Abstract
Control genes, commonly defined as genes that are ubiquitously expressed at stable levels in different biological contexts, have been used to standardize quantitative expression studies for more than 25 yr. We analyzed a group of large mammalian microarray datasets including the NCI60 cancer cell line panel, a leukemia tumor panel, and a phorbol ester induction time course as well as human and mouse tissue panels. Twelve housekeeping genes commonly used as controls in classical expression studies (including GAPD, ACTB, B2M, TUBA, G6PD, LDHA, and HPRT) show considerable variability of expression both within and across microarray datasets. Although we can identify genes with lower variability within individual datasets by heuristic filtering, such genes invariably show different expression levels when compared across other microarray datasets. We confirm these results with an analysis of variance in a controlled mouse dataset, showing the extent of variability in gene expression across tissues. The results show the problems inherent in the classical use of control genes in estimating gene expression levels in different mammalian cell contexts, and highlight the importance of controlled study design in the construction of microarray experiments.
[Supplemental material available online at http://genome.mcgill.ca/∼pdlee/control_genes and and http://www.genome.org.]
Although DNA microarrays open the door to large-scale expression experiments (Lander 1999; Young 2000), a major challenge facing these studies is the design of experimental controls that will permit comparison of quantitative expression profiles obtained from diverse biological contexts. In traditional assays, standardization of mRNA levels has been achieved by comparison to the level of a control gene, commonly defined as one that is ubiquitously expressed at stable levels across many biological contexts. Methods of standardization based on control genes have furthermore been used in microarray and genomic studies (Khan et al. 1998; Beger et al. 2001). We reexamine the traditional concepts of controls in expression experiments in the aim of determining appropriate measures for the control of microarray experiments.
In an attempt to identify genes that are expressed at constant levels across a wide range of biological contexts, we analyzed four published datasets that were prepared following similar methods based on a single microarray technology (Affymetrix oligonucleotide microarrays). The NCI60 dataset (Butte et al. 2000) consists of microarray measurements of gene expression in 60 cancer cell lines originating from nine tissue types. A dataset obtained from patients with hematologic malignancies (Golub et al. 1999) includes expression profiles for multiple homogeneous acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) tumor samples. Temporal and developmental fluctuations in control gene expression were assessed using a dataset obtained from four cell lines following treatment with phorbol 12-myristate 13-acetate (TPA) (Tamayo et al. 1999). Finally, the Huge Index dataset provides in vivo gene expression data for six human tissues (Warrington et al. 2000).
RESULTS AND DISCUSSION
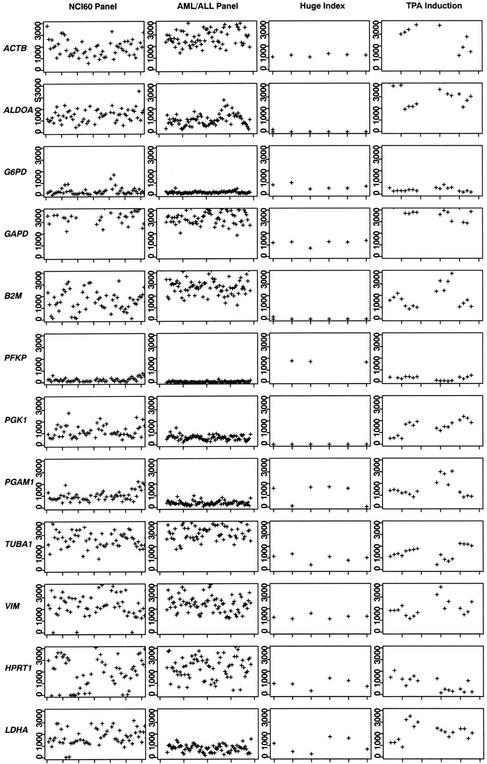
We initially studied the expression levels of 12 genes commonly used to normalize RNA levels measured by Northern blots or RT-PCR. The expression levels for many of these genes fluctuates dramatically both within and across datasets (Fig. 1). Within datasets, the maximum fold change (MFC, the ratio of the maximum and minimum values observed within a dataset) ranges from 1.3 for ACTB within the TPA induction dataset to >300 for VIM in the NCI60 dataset (Table 1). All commonly used control genes have an MFC of >2.0 in at least one dataset. In addition, the observed coefficients of variation (CVs) are frequently >0.5, reflecting the highly variable levels of expression of these genes within datasets.
