Abstract
A simple and efficient oligonucleotide array was developed to identify single nucleotide polymorphisms (SNPs) encoded within the highly polymorphic human major histocompatibility complex (MHC) using HLA-B as a model system. A total of 137 probes were designed to represent all known polymorphisms encoded in exons 2 and 3. PCR products were amplified from human genomic DNA and allowed to hybridize with the oligonucleotide array. Hybridization was detected by fluorescence scanning, and HLA-B alleles were assigned by quantitative analysis of the hybridization results. Variables known to influence the specificity of hybridization, such as oligonucleotide probe size, spacer length, surface density, hybridization conditions, and array uniformity and stability were studied. The efficiency and specificity of identifying HLA-B SNPs using the oligonucleotide arrays was evaluated by blinded analysis of 100 samples from unrelated individuals representing all HLA-B phenotypes. The oligonucleotide array method described in this paper provides unambiguous detection of complex heterozygous SNP combinations. This methodological approach may be applied to other highly polymorphic gene systems.
Human DNA sequence variation provides an invaluable tool for understanding linkage disequilibrium and recombination, the evolutionary and phylogenetic basis of human populations and the functional significance of diversity with respect to disease susceptibility (Collins et al. 1998; Shafer et al. 1998; Kruglyak 1999). The most common type of human genetic variation is the SNP, in which two alternative bases occur at appreciable frequency (>1%) in the population. Thirty-five percent of human mutations has been found to occur within CpG dinucleotides in genes causing human genetic disease (Cooper and Youssoufian 1998). Currently, >2,550,000 SNPs have been deposited into the central database for SNPs (dbSNP) established by the National Human Genome Research Institute (NHGRI), and the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/SNP) (Sherry et al. 1999). Large-scale surveys of human genetic variation have uncovered similar frequencies of SNPs in coding and noncoding regions (on average of 1 per 217 bp to 1 per 346 bp) (Wang et al. 1998; Cargill et al. 1999; Halushka et al. 1999).
Pedigree and association studies require an accurate, complete, and up-to-date collection of SNPs to facilitate research into the biological impact of human variation (Risch and Merikangas 1996; Collins et al. 1997; Kruglyak 1997, 1999). All of the well-established SNP scanning methods are gel-based assays: single-strand conformation polymorphism (SSCP) (Grompe 1993), heteroduplex analysis (HA) (White et al. 1992), denaturation gradient gel electrophoresis (DGGE) (Mashal and Sklar 1996), RNase and chemical cleavage mismatch (CCM) methods (Myers et al. 1985; Roberts et al. 1997), and DNA sequencing (Venter et al. 1996; Rowen et al. 1997). Newly developed scanning methods include T4 endonuclease VII cleavage (Babon et al. 1999), multiphoton detection (Hawkins 1997), Escherichia coli mismatch enzyme cleavage (Del Tito et al. 1998), denaturing high-performance liquid chromatography (DHPLC) (Underhill et al. 1996), mass spectrometry (Laken et al. 1998), and oligonucleotide arrays (Guo et al. 1994; Gilles et al. 1999). Methods for screening unknown DNA variation include single nucleotide primer extension (SnuPE) (Shumaker et al. 1996), oligonucleotide ligation (Tobe et al. 1996), and DNA arrays (Lipshutz et al. 1995).
Oligonucleotide array technology has been applied successfully for gene discovery, monitoring gene expression patterns, detecting mutations and polymorphisms, and mapping of genomic clones (Pease 1994; Lipshutz et al 1995; Lockhart et al. 1996; Sapolsky and Lipshutz 1996). With the potential to detect thousands of SNPs in parallel, oligonucleotide arrays offer a cost-effective approach for high-throughput polymorphism analysis.
The HLA Class I and II genes encode some of the most complex patterns of SNP substitutions in the human genome (So 1994). Related as a family through gene duplication, the class I HLA-A, B, and C genes are distinguished by locus-specific nucleotide substitutions (Robinson et al. 2000). Each Class I gene consists of eight exons; exons 2, 3, and 4 encode the α1, α2, and α3 domains of the Class I molecule, respectively. Exons 2 and 3 encode the majority of SNPs, which define the basis for the HLA phenotype (Robinson et al. 2000). Because of the extensive sharing of SNPs among HLA alleles of a given Class I gene, more than one combination of two alleles may yield the same collective pattern of SNPs (http://www.ebi.ac.uk/imgt/hla/intro.html).
Robust SNP-finding technology for the HLA system requires parallel analysis of multiple regions of variation and the power to distinguish alleles by their unique constellation of SNPs. In this paper, we use HLA-B as a model system to demonstrate how oligonucleotide arrays can be used to both identify SNPs and to genotype HLA-B alleles. Of the 381 known HLA-B alleles we studied, nucleotide substitutions occurred at 52 positions of exon 2 and at 42 positions of exon 3. We furthermore show how the majority of ambiguous heterozygous allele combinations can be surmounted through the judicious design of oligonucleotide probes representing all known polymorphisms in exons 2 and 3 of HLA-B. Proof of principle was established in a blinded testing of 100 unrelated human DNA samples.
RESULTS
Design of Oligonucleotide Probes and Probe Panels
A key feature of the oligonucleotide array assay is the high redundancy of probes. A panel of 68 20-mer oligonucleotide probes was designed for all polymorphisms in exon 2 and 69 20-mer probes were designed for exon 3. Probes were grouped into 15 regions for exon 2 and 13 regions for exon 3 (Table 1). The 20-mer probes in each group were descriptive for the identical base positions in exons 2 and 3. Of 137 probes, 16 probes were descriptive for a single nucleotide change.
Table 1.
