Abstract
Scaffold/matrix attachment regions (S/MARs) are essential regulatory DNA elements of eukaryotic cells. They are major determinants of locus control of gene expression and can shield gene expression from position effects. Experimental detection of S/MARs requires substantial effort and is not suitable for large-scale screening of genomic sequences. In silico prediction of S/MARs can provide a crucial first selection step to reduce the number of candidates. We used experimentally defined S/MAR sequences as the training set and generated a library of new S/MAR-associated, AT-rich patterns described as weight matrices. A new tool called SMARTest was developed that identifies potential S/MARs by performing a density analysis based on the S/MAR matrix library (http://www.genomatix.de/cgi-bin/smartest_pd/smartest.pl). S/MAR predictions were evaluated by using six genomic sequences from animal and plant for which S/MARs and non-S/MARs were experimentally mapped. SMARTest reached a sensitivity of 38% and a specificity of 68%. In contrast to previous algorithms, the SMARTest approach does not depend on the sequence context and is suitable to analyze long genomic sequences up to the size of whole chromosomes. To demonstrate the feasibility of large-scale S/MAR prediction, we analyzed the recently published chromosome 22 sequence and found 1198 S/MAR candidates.
Scaffold/matrix attachment regions (S/MARs) are abundant regulatory DNA elements of the eukaryotic genome. A proposed major function of S/MARs is the coordination of the expression of gene loci. Attachment of a genomic segment to the nuclear matrix places a gene in close proximity to its transcription factors, providing an essential step to expression (Bode et al. 1995, 2000; Boulikas 1995). S/MARs form the anchor points of loop domains with domain sizes ranging from a few kb to more than 100 kb (Bode et al. 1996). They can shield gene expression from position effects and increase transcription initiation levels (Mielke et al. 1990). It has been estimated that the human genome contains approximately 100,000 S/MARs (Boulikas et al. 1995; Bode et al. 1996), which demonstrates the functional importance of S/MARs.
With the huge amounts of sequence data available from the genome projects, the challenge is to extract functional information from genomic sequences. Experimental definition of S/MARs requires substantial effort (Kay and Bode 1995) and is not suitable for large-scale screening of genomic sequences. Therefore, bioinformatics methods are a prerequisite for the analysis of whole genomes. Two software tools for the prediction of S/MARs are currently available, demonstrating the feasibility of in silico methods. MAR-Finder (Singh et al. 1997) is based on the statistical occurrence of S/MAR motifs described as consensus sequences based on the International Union of Pure and Applied Chemistry (IUPAC) code for nucleotide sequences. These motifs are characteristic for origins of replication, TG-rich sequences, curved DNA, kinked DNA, topoisomerase II sites, and AT-rich sequences. The stress-induced duplex destabilization (SIDD) program (Benham et al. 1997) identifies regions of DNA unwinding associated with nuclear matrix binding using a statistical mechanical procedure. Both methods require a larger sequence context, and the results partially depend on the size of this context.
We developed a new algorithm called SMARTest which is based on a density analysis of S/MAR-associated patterns represented by a weight matrix library. The algorithm is independent of sequence context and is suitable for the analysis of genomic DNA sequences of unlimited length, for instance, the analysis of complete chromosomes. We show SMARTest to correctly identify 14 of 37 experimentally defined S/MARs in genomic sequences of 310 kb in length. SMARTest had only nine additional matches which, in the absence of additional evidence, are considered false positives. We analyzed the recently published 34.6 million bp sequence of chromosome 22 (Dunham et al. 1999) with SMARTest and identified 1198 S/MAR candidates.
RESULTS
A New In Silico S/MAR Prediction Software Program
S/MARs are known to have a minimum sequence length of 200 to 300 base pairs (Mielke et al. 1990). AT-rich patterns are present in S/MARs, and the number of these motifs will determine the stable and specific binding of S/MARs to the nuclear matrix (Romig et al. 1994). We used these experimental findings for the development of a new in silico S/MAR prediction tool called SMARTest. The approach is based on a library of S/MAR-associated, AT-rich patterns derived from comparative sequence analysis of experimentally defined S/MAR sequences. Density analysis of the matches of these S/MAR-associated weight matrices is used for the prediction of S/MARs in genomic DNA sequences.