Figure 1.
Gene expression profiles of classic control genes examined across multiple datasets: NCI60 cell line panel, ALL/AML tumor panel, Huge Index, and TPA cell-line induction. Gene expression levels uniformly rescaled are plotted on the Y-axis; samples (ordered according to their arrangement given in each respective study) are plotted on the X-axis. All datasets with the exception of the Huge Index were rescaled on the basis of mean intensity per scan.
Table 1.
Traditional control genes across four datasets
| Gene | Description | NC160 | AML/ALL | Huge index | TPA | ||||
|---|---|---|---|---|---|---|---|---|---|
| MFC | CV | MFC | CV | MFC | CV | MFC | CV | ||
| ACTB | Actin, beta | 7.8 | 0.39 | 3.5 | 0.32 | 1.3 | 0.11 | 4.6 | 0.35 |
| ALDOA | Aldolase A | 8.2 | 0.37 | 5.7 | 0.48 | 2.9 | 0.37 | 2.9 | 0.31 |
| G6PD | Glucose-6-phosphate dehydrogenase | 7.7 | 0.90 | 5.0 | 0.45 | 2 | 0.28 | 4.6 | 0.43 |
| GAPD | Glyceraldehyde-3-phosphate dehydrogenase | 5.3 | 0.31 | 12.3 | 0.33 | 2 | 0.19 | 1.9 | 0.17 |
| B2M | Beta-2-microglobulin | 25.6 | 0.58 | 14.0 | 0.42 | 3.4 | 0.27 | 4.6 | 0.49 |
| PFKP | Phosphofructokinase, platelet | 12.4 | 0.68 | >12 | 1.66 | >96 | 0.08 | 14.0 | 0.56 |
| PGK1 | Phosphoglycerate kinase 1 | 6.5 | 0.44 | 6.8 | 0.36 | N/A | N/A | 4.9 | 0.41 |
| PGAM1 | Phosphoglycerate mutase 1 (brain) | 5.2 | 0.40 | 12.4 | 0.48 | 1.6 | 0.19 | 3.6 | 0.47 |
| TUBA1 | Tubulin, alpha 1 (testis specific) | >50 | 1.09 | 53.7 | 0.78 | 3.1 | 0.84 | 6.7 | 0.46 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 10.7 | 0.45 | 43.9 | 0.53 | 1.6 | 0.42 | 5.6 | 0.49 |
| VIM | Vimentin | >300 | 0.68 | 12.7 | 0.42 | 3.8 | 0.41 | 28.9 | 0.94 |
| LDHA | Lactate dehydrogenase A | 91.0 | 0.38 | 9.2 | 0.40 | 5.5 | 0.60 | 4.1 | 0.34 |
CV, coefficient of variation; MFC, maximum fold change; AML/ALL, acute myeloid leukemia/acute lymphoblastic leukemia panels; TPA, phorbol 12-myristate 13-acetate induction.
We next used a simple heuristic filter to identify sets of genes showing lower variability. After excluding genes with signal intensities below threshold and with an MFC <2, genes were sorted and ranked according to their CVs. We use this measure of variability because it compensates for the apparent dependence of dispersion on signal (Novak et al. 2002). Similar results were obtained using alternate methods of estimating dispersion (data not shown). Of the housekeeping genes analyzed, only GAPD and ACTB rank among the 100 genes with the lowest variability; however, no traditional control genes display consistently low variability across the four datasets. Nine genes identified by filtering have CVs <0.7 across all four datasets (Table 2). These are not genes that have commonly been used as controls, but include several ribosomal protein (RP) genes (including RPS27A, RPL19, RPL11, RPS29, and RPS3). Even this set of genes shows differing amounts of variability across datasets (Supplementary Fig. 1, available online at http://genome.mcgill.ca/∼pdlee/control_genes and http://www.genome.org). For example, although RPS27A has the lowest CV in the NCI60 and leukemia datasets, its MFC ranges from 2.2 in the NCI60 dataset to 5.6 in the TPA induction dataset.
Table 2.