Sequences and Locations of Oligonucleotide Probes for HLA-B Exons 2 and 3
| Probe | Nucleotide position | Sequence |
|---|---|---|
| A. HLA-B Exon 2 | ||
| A1 | 01–20 | 5′GCTCCCACTTCATGAGGTAT3′ |
| A2 | 01–20 | 5′GCTCCCACTCCATGAGGTAT3′ |
| B1 | 01–20 | 5′TTCTACACCTCCGTGTCCCG3′ |
| B2 | 21–40 | 5′TTCTACACCGCCATGTCCCG3′ |
| B3 | 21–40 | 5′TTCGACACCGCCATGTCCCG3′ |
| B4 | 21–40 | 5′TTCCACACCTCCGTGTCCCG3′ |
| B5 | 21–40 | 5′TTCCACACCGCCATGTCCCG3′ |
| B6 | 21–40 | 5′TTCTACACCGCTATGTCCCG3′ |
| B7 | 21–40 | 5′TTCTACACCGCCGTGTCCCG3′ |
| C1 | 41–60 | 5′GCCCGTCCGCGGGGAGCCCC3′ |
| C2 | 41–60 | 5′GCCTGGCCGCGGGGAGCCCC3′ |
| C3 | 41–60 | 5′GCCCGGCCGCGGGGAGCCCC3′ |
| D1 | 61–80 | 5′GCTTCATCTCAGTGGGCTAC3′ |
| D2 | 61–80 | 5′GCTTCATCACCGTGGGCTAC3′ |
| D3 | 61–80 | 5′GCTTCATTGCAGTGGGCTAC3′ |
| D4 | 61–80 | 5′GCTTCATCGCAGTGGGCTAC3′ |
| E1 | 81–100 | 5′GTGGACGACACCCAGTTCGT3′ |
| E2 | 81–100 | 5′GTGGACGGCACCCAGTTCGT3′ |
| E3 | 81–100 | 5′GTGGACGACACGCTGTTCGT3′ |
| E4 | 81–100 | 5′GTGGACGACACGCAGTTCGT3′ |
| F1 | 111–130 | 5′AGCGACGCCACGAGTCCGAG3′ |
| F2 | 111–130 | 5′AGCGACGCCGCGAGTCCGAG3′ |
| G1 | 96–115 | 5′TTCGTGCGGTTCGACAGCGA3′ |
| G2 | 96–115 | 5′TTCGTGAGGTTCGACAGCGA3′ |
| H1 | 121–140 | 5′CGAGTCCGAGAGAGGAGCCG3′ |
| H2 | 121–140 | 5′CGAGTCCGAGGATGGCGCCC3′ |
| H3 | 121–140 | 5′CGAGTCCGAGGACGGAGCCC3′ |
| H4 | 121–140 | 5′CGAGTCCGAGGAAGGAGCCG3′ |
| I1 | 141–160 | 5′CGGGCGCCATGGATAGAGCA3′ |
| I2 | 141–160 | 5′CGGGCGCCGTGGGTGGAGCA3′ |
| I3 | 141–160 | 5′CGGGCGCCGTGGATAGAGCA3′ |
| J1 | 161–180 | 5′GGAGGGGCCGGAATATTGGG3′ |
| J2 | 161–180 | 5′AGAGGGGCCGGAGTATTGGG3′ |
| J3 | 161–180 | 5′GGAGGGGCCGGAGCATTGGG3′ |
| J4 | 161–180 | 5′GGAGGGGCCGGAGTATTGGG3′ |
| K1 | 181–200 | 5′ACCGGAACACACAGATCTAC3′ |
| K2 | 181–200 | 5′ACCGGAACACACAGATCTTC3′ |
| K3 | 181–200 | 5′ACCGGGAGACACAGATCTCC3′ |
| K4 | 181–200 | 5′ACCGGAACACACAGATCTGC3′ |
| K5 | 181–200 | 5′ACCGGAACACACAGATCTCC3′ |
| K6 | 181–200 | 5′ACCGGGAGACACGGAACATG3′ |
| K7 | 181–200 | 5′ACCGGGATACACAGATCTCC3′ |
| K8 | 181–200 | 5′ACCGGGAGACACAGATCTGC3′ |
| K9 | 181–200 | 5′ACCGGGAGACACAGAAGTAC3′ |
| K10 | 181–200 | 5′ACGGGGAGACACGGAACATG3′ |
| K11 | 181–200 | 5′ACCGGGAGACACAGATCTTC3′ |
| L1 | 201–220 | 5′AAGGCCCAGGCACAGACTGA3′ |
| L2 | 201–220 | 5′AAGACCAACACACAGACTTA3′ |
| L3 | 201–220 | 5′AAGGCCTCCGCGCAGACTTA3′ |
| L4 | 201–220 | 5′AAGGCCAAGGCACAGACTTA3′ |
| L5 | 201–220 | 5′AAGGCCAAGGCACAGACTGA3′ |
| L6 | 201–220 | 5′AAGCGCCAGGCACAGACTGA3′ |
| L7 | 201–220 | 5′AAGACCAACACACAGACTGA3′ |
| M1 | 216–235 | 5′ACTGACCGAGAGAGCCTGCG3′ |
| M2 | 216–235 | 5′ACTTACCGAGAGAACCTGCG3′ |
| M3 | 216–235 | 5′ACTTACCGAGAGAGCCTGCG3′ |
| M4 | 216–235 | 5′ACTGACCGAGAGGACCTGCG3′ |
| M5 | 216–235 | 5′ACTTACCGAGAGGACCTGCG3′ |
| M6 | 216–235 | 5′ACTGACCGAGTGAGCCTGCG3′ |
| M7 | 216–235 | 5′ACTGACCGAGTGGGCCTGCG3′ |
| M8 | 216–235 | 5′ACTGACCGAGAGAACCTGCG3′ |
| N1 | 231–250 | 5′CTGCGCACCGCGCTCCGCTA3′ |
| N2 | 231–250 | 5′CTGCGGATCGCGCTCCGCTA3′ |
| N3 | 231–250 | 5′CTGCGGACCCTGCTCCGCTA3′ |
| N4 | 231–250 | 5′CTGCGGAACCTGCTCCGCTA3′ |
| N5 | 231–250 | 5′CTGCGGAACCTGCGCGGCTA3′ |
| O1 | 251–270 | 5′CTACAACCAGAGCGAGGACG3′ |
| O2 | 251–270 | 5′CTACAACCAGAGCGAGGCCG3′ |
| B. HLA-B Exon 3 | ||
| A1 | 05–24 | 5′TCACACCCTCCAGAGCATGT3′ |
| A2 | 05–24 | 5′TCACATCATCCAGAGGATGT3′ |
| A3 | 05–24 | 5′TCACACTTGGCAGACGATGT3′ |
| A4 | 05–24 | 5′TCACACCCTCCAGTGGATGT3′ |
| A5 | 05–24 | 5′TCACACTTGGCAGAGGATGT3′ |
| A6 | 05–24 | 5′TCACACCCTCCAGACGATGT3′ |
| A7 | 05–24 | 5′TCACACCCTCCAGAATATGT3′ |
| A8 | 05–24 | 5′TCACATCATCCAGAGCATGT3′ |
| A9 | 05–24 | 5′TCACACCATCCAGAGGATGT3′ |
| A10 | 05–24 | 5′TCACATCATCCAGGTGATGT3′ |
| A11 | 05–24 | 5′TCACACCCTCCAGAGGATGT3′ |
| B1 | 21–40 | 5′ATGTACGGCTGCGACGTGGG3′ |
| B2 | 21–40 | 5′ATGTATGGCTGCGACCTGGG3′ |
| B3 | 21–40 | 5′ATGTTTGGCTGCGACGTGGG3′ |
| B4 | 21–40 | 5′ATGTAAGGCTGCGACGTGGG3′ |
| B5 | 21–40 | 5′ATGTCTGGCTGCGACGTGGG3′ |
| B6 | 21–40 | 5′ATGTACGGCTGCGACCTGGG3′ |
| B7 | 21–40 | 5′ATGTTTGGCTGCGACCTGGG3′ |
| B8 | 21–40 | 5′ATGTATGGCTGCGACATGGG3′ |
| B9 | 21–40 | 5′ATGTATGGCTGCGACGTGGG3′ |
| C1 | 41–60 | 5′GCCGGACGGGCGCCTCCTCC3′ |
| C2 | 41–60 | 5′GCCCGACGGGCGCTTCCTCC3′ |
| C3 | 41–60 | 5′GCCCGACGGGCGCCTCCTCC3′ |