S/MAR Matrix Library
Most S/MAR-associated patterns that have been published are defined solely as IUPAC descriptions (Sander and Hsieh 1985; Cockerill and Garrard 1986; Gasser and Laemmli 1986; Spitzner and Muller 1988; Mielke et al. 1990; Boulikas 1993, 1995; Bode et al. 1995; van Drunen et al. 1997). We decided to use weight matrices as descriptions of S/MAR-associated patterns because matrices can mirror a set of DNA training sequences more specifically than IUPAC consensus sequences, as was shown in studies describing promoter elements (Bucher 1990; Chen et al. 1995; Quandt et al. 1995).
We analyzed whether a part of the new S/MAR-associated matrices generated in our library is similar to known S/MAR-associated motifs. We compared the IUPAC representations of our matrices with published IUPAC descriptions of S/MAR-associated patterns. The motifs AATATT and ATATTT were part of the IUPAC representations of 13 and 17, respectively, of our S/MAR matrices. These motifs have been shown to function as core unpairing elements in S/MARs and to significantly contribute to the binding affinity of S/MARs (Cockerill and Garrard 1986; Mielke et al. 1990; Bode et al. 1995). The motif ATATTT also conforms to the core of the weakly defined consensus sequence for Drosophila topoisomerase II (GTN WAYATTNATNNR, Sander and Hsieh 1985; Mielke et al. 1990). The known core unpairing element AATATATTT (Bode et al. 1992) matches the IUPAC representations of 19 of our S/MAR matrices if one mismatch is tolerated. The motifs ATTA and ATTTA, which were found to be associated with S/MARs and origins of replication (Boulikas 1995), were contained in the IUPAC representations of 12 and 4, respectively, of our S/MAR matrices.
Accuracy of S/MAR Prediction
For the evaluation of the accuracy of SMARTest, we used six genomic sequences, three plant and three human sequences, for which experimentally determined S/MARs and non-S/MARs are available and that were not used for the generation of the matrix library (Table 1). A total of 310,151 bp of genomic sequences containing 37 experimentally verified S/MARs were analyzed. The results show a high degree of overlap of the SMARTest predictions with the experimentally defined S/MARs.
Table 1.
Evaluation of SMARTest Accuracy
| Sequence | Experimentally defined S/MARs Position (kb) | SMARTest predictions Position (kb) | MAR-Finder predictions Position (kb) | |
|---|---|---|---|---|
| Description | Length (kb) | |||
| Oryza sativa putative ADP-glucose pyrophosphorylase | 30.034 | 0.0–1.2 | — | — |
| subunit SH2 and putative NADPH-dependent | 5.4–7.4 | 6.5–7.0 | — | |
| reductase A1 genes (U70541; Avramova et al. | 15.2–15.7 | 15.7–15.9 | ||
| 1998) | 16.2–16.6 | |||
| 17.3–18.5 | 17.6–18.3 | 17.5–18.4 | ||
| 20.0–23.1 | 19.6–20.1 | 19.8–20.4 | ||
| 20.7–21.3 | 21.3–21.5 | |||
| 23.6–23.9 | 23.9–24.2 | |||
| 25.0–25.4 | 24.7–25.1 | |||
| 27.5–27.9 | ||||
| Sorghum bicolor ADP-glucose pyrophosphorylase | 42.446 | 0.0–1.5 | — | — |
| subunit SH2, NADPH-dependent reductase A1-a | 7.1–9.7 | — | — | |
| and NADPH-dependent reductase A1-b genes. | 21.3–21.9 | |||
| (AF010283; Avramova et al. 1998) | 22.4–24.7 | 22.9–24.0 | 23.2–24.2 | |
| 27.3–27.6 | 26.9–27.5 | |||
| 32.5–33.7 | — | — | ||