Genes identified by filtering with CV less than 0.7 across all four datasets
| Gene | Description | NC160 | AML/ALL | Huge index | TPA | ||||
|---|---|---|---|---|---|---|---|---|---|
| MFC | CV | MFC | CV | MFC | CV | MFC | CV | ||
| RPS27A | Ribosomal protein S27a | 2.2 | 0.17 | 3.2 | 0.19 | 3.1 | 0.37 | 5.6 | 0.61 |
| RPL19 | Ribosomal protein L19 | 3.2 | 0.24 | 2.6 | 0.19 | 2 | 0.21 | 2.6 | 0.29 |
| HSPCA | Heat shock 90kD protein 1, alpha | 9.8 | 0.25 | 7.7 | 0.44 | 7.3 | 0.47 | 4.8 | 0.32 |
| RPL11 | Ribosomal protein L11 | 3.4 | 0.25 | 3.8 | 0.2 | 2.8 | 0.3 | 11.8 | 0.54 |
| RPS29 | Ribosomal protein S29 | 3.1 | 0.26 | 4.2 | 0.25 | 1.6 | 0.16 | 3.3 | 0.33 |
| NONO | Non-POU-domain-containing, octamer-binding | 4 | 0.28 | 3.7 | 0.25 | 1.4 | 0.1 | 4.3 | 0.33 |
| AAMP | Angio-associated migratory cell protein | 4 | 0.31 | 8.4 | 0.34 | 2.2 | 0.31 | 4.2 | 0.43 |
| RPS3 | Ribosomal protein S3 | 4.2 | 0.32 | 3.5 | 0.24 | 3.9 | 0.35 | 2.7 | 0.26 |
| ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 7.6 | 0.38 | 7 | 0.29 | 1.4 | 0.15 | 3.8 | 0.31 |
CV, coefficient of variation; MFC, maximum fold change; AML/ALL, acute myeloid leukemia/acute lymphoblastic leukemia panels; TPA, phorbol 12-myristate 13-acetate induction.
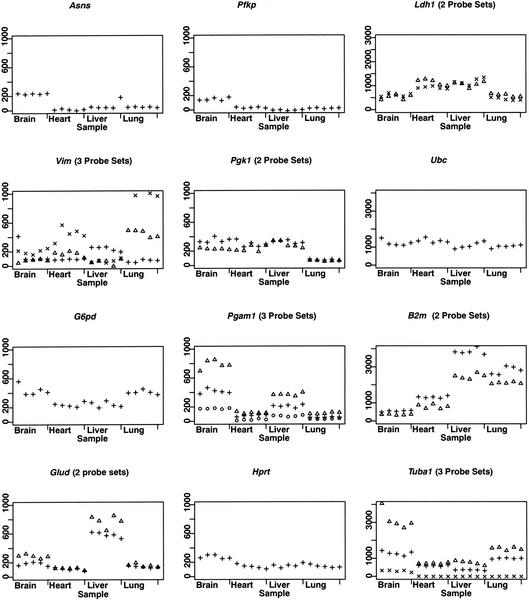
Our failure to identify control genes in the four expression datasets studied might occur if the microarray measurements were associated with high levels of technical variability. To assess whether the observed variation could be due to technical variability rather than biological context, we examined expression levels in triplicate for RNA samples obtained from liver, heart, lung, and brain of three male C57BL/6 mice reared under identical conditions using MU11KA and B arrays (Affymetrix) containing probe-sets for 11,000 mouse genes and ESTs. The expression levels of traditional control genes show greater variability among RNA samples obtained from different tissues than among RNA obtained from the same tissue harvested from different mice, or among identical RNA samples hybridized to replicate microarrays (Fig. 2). To determine whether other genes displayed similar behavior, we performed analysis of variance (ANOVA) on a per-gene basis to determine the amount of observed variability that could be attributed to differences among replicates, mice, or tissues. Technical replicates using identical RNA samples hybridized to three distinct arrays show the least amount of variability: only 3% of genes display significant differences across replicates (P < 0.05). Among biological replicates using RNA from three individual mice, 5%–10% of genes show significant differences (P < 0.05) after adjusting for variation between tissues, and between technical replicates. In contrast, 81%–99% of genes show significant variability (P < 0.05) among different tissues after adjusting for the variability between technical and biological replicates. This trend remains consistent regardless of the filtering criteria or procedure used to select genes (Table 3, Supplementary Fig. 3, available online at http://genome.mcgill.ca/∼pdlee/control_genes and http://www.genome.org). ANOVA performed on the TPA induction and NCI60 datasets similarly reveals greater variability in gene expression across different tissues than across different time points, cell lines, or datasets (Supplementary Fig. 2 and Supplementary Tables 1, 2, and 3, available online at http://genome.mcgill.ca/∼pdlee/control_genes and http://www.genome.org). Performing our analysis using multiple normalization methods did not impact our findings (Supplementary Fig. 2 and Supplementary Table 4, available online at http://genome.mcgill.ca/∼pdlee/control_genes and http://www.genome.org). These results indicate that the variability in gene expression detected in this experiment is not due to technical or intermouse variability, but rather due to the inherent differences in individual RNA levels present among different tissue types.