| D1 | 61–80 | 5′GCGGGCATGACCAGTACGCC3′ |
| D2 | 61–80 | 5′GCGGGCATAACCAGTACGCC3′ |
| D3 | 61–80 | 5′GCGGGCATAACCAGTTAGCC3′ |
| D4 | 61–80 | 5′GCGGGTATAACCAGTTCGCC3′ |
| D5 | 61–80 | 5′GCGGGCATGACCAGTCCGCC3′ |
| D6 | 61–80 | 5′GCGGGTATGACCAGTCCGCC3′ |
| D7 | 61–80 | 5′GCGGGTACCACCAGGACGCC3′ |
| D8 | 61–80 | 5′GCGGGCATGACCAGTTCGCC3′ |
| D9 | 61–80 | 5′GCGGGCATAACCAGTTCGCC3′ |
| D10 | 61–80 | 5′GCGGGTATGACCAGGACGCC3′ |
| D11 | 61–80 | 5′GCGGGTATAACCAGTTAGCC3′ |
| D12 | 61–80 | 5′GCGGGTATGACCAGTACGCC3′ |
| E1 | 81–100 | 5′TACGACGGCAAAGATTACAT3′ |
| E2 | 81–100 | 5′TACGACGGCAAGGATTACAT3′ |
| F1 | 111–130 | 5′GAGGACCTGAGCTCCTGGAC3′ |
| F2 | 111–130 | 5′GAGGACCTGCGCTCCTGGAC3′ |
| G1 | 131–150 | 5′CGCCGCGGACACGGCGGCTC3′ |
| G2 | 131–150 | 5′CGCGGCGGACACCGCGGCTC3′ |
| G3 | 131–150 | 5′CGCCGCGGACAAGGCGGCTC3′ |
| G4 | 131–150 | 5′CGCCGCGGACACGGCAGCTC3′ |
| G5 | 131–150 | 5′CGCCGCGGACACCGCGGCTC3′ |
| G6 | 131–150 | 5′CGCGGCGGACACGGCGGCTC3′ |
| H1 | 151–170 | 5′AGATCACCCAGCTCAAGTGG3′ |
| H2 | 151–170 | 5′AGATCTCCCAGCGCAAGTTG3′ |
| H3 | 151–170 | 5′AGATCACCCAGCGCAAGTGG3′ |
| I1 | 171–190 | 5′GAGGCGGCCCGTGAGGCGGA3′ |
| I2 | 171–190 | 5′GAGGCGGCCCGTGTGGCGGA3′ |
| J1 | 191–210 | 5′GCAGCGGAGAGCCTACCTGG3′ |
| J2 | 191–210 | 5′GCAGGACAGAGCCTACCTGG3′ |
| J3 | 191–210 | 5′GCAGTGGAGAGCCTACCTGG3′ |
| J4 | 191–210 | 5′GCAGCTGAGAACCTACCTGG3′ |
| J5 | 191–210 | 5′GCAGCGGAGAACCTACCTGG3′ |
| J6 | 191–210 | 5′GCAGCTGAGAGCCTACCTGG3′ |
| K1 | 211–230 | 5′AGGGCGAGTGCGTGGAGTGG3′ |
| K2 | 211–230 | 5′AGGGCCTGTGCGTGGAGTGG3′ |
| K3 | 211–230 | 5′AGGGCCTGTGCGTGGACGGG3′ |
| K4 | 211–230 | 5′AGGGCCTGTGCGTGGAGTCG3′ |
| K5 | 211–230 | 5′AGGGCACGTGCGTGGAGTCG3′ |
| K6 | 211–230 | 5′AGGGCCTGTGCGTGGAGGGG3′ |
| K7 | 211–230 | 5′AGGACCTGTGCGTGGAGTCG3′ |
| K8 | 211–230 | 5′AGGGCACGTGCGTGGAGTGG3′ |
| L1 | 231–250 | 5′CTCCGCAGACACCTGGAGAA3′ |
| L2 | 231–250 | 5′CTCCGCAGATACCTGGAGAA3′ |
| M1 | 257–276 | 5′GGACAAGCTGGAGCGCGCTG3′ |
| M2 | 257–276 | 5′GGACACGCTGGAGCGCGCGG3′ |
| M3 | 257–276 | 5′GGAGACGCTGCAGCGCGCGG3′ |
Each oligonucleotide probe has a 15 dT linker at its 5′-end. The polymorphic sites in each probe are in bold and underlined.
For heterozygous samples, two oligonucleotide probes in a group could be perfectly matched with the PCR product, each matched with a different allele. For homozygous samples, only one probe in each group would be perfectly matched with the PCR product. A theoretical hybridization pattern was generated for each allele by counting the probes matched with its sequence.
Each probe contained a 5′ amino group for immobilization chemistry, a 15-mer poly-dT spacer, followed by the 20-nucleotide hybridization sequence. The polymorphic sequence was situated near the center of each hybridization sequence. Melting temperatures of oligonucleotide probes were calculated using available software (http://www.genome.wi.mit.edu/genome_software/other/primer3.html). The 137 20-mer probes differed by a 5°C range in melting temperature.
Computer Simulation of HLA-B Heterozygous Allele Combinations
The majority of individuals are heterozygous for two different HLA-B alleles. Sequence polymorphisms or “motifs” can be shared among families of HLA-B alleles at a given locus (Robinson et al. 2000). Therefore, when both HLA-B alleles are coamplified in PCR, more than one combination of two alleles may produce identical patterns of hybridization to oligonucleotide probes.
Computer simulation of all potential heterozygous combinations of exon 2 and exon 3 polymorphisms was performed by translating the hybridization pattern of each allele into a table of binary data, with “1” representing a positive hybridization signal and “0” representing a negative hybridization signal with that probe. The resulting hybridization patterns of allele combinations were analyzed by summing the binary hybridization tables of their component alleles. The uniqueness of each allele combination was evaluated by comparing its hybridization pattern with hybridization patterns of all other potential allele combinations. A table then was generated for alleles of each unique hybridization pattern.