| 41.6–42.3 | — | — | ||
| Sorghum bicolor BAC clone 110K5 (AF124045; | 78.195 | ∼0.9 | — | — |
| Tikhonov et al. 2000) | ∼5.8 | — | — | |
| ∼6.3 | — | — | ||
| ∼9.3 | — | — | ||
| ∼15.0 | 15.1–15.8 | — | ||
| ∼18.5 | — | — | ||
| ∼21.9 | 21.7–22.0 | — | ||
| ∼23.3 | — | — | ||
| ∼25.6 | — | — | ||
| ∼29.1 | — | — | ||
| ∼34.6 | — | — | ||
| ∼44.1 | 44.1–44.5 | — | ||
| ∼48.5 | 47.9–49.5 | 47.9–49.4 | ||
| ∼57.9 | — | — | ||
| ∼62.9 | 63.1–63.7 | — | ||
| ∼67.1 | — | — | ||
| ∼69.3 | — | — | ||
| ∼73.7 | 74.3–74.7 | — | ||
| Human alpha-1-antitrypsin and corticosteroid | 30.461 | 2.6–6.3 | 5.5–6.0 | 3.0–3.2 |
| binding globulin intergenic region (AF156545; | 5.1–6.0 | |||
| Rollini et al. 1999) | 22.0–30.4 | 25.7–26.2 | 24.9–25.3 | |
| 27.5–27.8 | 25.5–25.8 | |||
| 26.2–26.4 | ||||
| 27.5–28.2 | ||||
| Human protamine locus (U15422 & AC00247; | 53.060 | 8.8–9.7 | — | 8.0–8.9* |
| Kramer et al. 1998) | 32.6–33.6 | — | 33.9–34.8* | |
| 37.2–39.4 | — | 37.7–38.6* | ||
| 51.8–53.0 | — | —* | ||
| Human beta-globin locus (22754 & U01317; Jarman | 75.955 | 1.5–3.0 | — | — |
| and Higgs 1988) | 15.6–19.0 | 18.0–18.4 | 15.5–16.0 | |
| 18.0–18.4 | ||||
| 34.4–34.9 | ||||
| 44.7–52.7 | — | 50.6–50.8 | ||
| 56.6–57.1 | 56.5–57.2 | |||
| 60.0–70.0 | 59.8–60.3 | 58.1–58.5 | ||
| 65.6–66.0 | 63.0–63.6 | |||
| 67.6–67.9 | 68.7–69.3 | |||
| 68.8–69.1 | ||||
| Sum (kb) | 310.151 | >56.1 | 14.5 | 13.8 |
| Total numbers: | 37 | 28 | 25 | |
| True positives [number of experimentally defined S/MARs found] | — | 19 [14] | 20 [12] | |
| False positives | — | 9 | 5 | |
| False negatives | — | 23 | 25 | |
| Specificity | — | 19/28 = >68% | 20/25 = >80% | |
| Sensitivity | — | 14/37 = >38% | 12/37 = >32% | |
Six different genomic sequences, three plant and three human sequences, for which experimentally defined S/MARs are known were analyzed with SMARTest to evaluate the accuracy of in silico predictions. For comparison, the same sequences were analyzed with the MAR-Finder program. True positive matches are printed in bold, minus (−) indicates false negative matches. Some of the longer (up to 10 kb) experimentally defined S/MARs contained more than one in silico prediction, each of them was counted as true positive match. Therefore, the number of true in silico predictions is higher than the number of experimentally defined S/MARs found. Specificity is defined as the ratio of true positive predictions, whereas sensitivity is defined as the ratio of experimentally defined S/MARs found.
A T-rich rule excluded using MAR-Finder.
SMARTest predicted 28 regions as S/MARs. Nineteen (68%) of these predictions correlate with experimentally defined S/MARs (true positives; bold letters in Table 1). Nine (32%) predictions are located in non-S/MARs (false positives). Note that the 19 true positive matches are located in only 14 of the experimentally defined S/MARs, as some of the long experimentally defined S/MARs have more than one SMARTest prediction. Twenty-three of the 37 experimentally defined S/MARs were not found by SMARTest (false negatives).
Using a different sequence dataset for the generation of the S/MAR matrix library, we obtained comparable results for the sensitivity and the specificity of SMARTest (Frisch et al. 2000).
