–36. doi: 10.1002/(SICI)1097-0061(200004)17:1<22::AID-YEA5>3.0.CO;2-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto MM, Slightom JL, Goodman M. Phylogenetic relations of humans and African apes from DNA sequences in the psi eta-globin region. Science. 1987;238:369–373. doi: 10.1126/science.3116671. [DOI] [PubMed] [Google Scholar]
- Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N. Prediction of the coding sequences of unidentified human genes, V: The coding sequences of 40 new genes (KIAA0161–KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1996;3:17–24. doi: 10.1093/dnares/3.1.17. [DOI] [PubMed] [Google Scholar]
- Orti R, Potier MC, Maunoury C, Prieur M, Creau N, Delabar JM. Conservation of pericentromeric duplications of a 200-kb part of the human 21q22.1 region in primates. Cytogenet Cell Genet. 1998;83:262–265. doi: 10.1159/000015201. [DOI] [PubMed] [Google Scholar]
- Patthy L. Genome evolution and the evolution of exon-shuffling: A review. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- Pujana MA, Nadal M, Gratacos M, Peral B, Csiszar K, Gonzalez-Sarmiento R, Sumoy L, Estivill X. Additional complexity on human chromosome 15q: Identification of a set of newly recognized duplicons (LCR15) on 15q11-q13, 15q24, and 15q26. Genome Res. 2001;11:98–111. doi: 10.1101/gr.155601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier V, Meddeb M, Lecointre G, Richard F, Duverger A, VanCong N, Dutrillaux B, Berheim A, Danglot G. Emergence and scattering of multiple neurofibromatosis (NF1)-related sequences during hominoid evolution suggests a process of pericentromeric interchromosomal transposition. Hum Mol Genet. 1997;6:9–16. doi: 10.1093/hmg/6.1.9. [DOI] [PubMed] [Google Scholar]
- Ritchie RJ, Mattei MG, Lalande M. A large polymorphic repeat in the pericentromeric region of human chromosome 15q contains three partial gene duplications. Hum Mol Genet. 1998;7:1253–1260. doi: 10.1093/hmg/7.8.1253. [DOI] [PubMed] [Google Scholar]
- Rogers J, Mahaney MC, Witte SM, Nair S, Newman D, Wedel S, Rodriguez LA, Rice KS, Slifer SH, Perelygin A, et al. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics. 2000;67:237–247. doi: 10.1006/geno.2000.6245. [DOI] [PubMed] [Google Scholar]
- Ruault M, Trichet V, Gimenez S, Boyle S, Gardiner K, Rolland M, Roizes G, De Sario A. Juxta-centromeric region of human chromosome 21 is enriched for pseudogenes and gene fragments. Gene. 1999;239:55–64. doi: 10.1016/s0378-1119(99)00381-9. [DOI] [PubMed] [Google Scholar]
- Sakoyama Y, Hong KJ, Byun SM, Hisajima H, Ueda S, Yaoita Y, Hayashida H, Miyata T, Honjo T. Nucleotide sequences of immunoglobulin epsilon genes of chimpanzee and orangutan: DNA molecular clock and hominoid evolution. Proc Natl Acad Sci. 1987;84:1080–1084. doi: 10.1073/pnas.84.4.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shaikh TH, Kurahashi H, Emanuel BS. Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: An update and literature review. Genet Med. 2001;3:6–13. doi: 10.1097/00125817-200101000-00003. [DOI] [PubMed] [Google Scholar]
- Seoighe C, Federspiel N, Jones T, Hansen N, Bivolarovic V, Surzycki R, Tamse R, Komp C, Huizar L, Davis RW, et al. Prevalence of small inversions in yeast gene order evolution. Proc Natl Acad Sci. 2000;97:14433–14437. doi: 10.1073/pnas.240462997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. Blast 2 sequences: A new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Trask BJ, Massa H, Brand-Arpon V, Chan K, Friedman C, Nguyen OT, Eichler E, van den Engh G, Rouquier S, Shizuya H, et al. Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum Mol Genet. 1998;13:2007–2020. doi: 10.1093/hmg/7.13.2007. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Willard HF. Evolution of centromeric alpha satellite DNA: molecular organisation within and between human and primate chromosomes. In: Jackson M, et al., editors. Human genome evolution. Oxford, UK: BIOS Scientific Publishers; 1997. pp. 121–145. [Google Scholar]
- Williams, G.W., Woollard, P.M., and Hingamp, P. 1998. NIX: A nucleotide identification system at the HGMP-RC. URL: http://www.hgmp.mrc.ac.uk/NIX/
- Zimonjic DB, Kelley MJ, Rubin JS, Aaronson SA, Popescu NC. Fluorescence in situ hybridization analysis of keratinocyte growth factor gene amplification and dispersion in evolution of great apes and humans. Proc Natl Acad Sci. 1997;94:11461–11465. doi: 10.1073/pnas.94.21.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]