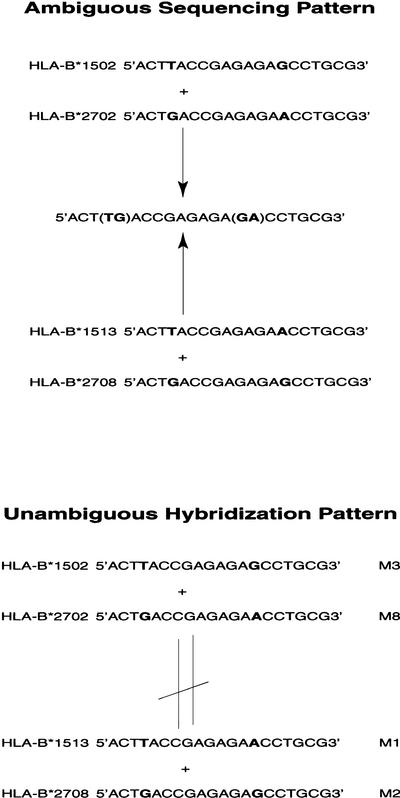
Of the 381 known HLA-B alleles, 467 ambiguous heterozygous combinations of alleles have been reported (http://www.ebi.ac.uk/imgt/hla/intro.html). To reduce the number of potential ambiguous hybridization results, the 20-mer oligonucleotide probes each were designed to represent combinations of SNPs, rather than one SNP. Computer simulation of all potential probe hybridization patterns reduced the total number of ambiguous combinations to 118. An example of how our probe panel can surmount ambiguous combinations is shown in Figure 1.
Figure 1.
Improved allele recognition by oligonucleotide array. Allele combination of HLA-B*1502 and B*2702 share the same polymorphisms with combinations of B*1513 and 2708 (polymorphic sites are shown in bold). Both allele combinations show the same sequence pattern (top) and cannot be distinguished by sequencing method. When hybridized with oligonucleotide arrays, these two combinations show different hybridization pattern (bottom). Combination of B*1502 and B*2702 is represented by probe M3 and M8, while combination of B*1513 and B*2708 is represented by probe M1 and M3 (see Table 1).
Construction of Oligonucleotide Arrays
HLA-B oligonucleotide arrays were constructed on treated microscopic slides by attaching presynthesized oligonucleotide probes. The arrays for exon 2 and exon 3 were fabricated on separate slides. Oligonucleotide probes were diluted to 500 pmol/μL, transferred into a 96-well microtiter plate, applied to glass slides by using a Molecular Dynamic spotter system and immobilized on glass supports by covalent binding (Guo et al. 1994).
The spotter system was designed to automatically collect samples from a 96-well microtiter plate with a 6-pen robot arm. Each pen collected from between 250 to 500 nL of solution per pen and deposited 0.25–1 nL (125–500 fmol) on each slide, creating spots that ranged from 100–150 μm in diameter. The robot was programmed so that adjacent spots were spaced to avoid contact with each other, with 400–500 μm separating the centers of each spot. The precision of this measurement is ∼10 μm. A total of 24 glass slides, each having capacity of 3,072 different oligonucleotides, were arrayed at one time.
The intensity of the hybridization signal can differ substantially from one spot to another because of variability of the concentration of spotted oligonucleotide probe, concentration of template DNA across the glass slide, and surface conditions. These variables can complicate the interpretation of the hybridization signals and potentially cause false positive or false negative results (Lee et al. 2000). To reduce these variables, we spotted each oligonucleotide probe in quadruplicate and averaged the intensities for each probe.
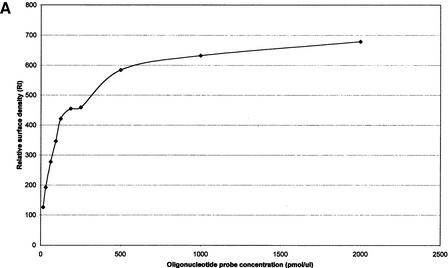
The surface density of the oligonucleotide probes on the glass surface has proven to be an important parameter for hybridization efficiency of the oligonucleotide array (Guo et al. 1994). The surface density of each oligonucleotide probe can be adjusted easily by changing the concentration of the oligonucleotide solution during the application step. Experiments were conducted to optimize the surface density (Fig. 2A,B). Different concentrations of an oligonucleotide, labeled with an amino group at its 5′-end and a Rhodamine dye at its 3′-end, were applied to the glass surface for immobilization. The surface densities of immobilized oligonucleotide probes were quantified by fluorescence scanning after the excess oligonucleotides were washed away (Fig. 2A). These results indicate a near linear relationship between the oligonucleotide concentration and the surface density below a DNA concentration of 500 pmol/μL. Above 500 pmol/μL, the surface density increased slightly with increased oligonucleotide concentration.
Figure 2.
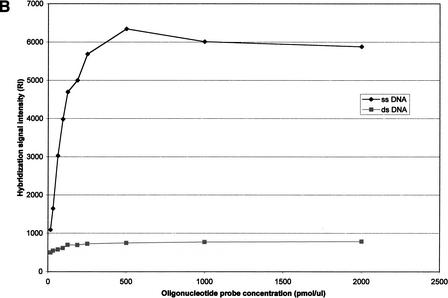
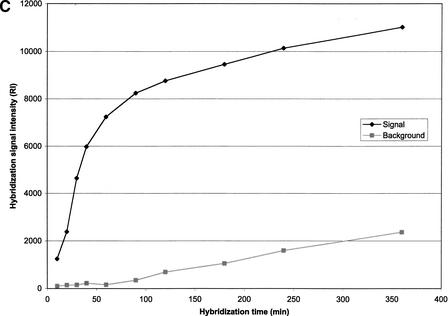
(A) Effect of oligonucleotide probe concentration on surface density of immobilized oligonucleotides. The K5 oligonucleotide probe (Table 1) was labeled with an amino group at its 5′-end and a Rhodamine dye at its 3′-end, and mixed with an oligonucleotide of the same sequence having an amino group labeled at its 5′-end (ratio of 1:10). Mixtures of oligonucleotides ranging from 15.65 to 2000 pmol/mL were spotted on activated glass slides, incubated at 37°C for 2 h and then washed with 1% NH4OH and water. (B) Effect of oligonucleotide surface density on hybridization signal intensity. A mixture of 50μL double-stranded or single-stranded Rhodamine-labeled PCR product of HLA-B exon 2 in 5×SSPE and 0.5% SDS was hybridized to the array using the K5 oligonucleotide probe (Table 1) at various surface densities. (C) Hybridization kinetics. Fluorescence intensities of specific hybridization signals and nonspecific background both increase with longer hybridization time.
To determine the optimal surface density, hybridization experiments were performed using Rhodamine-labeled, double-stranded or single-stranded PCR product amplified from exon 2 as described in the Experimental Protocol section. The results, shown in Fig. 2B, demonstrate that the strongest hybridization signal was achieved at a probe concentration of 500 pmol/μL, which corresponds to a surface density of ∼350 Å2/molecule. The different signal intensities of single-stranded PCR products and double-stranded products also are shown in Fig. 2B. At each surface density, single-stranded products always generated much higher signals than double-stranded DNA.