Figure 2.
Replicate samples from four mouse tissues. RNA was extracted from the liver, heart, lung, and brain of three adult male C57BL/6 mice. To assess technical variability, we divided the RNA from each tissue of one mouse and hybridized it in replicate to three separate arrays. To assess biological variability, we hybridized RNA from identical tissues of three individual mice to three separate arrays. Points are arranged in the following order for each tissue: mouse1-replicate1, mouse1-replicate2, mouse1-replicate3, mouse2-replicate1, mouse3-replicate1. Multiple probe sets, present for Glud, Pgk1, Pgam1, and Ldh1, show consistency in measurements of expression levels across tissues. Other probe sets for Tuba1, Vim, and B2m show a higher degree of variability, indicating issues inherent in probe design. Samples were normalized by mean intensity per scan.
Table 3.
Summary of ANOVA conducted on mouse dataset
| Threshold | Number of genes with expression > threshold | Proportion of genes with P-value < 0.05 when following variables were tested individually* | ||
|---|---|---|---|---|
| Replicate (n = 3) | Mouse (n = 3) | Tissue (n = 4) | ||
| >0 | 6016 | 0.035 | 0.066 | 0.814 |
| >20 | 963 | 0.031 | 0.086 | 0.978 |
| >50 | 773 | 0.031 | 0.082 | 0.978 |
| >100 | 440 | 0.030 | 0.098 | 0.980 |
| >200 | 220 | 0.023 | 0.055 | 0.990 |
General linear model tested on a per-gene basis (expression = replicate + mouse + tissue). P values considered were calculated for each variable individually having adjusted for the variation due to the remaining variables (added-last test).
It is possible that our failure to identify control genes may result from data-filtering techniques that excluded RNA species expressed at low copy number across a wide range of tissues, or genes that are simply not present on the microarrays used in these studies. These issues may be addressed by the future development of more sensitive complete genome arrays. Despite this, our results clearly show that the expression levels of genes that have been commonly used as controls in classical experiments vary significantly among different cellular and experimental contexts. Furthermore, we fail to identify mammalian genes that qualify as “control genes” on the basis of a definition of ubiquitous and stable expression. Although some genes do appear quite stable in expression level within any one experiment, there do not appear to be any genes expressed at stable levels across all four datasets studied in this paper. Hence, the traditional use of individual genes as normalization controls in experiments that compare diverse biological tissues would lead to substantial errors in the derived estimates of fold change in gene expression levels. From inspection of the data, it is apparent that some transcripts may serve as control genes for studies performed in a single tissue context; however, these conclusions are limited by a study design that does not address the effects of physiologic regulation on the expression of these genes.
The unproven existence of control genes seems to have achieved acceptance in part because of its conceptual simplicity and practical limitations of the past. Recent studies have expressed concern that individual genes or groups of genes may serve as inadequate internal standards for measuring RNA expression levels (Souaze et al. 1996; Savonet et al. 1997; Ivell 1998; Serazin-Leroy et al. 1998; Oliveira et al. 1999; Thellin et al. 1999; Suzuki et al. 2000; Wu and Rees 2000 ); Measures for data standardization and quality control in microarray databases are currently being reviewed by the MGED working group on Microarray Data Annotations (www.mged.org). The establishment of common frames of reference requires a reexamination of assumptions inherent in the design of biological experiments. From these findings, we propose that all genes are differentially expressed in at least one biological context and that the expression of every gene is therefore context dependent. Given the absence of ubiquitous control genes, variation in microarray expression studies must instead be interpreted using statistical characteristics of the data without preconceptions arising from the traditional notions of internal control genes.
METHODS
Public Microarray Datasets
Microarray datasets for the NCI60 cancer cell line panel, the ALL/AML tumors, and the TPA treatment in HL60, U937, NB4, and Jurkat cell lines are available at http://www.genome.wi.mit.edu/MPR/datasets. The human tissue expression profiles contained in the Huge Index dataset were obtained at http://www.hugeindex.org/.