S/MAR Prediction on the Complete Chromosome 22
SMARTest is the first tool available that is able to scan complete chromosomes for S/MAR candidates. We analyzed the recently published chromosome 22 sequence (34.6 million bp, Dunham et al. 1999) with SMARTest and obtained 1198 S/MAR candidates (Fig. 1). We correlated the location of the 1198 predicted S/MARs with the location of the 545 genes and 134 pseudogenes annotated (a total of 679 genes) (Dunham et al. 1999). Of the 1198 predicted S/MARs, 412 (34%) were included in or were overlapping with regions annotated as genes, and 786 (66%) of the S/MARs were located in intergenic or unannotated regions. Nearly all predicted S/MARs that were overlapping with genes were located in introns; only 28 (about 2%) of the 1198 predicted S/MARs were overlapping with annotated exons (a total of 3380 exons were annotated).
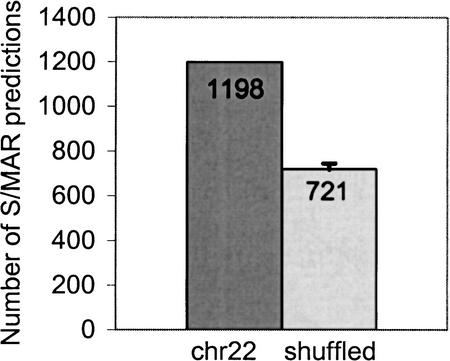
Figure 1.
Number of SMARTest predictions on chromosome 22 and on shuffled sequences with a conserved local nucleotide composition (mean value and standard deviation: average of 10 experiments).
The length of the 1198 predicted S/MARs in chromosome 22 ranged from 299 bp to 2144 bp; the average length was 484 bp. The AT-content of the predicted regions ranged from 45.4% to 88.9%; the average AT-content was 71.3%. Thus, most of the fragments predicted were AT-rich, whereas chromosome 22 is not AT-rich in total (52.2% AT). To evaluate whether the 1198 regions were identified by their high AT content only or by the specificity of the patterns of the S/MAR matrix library, we performed SMARTest analyses using randomly shuffled sequences. A shuffled sequence was generated by segmentation of the chromosome 22 sequence into nonoverlapping windows of 10 bp and by separately shuffling the nucleotides in each window. This way, all potential signals should be destroyed, whereas the local nucleotide composition is preserved. SMARTest predicted only 721 S/MAR candidates in the shuffled sequences (average of 10 experiments, Fig. 1), which is 60% of the 1198 predictions in the original sequence. Therefore, at least 40% of the SMARTest predictions are assumed to be due to specific recognition of patterns occurring in genomic sequences which are represented in the S/MAR matrix library.
Comparison with MAR-Finder
For comparison, we analyzed the same six genomic sequences from Table 1 using the software program MAR-Finder (http://www.futuresoft.org/MarFinder/; Singh et al. 1997). The cut-off threshold was set to 0.4, and all other parameters were set to default except for the analysis of the protamine locus, where the AT-richness rule was excluded (to detect the non-AT-rich S/MARs as was done for the protamine locus in Singh et al. 1997). MAR-Finder predicted 25 regions as S/MARs. Twenty (80%) of these predictions correlate with experimentally defined S/MARs (true positives). Five (20%) predictions are located in non-S/MARs (false positives). Note that the 20 true positive matches are located in only 12 of the experimentally defined S/MARs, as some of the long experimentally defined S/MARs have more than one MAR-Finder prediction. Twenty-five of the 37 experimentally defined S/MARs were not found by MAR-Finder (false negatives).
Analysis of chromosome 22 sequences was also performed with MAR-Finder (MAR-Finder cut-off threshold: 0.4, AT-richness rule excluded, otherwise default parameters). A complete analysis of the 34.6 million bp was not possible, as the web version of MAR-Finder is restricted to a maximum sequence length of 500 kb. Therefore, we used five different randomly selected 500 kb fragments from chromosome 22 and the respective shuffled sequences. MAR-Finder found a total of 59 S/MAR candidates in the five chromosome 22 sequence fragments and 47.9 S/MARs in the shuffled sequences (average of 10 experiments), which is 81% of the number of predictions from the original sequences (Fig. 2). SMARTest found a total of 98 S/MAR candidates in the five chromosome 22 sequence fragments and 58.7 S/MARs in the shuffled sequences (average of 10 experiments), which is only 60% of the number of predictions from the original sequences (Fig. 2). SMARTest predicted a significantly smaller amount of S/MARs in the shuffled sequences compared to the original sequences (Fig. 2), suggesting a more specific recognition.