Preparation of Single-Stranded Template
Although it is simpler to prepare double-stranded rather than single-stranded PCR products, hybridization of the double-stranded DNA to the support-bound oligonucleotide array will necessarily suffer from competition of the complementary strand with the oligonucleotide probes (Guo et al. 1994).
Double-stranded PCR product was first generated as previously described (Petersdorf et al. 1995) and purified to remove the excess primers. Single-stranded DNA molecules then were generated from asymmetric PCR using one 5′ fluorescent-labeled primer. Single-stranded template generated from asymmetric PCR demonstrated very high hybridization efficiency when applied to the oligonucleotide array (Fig. 2B).
Hybridization Kinetics
A key parameter of the hybridization process is hybridization speed. Hybridization kinetics was evaluated by varying hybridization time (Fig. 2C). The specific hybridization signal increased with increasing hybridization time, while a substantial nonspecific background became evident at ∼3 h. Therefore, a hybridization period of 3 h was routinely employed for hybridization with exon 2 and 3 oligonucleotide arrays, producing a consistent and reproducible hybridization signal without background interference.
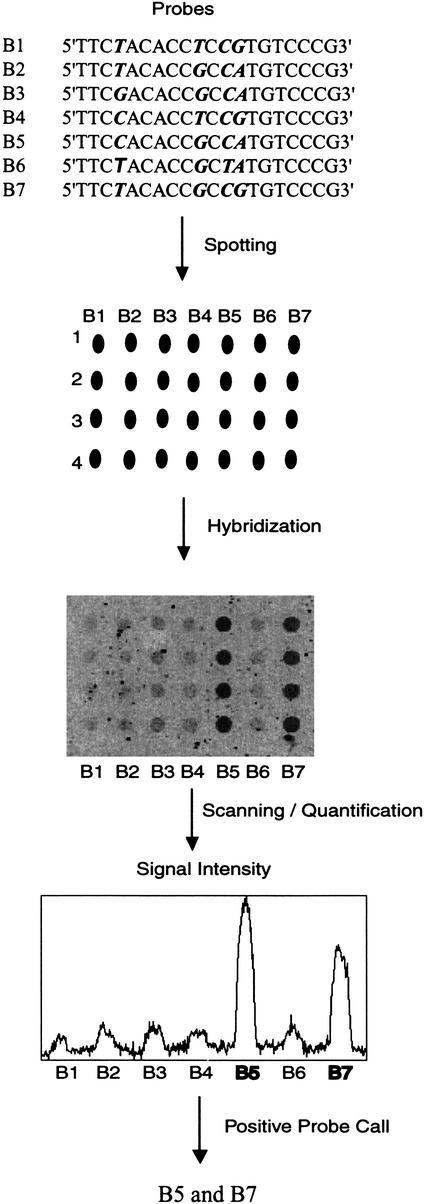
Hybridization signal intensities were quantified by summing the intensities of all pixels located inside the image spot. Each signal was ranked from highest to lowest in each probe group according to their intensities. The highest ranked signal(s) in each group was chosen as the positive signal(s) (Fig. 3). When hybridized to homozygous samples, only one probe in each group produced a positive signal as the perfect match. In samples heterozygous for two different HLA-B alleles, two probes with highest signal intensities in a group were selected as positive, unless only one positive signal was obtained in that group.
Figure 3.
Procedure for analysis of hybridization data is illustrated for position 21–40 of exon 2. All oligonucleotide probes were assigned into one group and named B1 to B7 (refer to Table 1 for probes). The seven probes were spotted in quadruplicate. After hybridization, signal intensities were quantified, and signals with the highest intensities in this group (B5 and B7) were identified as positive.
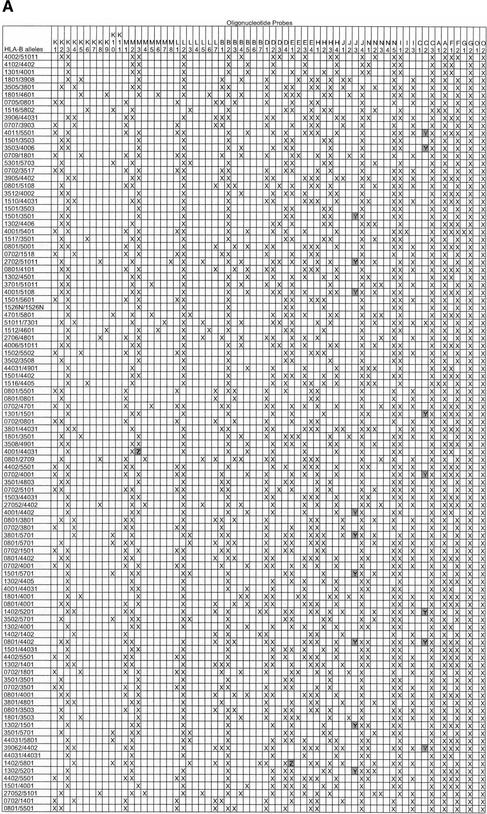
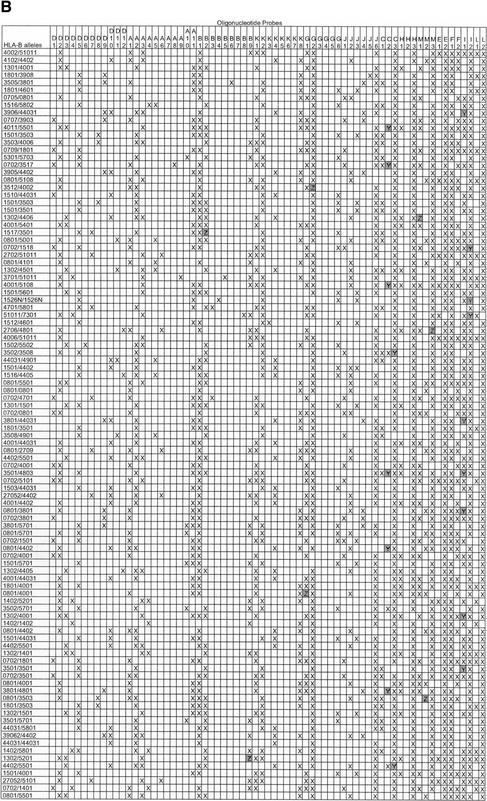
We employed an intensity ratio of 3:1 as the threshold to categorize the positive and negative signals. In each probe group, signals with intensities more than one-third of the strongest signal in that group were considered positive signals, while signals with intensities less than one-third of the strongest signal were considered negative. In most cases, the ratio between the intensities of positive signals and those of negative signals in each group varied from 42:1 to 6:1 (Fig. 3), depending on the degree of mismatches among probes. The stringent conditions described above enabled single nucleotide mismatches between probe sequence and sample sequence to be unequivocally distinguished. Exceptions to this occurred with four pairs of probes (C2 and C3 for exon 2, J3 and J4 for exon 2, I1 and I2 for exon 3, and C2 and C3 for exon 3). The C2 and C3 exon 2 probes involved a C/T change four nucleotides from the 5′-end; the J3 and J4 probes described a C/T change six nucleotides from the 3′-end; the I1 and I2 involved an A/T change six nucleotides from 3′-end; and the C2 and C3 exon 3 probes encoded a C/T change six nucleotides from the 3′-end. The positive-to-negative ratios for these pairs declined to 2:1 yielding a false-positive signal at low frequency (32 cases in 13,700 hybridization signals among 100 blinded samples in the equivalency study; Table 2). The polymorphisms defined by the C2/C3 exon 2, J3/J4, I1/I2, and C2/C3 exon 3 probe pairs described a single base change each and were included for this reason, although information from these eight probes was not mandatory for allele assignment.