Mouse Microarray Dataset
Mouse tissues were obtained from three adult male C57BL/6 littermates. Mice were killed by cervical dislocation and the tissues rapidly dissected and homogenized in Trizol reagent (Life Technologies). Total cellular RNA was prepared according to the manufacturer's instructions and analyzed by nondenaturing (1% agarose-1 × TBE) gel electrophoresis. Probes for the microarray studies were prepared by priming 20 μg of total RNA with 100 pmole of T7– (T) 24 primer (Genosys). The RNA-primer mixture was denatured for 10 min at 70°C, and then chilled on ice. First-strand cDNA was synthesized using Superscript II reverse transcriptase (Life Technologies). Second-strand synthesis was performed using RNAse H, DNA polymerase I, and Escherichia coli DNA ligase (Life Technologies). Biotinylated riboprobes were prepared from the entire cDNA reaction using the ENZO Bioarray High Yield RNA Transcript Labeling Kit (ENZO Diagnostics). The average probe length was reduced by incubating the probe in 1X Fragmentation Buffer for 35 min at 95°C. Hybridization was performed at 45°C for 16–20 h using 15 μg of biotinylated probe. Following hybridization, the arrays were subjected to 10 low-stringency washes and 4 high-stringency washes using a GeneChip Fluidics Station 400 (Affymetrix). Bound probe was detected by incubating arrays with SAPE (streptavidin phycoerthryin, Molecular Probes) and scanning the chips using a GeneArray Scanner (Agilent). Scanned images were analyzed using the GeneChip Analysis Suite 3.3 (Affymetrix). Full details of the microarray methods have been described previously (Novak et al. 2002).
Data Analysis
Traditional control genes analyzed in human datasets included: β-actin (ACTB), β-2-microglobulin (B2M), phosphofructokinase (PFKP), phosphoglycerate kinase (PGK1), aldolase A (ALDOA), phosphoglycerate mutase (PGAM), α-tubulin (TUBA), glyceraldehyde-3 phosphate dehydrogenase (GAPD), glucose-6 phosphate dehydrogenase (G6PD), lactate dehydrogenase A (LDHA), hypoxanthine phosphoribosyltransferase (HPRT), and vimentin (VIM). Traditional control genes analyzed in mouse datasets included asparagine synthetase (Asns), phosphofructokinase (Pfkp), lactate dehydrogenase A (Ldh1), vimentin (Vim), phosphoglycerate kinase (Pgk1), ubiquitin (Ubc), glucose-6 phosphate dehydrogenase (G6pd), phosphoglycerate mutase (Pgam1), β-2-microglobulin (B2m), glutamate dehydrogenase (Glud), hypoxanthine phosphoribosyltransferase (Hprt), and α-tubulin (Tuba1). For accession numbers, see Supplementary Table 5 (available online at http://genome.mcgill.ca/∼pdlee/control_genes and http://www.genome.org).
Regression scaling was performed only on datapoints assigned a ‘P’ absolute call by the Affymetrix GeneChip software: the absolute call estimates the hybridization quality for an individual probe set on the basis of measures of background and signal dispersion. The regression scaling algorithm has been described previously (Novak et al. 2002): it uses normalization to the regression coefficient of the first sample in each dataset. We rescaled datasets on the basis of mean overall intensity per scan. Mean intensity was calculated on the genes with a minimum average difference of 50 and an absolute call of ‘P’ by the GeneChip algorithm.
Data manipulation and analysis was accomplished using a variety of Perl and VBScripts in Microsoft Excel. Graphs were created using R (http://www.r-project.org). ANOVA was performed using SAS (SAS Institute Inc), testing the amount of observed variability in expression of each gene resulting from replicate (repeat hybridizations of the same RNA sample), mouse (samples from three individual mice), or tissue (samples from four different tissues); a general linear model was used on a per-gene basis (PROC GLM). P values considered were calculated for each variable individually, having adjusted for the variation resulting from remaining variables (added-last test / SAS Type III F-Test). We conducted ANOVA separately on subsets of the data meeting initial filtering criteria of minimum expression levels of greater than 20, 50, 100, or 200 units across all 12 experiments. ANOVA results must be interpreted with caution because the small sample size makes assessments of normality and homoscedasticity difficult. P values considered were for the added-last F-test (testing each variable individually, having adjusted for all other variables). Datasets, supplementary figures, tables, and analytical scripts are available at http://genome.mcgill.ca/∼pdlee/control_genes.