Figure 2.
Number of SMARTest and MAR-Finder predictions on five different 500 kb fragments from chromosome 22 and on shuffled sequences with a conserved local nucleotide composition. The number of predictions in the five original chromosome 22 fragments was set to 100% for both SMARTest and MAR-Finder. Mean value and standard deviation are the average of 10 experiments.
DISCUSSION
Although several S/MAR-binding proteins are known (Bode et al. 2000), biological data of S/MAR-associated protein binding sites are limited. Examples are SATB1 (Dickinson et al. 1992, 1997; Banan et al. 1997; Liu et al. 1997), NFμNR (Zong and Scheuermann 1995), Bright (Herrscher et al. 1995), and topoisomerase II (Käs and Laemmli 1992). Development of models suitable for the prediction of S/MARs similar to our approaches describing Lentivirus LTRs (Frech et al. 1996) and actin promoters (Frech et al. 1998) was not possible due to the lack of a sufficient number of specific elements. Therefore, a new in silico approach to define S/MAR patterns directly from the sequences became a prerequisite. This approach resulted in a library of 97 S/MAR-associated weight matrices.
Known S/MAR-associated motifs were represented by our new S/MAR matrix library. This was shown for three core unpairing elements, AATATT, ATATTT, and AATATATTT. Core unpairing elements contribute to the function and binding affinity of S/MARs (Cockerill and Garrard 1986; Mielke et al. 1990; Bode et al. 1992, 1995).
The selectivity of each S/MAR-associated matrix in our library is similar to the selectivity of the bipartite MAR recognition signature (MRS) published by van Drunen et al. (1999). The single IUPAC elements of the bipartite MRS are both represented by our matrix library if one mismatch is tolerated. The bipartite MRS matches nine of the 34 S/MAR sequences used and has about one match per 10,000 bp in human genomic sequences, which is the same order of magnitude as for each of our matrices. The selectivity of a single S/MAR-associated matrix appears too low for the prediction of S/MARs in genomic sequences. Therefore, we compiled a large library of S/MAR-associated matrices to compensate for the low selectivity.
The evaluation of SMARTest on six genomic sequences shows a good correlation of the SMARTest results with the experimentally defined S/MARs (Table 1). The sensitivity of SMARTest was 38%, and 68% of the SMARTest predictions were true positives. A reason for SMARTest not finding a number of experimentally verified S/MARs may be that the current S/MAR matrix library was derived from AT-rich S/MARs that were used as the training dataset. Other S/MAR classes divergent from the AT-rich class exist (Boulikas and Kong 1993; Bode et al. 1996) which are probably not represented by our current library. For instance, the experimentally verified S/MARs in the protamine locus (Table 1; Singh et al. 1997) are not AT-rich and were not found by SMARTest. The protamine locus S/MARs were found by MAR-Finder, but only if appropriate parameter settings were used to mask the AT-rich classifier. Some other experimentally defined S/MARs were not detected by MAR-Finder but were found by SMARTest (six S/MARs from Table 1). Therefore, MAR-Finder and SMARTest may complement one another in S/MAR prediction.
The results in Table 1 show that MAR-Finder has a higher specificity than SMARTest (80% and 68%, respectively), whereas SMARTest has a higher sensitivity than MARFinder (38% and 32%, respectively). Important advantages of SMARTest are: (1) its suitability for large-scale analyses as demonstrated for chromosome 22 (Fig. 1); (2) its results are independent of the sequence context, and (3) there are no sequence-dependent parameter settings.
Additional weight matrices derived from new experimental data can be used immediately to improve the library of weight matrices continuously without changing the SMARTest algorithm. This feature will also be useful to improve the specificity of SMARTest. However, the availability of experimentally well defined S/MARs and non-S/MARs required as training and evaluation data is a significant problem. A major obstacle in generating a library of S/MAR-associated patterns is the fact that S/MARs are not well defined, and there is even an example where different experimental assays led to different assertions regarding the S/MAR or non-S/MAR character of a sequence (Razin 1996). Further improvement of the sensitivity and specificity of SMARTest is possible by extending the matrix library. However, this will definitely require additional experimental data.