Table 2.
Hybridization Results of Equivalency Study of (A) 68 Exon 2 Probes and (B) 69 Exon 3 Probes
Probes are named (A) K1-O2 and (B) D1-L2. HLA-B alleles are designated according to standard nomenclature (Robinson et al. 2000). An “X” indicates a signal consistent with predicted pattern, a “Y” indicates a false positive, and a “Z” indicates a false negative.
Allele Assignment
HLA-B alleles were assigned by quantitative analysis of the hybridization pattern of the DNA sample (Fig. 3). Assignment of the alleles was accomplished by comparing the detected hybridization pattern, with the theoretical patterns of all known HLA-B allele combinations.
All probes in each group served as the control for each other, offering unequivocal and specific discrimination of each SNP.
In the first step, the hybridization signals were separated into groups according to the locations they hybridized with (Fig. 3), and their intensities were integrated. In the second step, hybridization signals were categorized into positive signals and negative signals by comparing signal intensities in each group. The strongest one or two signals in each group were considered positive and the rest of signals negative. When compared with theoretical hybridization patterns, the probes were analyzed in the order of numbers of probes in that group. Groups with the most probes were analyzed first (group K in exon 2 and group D in exon 3), and the groups with only two probes were analyzed last. When the signals for probes in groups K, M, L, and B of exon 2 and D, A, B, and K of exon 3 were compared with their corresponding theoretical hybridizations, the allele family could be assigned from this information alone. The information from other positive hybridization signals then was added to refine the results from allele families into single allele types.
In the course of our studies, we encountered two instances where the predicted and actual probe hybridizations were discrepant. As described above, 32 false-positive or false-negative signals were encountered for the four pairs of probes; in this situation, the alleles consistent with the other probe hybridizations were considered to be the correct interpretation. The second category occurred in 17 samples and involved inconsistent hybridization results for more than two probes. All 17 samples were rehybridized, and the rehybridization results were internally consistent.
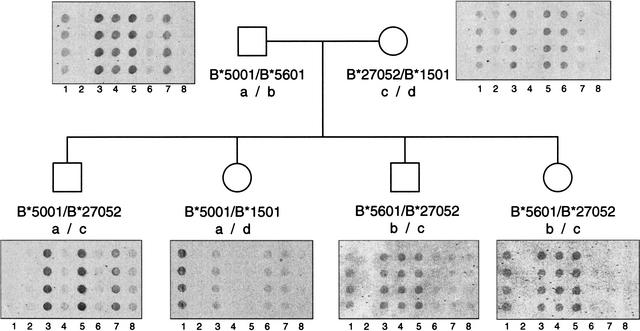
Figure 4 demonstrates the range of hybridization results for the various groups of HLA-B probes. Although pedigree analysis is not necessary to assign HLA-B genotypes, this family study demonstrates the power of our system to haplotype the HLA-B alleles.
Figure 4.
Segregation of HLA-B SNPs in a family study. Probes descriptive for positions 154–173 (lanes 1–4) and positions 194–213 (lanes 5–8) of exon 2 are illustrated for HLA-B*5001 (paternal “a” haplotype, lanes 3 and 7), HLA-B*5601 (paternal “b” haplotype, lanes 4 and 5), B*27052 (maternal “c” haplotype, lanes 3 and 5) and B*1501 (maternal “d” haplotype, lanes 1 and 6). Positions of the probes are numbered according to the HLA/IMGT database. Cross-hybridization is observed for eight of the 137 (6%) probes, however, these spurious signals can be identified by analyzing the hybridization results of the other probes in the group. For example, the two family members with the “b/c” haplotype in this figure demonstrate a false-positive hybridization with the probe in lane 1. Despite the signal from this probe, the correct allele was called because of the lack of signal from the probe in lane 6.
Equivalency Study
To demonstrate how the oligonucleotide array could be used to genotype blinded samples without the need for pedigree analysis, we conducted an equivalency study of 100 unrelated individuals whose HLA-B alleles were typed previously using sequencing methods (Petersdorf and Hansen 1995). The 100 samples represented all serologically defined HLA-B phenotypes and 65 unique HLA-B alleles, and included 97 heterozygous samples and three homozygous samples (Table 2). Additionally, three of the 97 heterozygous samples were known to generate ambiguous probe hybridization patterns (B*1501, 3501 in one sample and B*1501, 3503 in two samples). HLA-B allele assignments were made by an independent observer who had no knowledge of the samples' genotypes. Among the 100 hybridizations to the 137 probes, 32 false positives were observed (32/13,700, 0.2%) in the C and J group of exon 2 and the C and I group of exon 3, and were related to the difficulty in discriminating a single SNP near the end of these probes (refer to Hybridization Kinetics, above). These probes described four out of the 94 total polymorphisms (4%) and were located in regions that were not essential for the assignment of the HLA-B alleles. Hybridization data from the remaining probes was more than sufficient to deduce the genotypes. When the genotypes of the 100 samples were unblinded and compared to sequencing results, the correct interpretation of the array hybridization patterns was made for all three homozygous samples and all 97 heterozygous samples. Furthermore, the three heterozygous samples encoding ambiguous allele combinations were correctly recognized and each interpreted as HLA-B*15, 35. This equivalency study demonstrates that the oligonucleotide probe array can discriminate the presence of unique SNPs and heterozygous combinations.
DISCUSSION
Compared with conventional sequencing-based methods, oligonucleotide arrays afford a much higher throughput, by virtue of parallel analysis of multiple genetic regions. In this way, arrays are ideally suited for the analysis of MHC genes. Arrays can be used for both the detection of new polymorphisms, and for the typing of known alleles. When employed as a SNP typing tool, our rate of false-positive/false-negative hybridizations compares favorably with that observed by other investigators (Wang et al. 1998). When used as an allele genotyping system, the high redundancy of probes provides unequivocal allele assignments and obviates the need for pedigree analysis.
A unique feature of the method we describe is the requirement for only one or two positive signals in each group of probes. This approach eliminates the need for all negative signals to disappear. Negative signals can be determined easily by their lower signal intensities when compared with other signals in its group rather than by their disappearance.