WEB SITE REFERENCES
http://www-genome.wi.mit.edu/MPR, Whitehead/MIT Center for Genome Research.
http://www.hugeindex.org, The Human Gene Expression Index.
http://www.mged.org, Microarray Gene Expression Data Group.
http://www.r-project.org, The R Project.
http://genome.mcgill.ca/∼pdlee/control_genes, Supplementary Data and Figures (Montreal Genome Centre).
Acknowledgments
We thank J. Novak, B. Ge, J. Engert, C. Loredo-Osti, T. Golub, J. Staunton, D. DeGraaf, P. Tamayo, and T. Maniatis for their useful discussions. This research was supported by grants from the Canadian Institutes for Health Research (Grant number GOP 36056), the Canadian Genetics Diseases Network and the Mathematics of Information Technology and Complex Systems (Networks of Centres of Excellence Program), and a research contract from Bristol Myers Squibb, Millennium Pharmaceuticals Inc., and Affymetrix. R.S. and T.J.H. are respectively the recipients of a fellowship and a Clinician-Scientist award from the Canadian Institutes of Health Research. C.G. is a Chercheur-Boursier of Fonds de la Recherche en Santé du Québéc.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tjhudson@med.mcgill.ca; FAX (514) 933-7146.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.217802.
REFERENCES
- Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P, Welch PJ, Welte K, King MC, Barber JR, et al. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci. 2001;98:130–135. doi: 10.1073/pnas.98.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butte AJ, Tamayo P, Slonim D, Golub TR, Kohane IS. Discovering functional relationships between RNA expression and chemotherapeutic susceptibility using relevance networks. Proc Natl Acad Sci. 2000;97:12182–12186. doi: 10.1073/pnas.220392197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Ivell R. A question of faith—or the philosophy of RNA controls. J Endocrinol. 1998;159:197–200. doi: 10.1677/joe.0.1590197. [DOI] [PubMed] [Google Scholar]
- Khan J, Simon R, Bittner M, Chen Y, Leighton SB, Pohida T, Smith PD, Jiang Y, Gooden GC, Trent JM, et al. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- Lander ES. Array of hope. Nat Genet. 1999;21:3–4. doi: 10.1038/4427. [DOI] [PubMed] [Google Scholar]
- Novak, J.P., Sladek, R., and Hudson, T.J. 2002. Characterization of variability in large-scale gene expression data: Implications for study design. Genomics 79: (in press). [DOI] [PubMed]
- Oliveira JG, Prados RZ, Guedes AC, Ferreira PC, Kroon EG. The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase is inappropriate as internal control in comparative studies between skin tissue and cultured skin fibroblasts using Northern blot analysis. Arch Dermatol Res. 1999;291:659–661. doi: 10.1007/s004030050471. [DOI] [PubMed] [Google Scholar]
- Savonet V, Maenhaut C, Miot F, Pirson I. Pitfalls in the use of several ‘housekeeping‘ genes as standards for quantitation of mRNA: The example of thyroid cells. Anal Biochem. 1997;247:165–167. doi: 10.1006/abio.1997.2055. [DOI] [PubMed] [Google Scholar]
- Serazin-Leroy V, Denis-Henriot D, Morot M, de Mazancourt P, Giudicelli Y. Semi-quantitative RT-PCR for comparison of mRNAs in cells with different amounts of housekeeping gene transcripts. Mol Cell Probes. 1998;12:283–291. doi: 10.1006/mcpr.1998.0182. [DOI] [PubMed] [Google Scholar]
- Souaze F, Ntodou-Thome A, Tran CY, Rostene W, Forgez P. Quantitative RT-PCR: Limits and accuracy. Biotechniques. 1996;21:280–285. doi: 10.2144/96212rr01. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc Natl Acad Sci. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: Use and limits. J Biotechnol. 1999;75:291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- Warrington JA, Nair A, Mahadevappa M, Tsyganskaya M. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol Genomics. 2000;2:143–147. doi: 10.1152/physiolgenomics.2000.2.3.143. [DOI] [PubMed] [Google Scholar]
- Wu YY, Rees JL. Variation in epidermal housekeeping gene expression in different pathological states. Acta Derm Venereol. 2000;80:2–3. doi: 10.1080/000155500750012397. [DOI] [PubMed] [Google Scholar]
- Young RA. Biomedical discovery with DNA arrays. Cell. 2000;102:9–15. doi: 10.1016/s0092-8674(00)00005-2. [DOI] [PubMed] [Google Scholar]