To demonstrate the feasibility of large-scale S/MAR prediction, we analyzed the 34.6 million bp sequence of chromosome 22. Only 2% of the 1198 S/MARs predicted were overlapping with the 3380 exons annotated for chromosome 22. This is consistent with the observation that S/MARs are found in nontranscribed regions or within transcription units, but rarely in coding regions (Bode et al. 2000). However, the annotated exons in chromosome 22 have an AT content of only 44.6% on average, and thus it is a priori unlikely that SMARTest predicts AT-rich S/MARs in exons.
SMARTest predicted about 40% more S/MARs in the original chromosome 22 sequence than in shuffled sequences with the same AT profile (Fig. 1). This implies that SMARTest is not a simple AT cluster finder, but that a considerable part of the predictions are based on specific sequence recognition. It cannot be ruled out that shuffling of known S/MARs may sometimes also generate new artificial S/MARs in the shuffled sequences, particularly when the local nucleotide composition of the sequences is preserved. Therefore, a number of SMARTest predictions in the shuffled sequences may also be “true.” However, there is no way to sort those “true” matches out without experimental verification. We assume the current version of SMARTest will be a valuable tool for the prediction of matrix attachment regions because it is applicable to megabases of genomic sequences. One important feature of SMARTest is the capability to automatically update the matrix library upon availability of new data, whereby we can take full advantage of the highly dynamic situation of current molecular genomics.
METHODS
Definition of S/MAR-Associated Motifs
Training sequences were selected from the EMBL database, from literature, and from the S/MAR database S/MARt DB (http://transfac.gbf.de/SMARtDB/index.html, Wingender et al. 2000; Liebich et al. 2002). Thirty-four AT-rich (<60%) S/MARs [18 animal S/MARs (human, rodent, chicken) and 16 plant S/MARs] were used to define the motifs. The program DiAlign (Morgenstern et al. 1996) was used for alignment of subgroups of the 34 S/MARs and for detection of DNA fragments common to the subgroups. These regions were used for the definition of weight matrices (GEMS Launcher software package, Genomatix Software, Munich, Germany). The resulting weight matrices were selected for two-fold overrepresentation in the 34 training S/MAR sequences compared to shuffled sequences with the same nucleotide content. In addition, the matrices were required to have less than 0.4 matches per 1000 bp in the shuffled sequences. Ninety-seven weight matrices fulfilled these criteria, all describing short (10 to 21 base pairs in length), AT-rich DNA motifs.
Identification of S/MAR Candidates
Based on this library of S/MAR-associated DNA patterns, we developed a new tool, SMARTest (http://www.genomatix.de/cgi-bin/smartest_pd/smartest.pl) that searches for clusters of these patterns in genomic DNA sequences to identify potential S/MARs. SMARTest scans DNA sequences for matches to the S/MAR-associated weight matrix library and determines the number of matches in a sliding window of 300 nucleotides. We chose 300 bp as the window size because this is assumed to be the minimum length of a S/MAR. The sliding window is shifted by five nucleotides in each step of the analysis, which is less than half of the length of a weight matrix. If the number of base pairs covered by S/MAR matrices in a window exceeds a defined threshold, this region is reported as a S/MAR candidate. The threshold was derived from the analysis of the 34 S/MAR training sequences and two genomic sequences with experimentally mapped S/MARs and non-S/MARs (Cockerill et al. 1987; Jarman and Higgs 1988). Using the default threshold, SMARTest found 27 of the 34 S/MARs in the training dataset.
Accession Numbers
Oryza sativa sequence, EMBL U70541; Sorghum bicolor sequence, EMBL AF010283; Sorghum bicolor BAC clone 110K5, EMBL AF124045; human sequences, EMBL AF156545, U15422 and AC00247, L22754 and U01317; mouse sequence, EMBL J00440.
WEB SITE REFERENCES
http://transfac.gbf.de/SMARtDB/index.html, S/MAR database.
http://www.futuresoft.org/MarFinder/, MAR-Finder software program.
http://www.genomatix.de/cgi-bin/smartest_pd/smartest.pl, S/MAR matrix library.