New SNPs and alleles in the HLA and other genomic regions continue to be discovered at a rapid pace. The addition of new probe sequences descriptive of novel polymorphisms is a necessary and anticipated modification of this first generation array. The flexible nature of our array system allows probes to be easily added or modified without significantly increased costs. Our oligonucleotide array system provides information on both SNP identity and SNP linkage and can be adapted easily for SNPs detection in other genomic regions.
METHODS
Oligonucleotides
All oligonucleotides were synthesized by Life Technologies, Inc. Each oligonucleotide probe used in the manufacture of oligonucleotide arrays contained a 5′ amino group. For fluorescence detection, the 3′ primer employed in single-stranded PCR amplification was fluorescently labeled with 6-Rhodamine group (Applied Biosystems, Inc.) at its 5′-end and purified by reverse-phase, high-performance, liquid chromatography. The oligonucleotide probe used in the surface density study was labeled with an amino group at its 5′-end and a Rhodamine dye at its 3′-end. Concentrations of all oligonucleotides were determined by ultraviolet spectrophotometry at 260 nm.
Preparation of Oligonucleotide Arrays on Glass Supports
Precleaned microscope slides (Becton, Dickinson and Co.) were immersed in concentrated HCl for 2 h, then washed 10 times with distilled water, 5 min per wash, and air dried. The cleaned slides were placed in a vacuum chamber with 700 μL of 3-aminopropyltrimethoxysilane (Aldrich Chemical) and the vacuum chamber was maintained at 160°C and 30 ppm Hg pressure for 3 h. The slides were subsequently washed five times with acetone, 5 min per wash, and then treated for 2 h with a solution of 0.2% 1,4-phenylene diisothiocyanate (Aldrich) in 10% pyridine/dimethyl formamide, and washed with acetone then methanol for five times, 5 min per wash. The activated glass slides were stored at 4°C in a vacuum desiccator containing anhydrous calcium.
Oligonucleotide probes, labeled with a 5′ amino group, were dissolved at a concentration of 500 pmol/μL in water. Twenty-five microliters of the resultant solutions were transferred into a 96-well microtiter plate. The plate was placed into a Molecular Dynamic Generation II array spotter, along with the chemically activated glass slides. The transfer of oligonucleotide solution from microtiter plate to glass slides was conducted at 50% humidity, and each oligonucleotide solution was spotted in quadruplicate. The spotting step was repeated three times for each glass slide. Slides spotted with oligonucleotide probes then were incubated at 37°C in a covered Petri dish containing 1 mL of water for 2 h, removed, washed once with 1% NH4OH, three times with water, and air dried at room temperature.
DNA Sample Preparation
Human genomic DNA was extracted using procedures described elsewhere (QIAGEN 1999). The quality of DNA samples was tested by OD 260/280 measurement. Amplification of double-stranded exon 2 product utilized a 5′ primer 5′-GCTCCCACTCCATGAGGTAT-3′ and 3′ primer 5′-AAATGAAACCGGGTAAAC-3′ to generate a 456 bp fragment from position 1 of exon 2 to position 186 of intron 2. The amplification reaction (100 μL) contained 50 mM KCl, 10 mM Tris-HCl, 1.5 mg MgCl2, 10 mg of gelatin, 20 ng of genomic DNA, 2 μM of each primer, 200 μM each dATP, dCTP, dTTP and dGTP, and 2.5 U of Taq DNA polymerase. PCR was performed in a Perkin-Elmer Cetus 9600 thermal cycler using 35 cycles of the following profile: 96°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min. The PCR mixture then was purified using a QIAGEN PCR purification kit (QIAGEN Inc.). Single-stranded exon 2 was generated using the 3′ primer 5′-R-CGGCCTCGCTCTGGTTGTAG-3′ (positions 270–251 of exon 2) (R, Rhodamine label). The PCR was performed in 30 cycles using the following profile: 96°C for 30 sec, 60°C for 30 sec and 72°C for 1 min.
Amplification of exon 3 of HLA-B was accomplished using a 5′ primer 5′-ACCCGGTTTCATTTTCAGTTG-3′ (intron 2, position 184–164) and a 3′ primer 5′-CCCACTGCCCCTGG TACC-3′ (intron 3, position 17–exon 3, position 276) to generate a 396-bp fragment. The PCR was performed in 35 cycles of the following profile: 96°C for 30 sec, 60°C for 30 sec and 72°C for 2 min. To generate single-stranded exon 3 product, the second PCR employed 3′ primer 5′-R-CCCACTGC CCCTGGTACC-3′ (intron 3, position 17–exon 3, position 276), in 30 cycles of the following profile: 96°C for 30 sec, 60°C for 30 sec, and 72°C for 2 min.
Oligonucleotide Immobilization Studies
The oligonucleotide probe K5 (Table 1A), labeled with an amino group at its 5′-end and a Rhodamine dye at its 3′-end was mixed with an oligonucleotide of the same sequence labeled with an amino group at its 5′-end, at a ratio of 1:10. Twenty-five microliters of mixed oligonucleotides at a total concentration of 15.65, 31.25, 62.5, 93.75, 125, 187.5, 250, 500, 1000, and 2000 pmol/mL were placed into a 96-well microtiter plate and spotted on the activated glass slides using the Molecular Dynamic Generation II array spotter. The oligonucleotide mixtures were incubated on the glass slide at 37°C for 2 h in an incubation chamber. The glass slides were washed with 1% NH4OH followed by water, 15 min each time. The glass slides then were scanned using Molecular Dynamic fluorescence scanner to quantify the amount of oligonucleotide immobilized on the surface (Fig. 2A).
To study the effect of oligonucleotide surface density on hybridization efficiency, the 5′ amino-labeled oligonucleotide probe K5 was immobilized on the glass slide at concentrations of 15.65, 31.25, 62.5, 93.75, 125, 187.5, 250, 500, 1000, and 2000 pmol/mL as described above. Hybridization mixtures (50 μL of double-stranded or single-stranded Rhodamine-labeled PCR product of exon 2 in 5xSSPE, 0.5% SDS) were applied to each slide. The glass slide then was covered with a cover glass and incubated for 3 h at 30°C in a closed Petri dish over water. Two 15-min washes were performed in a shaking incubator at 37°C using 20 mL of washing buffer (2xSSPE, 0.2% SDS) each. Hybridization signals were quantified by fluorescence scanning (Fig. 2B).
Hybridization Conditions
To study hybridization kinetics, 50 μL of single-stranded Rhodamine-labeled PCR products of exon 2 were hybridized with the exon 2 oligonucleotide arrays as described above for 10, 20, 30, 40, 60, 90, 120, 180, 240, and 360 min, respectively. After hybridization, the oligonucleotide arrays were washed with washing buffer (conditions described above) and scanned with the fluorescence scanner to quantify both the hybridization signal intensity and the corresponding background level (Fig. 2C).
At optimal conditions, single-stranded, Rhodamine-labeled exon 2 PCR product (50 μL in 5xSSPE, 0.5% SDS) was applied to the array slide, covered with a cover glass, and incubated at 37°C for 2 h. The glass slide then was washed twice with 20 mL washing buffer (20xSSPE, 0.2% SDS) at 30°C for 15 min each.
For hybridization with exon 3 oligonucleotide arrays, 50 μL solution (5xSSPE, 0.5% SDS) of the single-stranded, Rhodamine-labeled PCR product of exon 3 was added to the glass slides and incubated at 30°C for 2 h and then washed twice at room temperature.
Fluorescence Detection
After hybridization and washing, 50 μL of washing buffer (2xSSPE, 0.2% SDS) was applied to the glass slide, and the slide was covered with a cover glass to provide an aqueous environment for the fluorescence scanning. The slide was scanned using a Molecular Dynamic array scanner. After scanning, the fluorescence image was analyzed using the ImageQuant software provided with the array scanner. For each spot, pixel intensities within the spot image were summed. The average value and standard deviation of pixel intensities for each spot was calculated and the local background level was subtracted from the sum of the signal intensity.
Acknowledgments
We thank Dr. Leonid Kruglyak and Dr. Mari Malkki for their invaluable help with this manuscript. This work was supported by grants CA18029, CA72978, and AI33484 (EWP) from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL epetersd@fhcrc.org; FAX (206) 667-5255.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.206402. Article published online before print in February 2002.
REFERENCES
- Babon JJ, McKenzie M, Cotton RG. Mutation detection using fluorescent enzyme mismatch cleavage with T4 endonuclease VII. Electrophoresis. 1999;20:1162–1170. doi: 10.1002/(SICI)1522-2683(19990101)20:6<1162::AID-ELPS1162>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Charkravarti A. Variations on a theme: Cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1998;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Del Tito BJ, Jr, Poff HE, 3rd, Novotny MA, Cartledge DM, Walker RI, 2nd, Earl CD, Bailey AL. Automated fluorescent analysis procedure for enzymatic mutation detection. Clin Chem. 1998;44:731–739. [PubMed] [Google Scholar]
- Gilles PN, Wu DJ, Foster CB, Dillon PJ, Chanock SJ. Single nucleotide polymorphic discrimination by an electronic dot blot assay on semiconductor microchips. Nature Biotech. 1999;17:365–370. doi: 10.1038/7921. [DOI] [PubMed] [Google Scholar]
- Grompe M. The rapid detection of unknown mutations in nucleic acids. Nat Genet. 1993;5:111–117. doi: 10.1038/ng1093-111. [DOI] [PubMed] [Google Scholar]
- Guo Z, Guilfoyle RA, Thiel AJ, Wang R, Smith LM. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 1994;22:5456–5465. doi: 10.1093/nar/22.24.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Hawkins JR. Finding mutations. Oxford, UK: IRL Press; 1997. [Google Scholar]
- Kruglyak L. The use of a genetic map of biallelic markers in linkage studies. Nat Genet. 1997;17:21–24. doi: 10.1038/ng0997-21. [DOI] [PubMed] [Google Scholar]
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- Laken SJ, Jackson PE, Kinzler KW, Vogelstein B, Strickland PT, Groopman JD, Friesen MD. Genotyping by mass spectrometric analysis of short DNA fragments. Nat Biotechnol. 1998;16:1352–1356. doi: 10.1038/4333. [DOI] [PubMed] [Google Scholar]
- Lee MT, Kuo F, Whitmore GA, Sklar J. Importance of replication in microarray gene expression studies: Statistical methods and evidence from repetitive cDNA hybridizations. Proc Natl Acad Sci. 2000;97:9834–9839. doi: 10.1073/pnas.97.18.9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshutz RJ, Morris D, Chee M, Hubbell E, Kozal MJ, Shah N, Shen N, Yang R, Fodor SP. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques. 1995;19:442–447. [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Mashal RD, Sklar J. Practical methods of mutation detection. Curr Opin Genet Dev. 1996;6:275–280. doi: 10.1016/s0959-437x(96)80002-5. [DOI] [PubMed] [Google Scholar]
- Myers RM, Larin Z, Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA: DNA duplexes. Science. 1985;230:1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SP. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersdorf EW, Hansen JA. A comprehensive approach for typing the alleles of the HLA-B locus by automated sequencing. Tissue Antigens. 1995;46:73–85. doi: 10.1111/j.1399-0039.1995.tb02482.x. [DOI] [PubMed] [Google Scholar]
- QIAGEN. QIAamp® DNA mini kit and QIAamp DNA blood mini kit handbook. Valencia, CA: QIAGEN; 1999. [Google Scholar]
- Risch N, Merikangas K. The future of genetic study of complex human disease. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Roberts E, Deeble VJ, Woods CG, Taylor GR. Potassium permanganate and tetraethylammonium chloride are a safe and effective substitute for osmium tetroxide in solid phase fluorescent chemical cleavage of mismatch. Nucleic Acids Res. 1997;25:3377–3378. doi: 10.1093/nar/25.16.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Malik A, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA database — A sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- Rowen L, Mahairas G, Hood L. Sequencing the human genome. Science. 1997;278:605–607. doi: 10.1126/science.278.5338.605. [DOI] [PubMed] [Google Scholar]
- Sapolsky RJ, Lipshutz RJ. Mapping genomic library clones using oligonucleotide array. Genomics. 1996;33:445–456. doi: 10.1006/geno.1996.0219. [DOI] [PubMed] [Google Scholar]
- Schafer AL, Hawkins JR. DNA variation and the future of human genetics. Nat Biotechnol. 1998;16:33–39. doi: 10.1038/nbt0198-33. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K. dbSNP-database for single-nucleotide polymorphisms and other class of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- Shumaker JM, Metspalu A, Caskey CT. Mutation detection by solid phase primer extension. Hum Mutat. 1996;7:346–354. doi: 10.1002/(SICI)1098-1004(1996)7:4<346::AID-HUMU9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- So A. Genetics, polymorphism and regulation of expression of HLA region genes. In: Lecher A, editor. HLA and disease. San Diego, CA: Academic Press Inc.; 1994. pp. 1–34. [Google Scholar]
- Tobe VO, Taylor SL, Nickerson DA. Single-well genotyping of diallelic sequence variations by a two-color ELISA-based oligonucleotide ligation assay. Nucleic Acids Res. 1996;24:3728–3732. doi: 10.1093/nar/24.19.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Zemans R, Oefner PJ, Cavalli-Sforza LL. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci. 1996;93:196–200. doi: 10.1073/pnas.93.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Smith HO, Hood L. A new strategy for genome sequencing. Nature. 1996;381:364–366. doi: 10.1038/381364a0. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- White MB, Carvalho M, Derse D, O'Brien SJ, Dean M. Detecting single base substitution as heteroduplex polymorphisms. Genomics. 1992;12:301–306. doi: 10.1016/0888-7543(92)90377-5. [DOI] [PubMed] [Google Scholar]