Acknowledgments
We thank Edgar Wingender for helpful discussions and for privileged access to S/MARt DB.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article published online before print in January 2002
E-MAIL frisch@genomatix.de; FAX 49-(0)89-599766-55.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.206602. Article published online before print in January 2002.
REFERENCES
- Avramova Z, Tikhonov A, Chen M, Bennetzen JL. Matrix attachment regions and structural colinearity in the genomes of two grass species. Nucleic Acids Res. 1998;26:761–767. doi: 10.1093/nar/26.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banan M, Rojas IC, Lee WH, King HL, Harriss JV, Kobayashi R, Webb CF, Gottlieb PD. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J Biol Chem. 1997;272:18440–18452. doi: 10.1074/jbc.272.29.18440. [DOI] [PubMed] [Google Scholar]
- Benham C, Kohwi-Shigematsu T, Bode J. Stress-induced duplex DNA destabilization in scaffold/matrix attachment regions. J Mol Biol. 1997;274:181–196. doi: 10.1006/jmbi.1997.1385. [DOI] [PubMed] [Google Scholar]
- Bode J, Benham C, Knopp A, Mielke C. Transcriptional augmentation: Modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements) Crit Rev Eukaryot Gene Expr. 2000;10:73–90. [PubMed] [Google Scholar]
- Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- Bode J, Schlake T, Rios-Ramirez M, Mielke C, Stengert M, Kay V, Klehr-Wirth D. Scaffold/matrix-attached regions: Structural properties creating transcriptionally active loci. Int Rev Cytol. 1995;162A:389–454. doi: 10.1016/s0074-7696(08)61235-8. [DOI] [PubMed] [Google Scholar]
- Bode J, Stengert-Iber M, Kay V, Schlake T, Dietz-Pfeilstetter A. Scaffold/matrix-attached regions: Topological switches with multiple regulatory functions. Crit Rev Eukaryot Gene Expr. 1996;6:115–138. doi: 10.1615/critreveukargeneexpr.v6.i2-3.20. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Nature of DNA sequences at the attachment regions of genes to the nuclear matrix. J Cell Biochem. 1993;52:14–22. doi: 10.1002/jcb.240520104. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Chromatin domains and prediction of MAR sequences. Int Rev Cytol. 1995;162A:279–388. doi: 10.1016/s0074-7696(08)61234-6. [DOI] [PubMed] [Google Scholar]
- Boulikas T, Kong CF. Multitude of inverted repeats characterizes a class of anchorage sites of chromatin loops to the nuclear matrix. Cell Biochem. 1993;53:1–12. doi: 10.1002/jcb.240530802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- Chen QK, Hertz GZ, Stormo GD. MATRIX SEARCH 1.0: A computer program that scans DNA sequences for transcriptional elements using a database of weight matrices. Comput Appl Biosci. 1995;11:563–566. doi: 10.1093/bioinformatics/11.5.563. [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Yuen MH, Garrard WT. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987;262:5394–5397. [PubMed] [Google Scholar]
- Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Dickinson LA, Dickinson CD, Kohwi-Shigematsu T. An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J Biol Chem. 1997;272:11463–11470. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Frech K, Brack-Werner R, Werner T. Common modular structure of lentivirus LTRs. Virology. 1996;224:256–267. doi: 10.1006/viro.1996.0527. [DOI] [PubMed] [Google Scholar]
- Frisch M, Frech K, Klingenhoff A, Quandt K, Liebich I, Werner T. A new tool for the in silico prediction of matrix attachment regions in large genomic sequences. In: Bornberg-Bauer E, Rost U, Stoye J, Vingron M, editors. Proceedings of the German Conference on Bioinformatics. Berlin: Logos Verlag; 2000. pp. 27–34. [Google Scholar]
- Frech K, Quandt K, Werner T. Muscle actin genes: A first step towards computational classification of tissue specific promoters. In Silico Biol. 1998;1:29–38. [PubMed] [Google Scholar]
- Gasser SM, Laemmli UK. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986;46:521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R, Tucker PW. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: A B cell-specific trans-activator that describes a new DNA-binding protein family. Genes & Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Higgs DR. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 1988;7:3337–3344. doi: 10.1002/j.1460-2075.1988.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay V, Bode J. Detection of scaffold-attached regions (SARs) by in vitro techniques; Activities of these elements in vivo. In: Papavassiliou AG, King SL, editors. Methods for studying DNA-protein interactions—An overview. Wiley-Liss; 1995. pp. 186–194. [Google Scholar]
- Käs E, Laemmli UK. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992;11:705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JA, Adams MD, Singh GB, Doggett NA, Krawetz SA. Extended analysis of the region encompassing the PRM1–→PRM2–→TNP2 domain: Genomic organization, evolution and gene identification. J Exp Zool. 1998;282:245–253. [PubMed] [Google Scholar]
- Liebich, I., Bode, J., Frisch, M., and Wingender, E. 2002. S/MARt DB: A database on scaffold/matrix attached regions. Nucleic Acids Res. (in press). [DOI] [PMC free article] [PubMed]
- Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross SR, Dudley JP. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol Cell Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke C, Kohwi Y, Kohwi-Shigematsu T, Bode J. Hierarchical binding of DNA fragments derived from scaffold-attached regions: Correlation of properties in vitro and function in vivo. Biochemistry. 1990;29:7475–7485. doi: 10.1021/bi00484a017. [DOI] [PubMed] [Google Scholar]
- Morgenstern B, Dress A, Werner T. Multiple DNA and protein sequence alignment based on segment-to-segment comparison. Proc Natl Acad Sci. 1996;93:12098–12103. doi: 10.1073/pnas.93.22.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin SV. Functional architecture of chromosomal DNA domains. Crit Rev Eukaryot Gene Expr. 1996;6:247–269. doi: 10.1615/critreveukargeneexpr.v6.i2-3.70. [DOI] [PubMed] [Google Scholar]
- Rollini P, Namciu SJ, Marsden MD, Fournier RE. Identification and characterization of nuclear matrix-attachment regions in the human serpin gene cluster at 14q32.1. Nucleic Acids Res. 1999;27:3779–3791. doi: 10.1093/nar/27.19.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romig H, Ruff J, Fackelmayer FO, Patil MS, Richter A. Characterisation of two intronic nuclear-matrix-attachment regions in the human DNA topoisomerase I gene. Eur J Biochem. 1994;221:411–419. doi: 10.1111/j.1432-1033.1994.tb18753.x. [DOI] [PubMed] [Google Scholar]
- Sander M, Hsieh TS. Drosophila topoisomerase II double-strand DNA cleavage: Analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985;13:1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh GB, Kramer JA, Krawetz SA. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 1997;25:1419–1425. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzner JR, Muller MT. A consensus sequence for cleavage by vertebrate DNA topoisomerase II. Nucleic Acids Res. 1988;16:5533–5556. doi: 10.1093/nar/16.12.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov AP, Bennetzen JL, Avramova ZV. Structural domains and matrix attachment regions along colinear chromosomal segments of maize and sorghum. Plant Cell. 2000;12:249–264. doi: 10.1105/tpc.12.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drunen CM, Oosterling RW, Keultjes GM, Weisbeek PJ, van Driel R, Smeekens SC. Analysis of the chromatin domain organisation around the plastocyanin gene reveals an MAR-specific sequence element in Arabidopsis thaliana. Nucleic Acids Res. 1997;25:3904–3911. doi: 10.1093/nar/25.19.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drunen CM, Sewalt RG, Oosterling RW, Weisbeek PJ, Smeekens SC, Driel R. A bipartite sequence element associated with matrix/scaffold attachment regions. Nucleic Acids Res. 1999;27:2924–2930. doi: 10.1093/nar/27.14.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Prüß M, Reuter I, Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong RT, Scheuermann RH. Mutually exclusive interaction of a novel matrix attachment region binding protein and the NF-muNR enhancer repressor. Implications for regulation of immunoglobulin heavy chain expression. J Biol Chem. 1995;270:24010–24018. doi: 10.1074/jbc.270.41.24010. [DOI] [PubMed] [Google Scholar]