Abstract
A major interest in human genetics is to determine whether a nonsynonymous single-base nucleotide polymorphism (nsSNP) in a gene affects its protein product and, consequently, impacts the carrier's health. We used the SIFT (Sorting Intolerant From Tolerant) program to predict that 25% of 3084 nsSNPs from dbSNP, a public SNP database, would affect protein function. Some of the nsSNPs predicted to affect function were variants known to be associated with disease. Others were artifacts of SNP discovery. Two reports have indicated that there are thousands of damaging nsSNPs in an individual's human genome; we find the number is likely to be much lower.
A major interest in human genetics is to distinguish mutations that are functionally neutral from those that contribute to disease. Amino acid substitutions currently account for approximately half of the known gene lesions responsible for human inherited disease (Cooper et al. 1998). Therefore, it is important to determine whether a nonsynonymous single nucleotide polymorphism (nsSNP) that affects the amino acid sequence of a gene product can alter protein function and contribute to disease.
The number of potentially damaging nsSNPs in a human individual is also of major interest because if the number is high, it can affect human welfare. Two groups, Sunyaev et al. (2001) and Chasman and Adams (2001), have applied computational tools that predict the effect of an amino acid substitution on protein function to nsSNPs. These groups estimated that ∼20% and 30%, respectively, of nsSNPs damage protein function. Based on these estimates, they proposed that each individual has on average 2000 (Sunyaev et al. 2001) and 9500 nsSNPs (Chasman and Adams 2001) that affect protein function and may contribute to health ailments.
Previously, we introduced SIFT, which uses sequence homology to predict whether an amino acid substitution in a protein will affect protein function (Ng and Henikoff 2001). SIFT is based on the premise that important amino acids will be conserved among sequences in a protein family, so changes at amino acids conserved in the family should affect protein function. Given a protein sequence, SIFT chooses related proteins, obtains an alignment of these proteins with the query, and, based on the amino acids appearing at each position in the alignment, makes a prediction as to whether a substitution will affect protein function. A position in the protein query that is conserved in the alignment will be scored by SIFT as intolerant to most changes; a position that is poorly conserved will be scored by SIFT as tolerating most changes. Unlike the tools of Sunyaev et al. (2001) and Chasman and Adams (2001), SIFT does not require structural information and therefore can be applied to a much larger number of proteins.
Here we apply SIFT to human disease and polymorphism databases. We find that SIFT's prediction ability is similar to that of tools that require structural information. However, we do not arrive at a similar conclusion concerning the number of damaging nsSNPs in the human genome. Rather, our detailed examination of the source of nsSNPs in current databases reveals biases that inflated the other groups' estimates.
RESULTS
We define a damaging nsSNP as a mutation whose resulting amino acid substitution in the corresponding protein affects protein function. We define an nsSNP as tolerated or neutral if the resulting amino acid substitution in the protein does not detectably alter protein phenotype. These definitions exclude mutations that affect transcription, translation, splicing, and other possible pretranslational alterations. Because SIFT predicts on amino acid substitutions in the protein product, it does not take into account these factors.
SIFT Analysis of Human Variant Databases
SIFT was applied to three different datasets of human variants and a summary of the prediction results is shown in Table 1. The first dataset consisted of substitutions annotated as involved in disease according to SWISS-PROT/TrEMBL (Bairoch and Apweiler 2000). SIFT predicted 69% (3626/5218) of these substitutions as damaging. Some of these substitutions may be functionally neutral but incorrectly annotated as causing disease if they were observed in patients or are in linkage disequilibrium with another mutation that is causing the disease phenotype. Thus, 69% is a lower bound of prediction accuracy on damaging substitutions.
Table 1.
SIFT Prediction on Human Variant Databases
| Datasets | Sequence coverage | Amino acid substitution coverage | % Predicted as damaging | Weighted false positive error |
|---|---|---|---|---|
| Amino acid substitutions annotated to be involved in disease | 76% (462/606) | 71% (5218/7397) | 69% (3626/5218) | Hypothesis not applicable |
| Polymorphisms detected in normal individuals (WI-nsSNPs) | 77% (53/69) | 62% (115/185) | 19% (22/115) | 20% |
| Nonsynonymous changes from dbSNP | 60% (1789/3005) | 53% (3084/5780) | 25% (757/3084) | 19% |
Sequence coverage is the percentage of proteins whose alignments contained enough diverse homologues so that predictions could be made. Amino acid substitution coverage is the percentage of substitutions predicted on. The weighted false positive error is the percentage of substitutions shown experimentally to be neutral that were incorrectly predicted as damaging.
A second dataset consisted of nonsynonymous polymorphisms in normal individuals detected by the Whitehead Institute (Cargill et al. 1999). These nsSNPs (referred to as WI-nsSNPs) represent an unbiased set of nsSNPs because they were systematically detected and confirmed across many genes in control individuals. Some of the WI-nsSNPs may affect protein function, even though they were detected in control individuals if the altered phenotype was recessive or undiagnosed. Of the WI-nsSNPs, 19% (22/115) were predicted by SIFT to be damaging (Table 1). However, these may be neutral because there was no apparent difference between this value and SIFT's 20% weighted false positive error. Because SIFT predicted most (69%) of the substitutions involved in disease as damaging and most (81%) of the known polymorphisms as neutral, the results from these two datasets indicate that SIFT can distinguish between damaging and neutral human nsSNPs.
A third dataset consisted of putative nsSNPs in dbSNP (Sherry et al. 2001), one of the largest public SNP databases. Of the proteins containing nsSNPs from dbSNP, 60% (1789/3005) had enough homologs for SIFT prediction (Table 1). For these proteins, 25% (757/3084) of the substitutions were predicted to be damaging by SIFT. The weighted false positive error was calculated and indicated that if all of the nsSNPs from dbSNP were functionally neutral, only 19% should have been predicted as damaging.
We investigated the difference between the percentage predicted to be damaging for dbSNP variants (25%) and that expected if dbSNP contains only functionally neutral substitutions (19%). Sixteen genes were chosen because they had a high fraction of nsSNPs from dbSNP predicted to affect protein function. In the following sections, we show that the apparent polymorphisms in these genes could be explained by reasons other than SIFT prediction error (Table 2 for summary).
Table 2.
List of the 16 Genes that Had the Highest Fraction of dbSNP Changes Predicted to Be Damaging by SIFT
| Genes with nsSNPs that had previously shown to be involved in disease |
| MSHR, encodes melanocyte stimulating hormone receptor |
| PPARα, encodes peroxisome proliferator activated receptor α |
| MTHFR, encodes methylenetetrahydrofolate reductase |
| FBN1, encodes fibrillin |
| APOA1, encodes apolipoprotein A-I |
| Genes containing dbSNP entries erroneously mapped from pseudogenes |
| AGP1, encodes α1-acid glycoprotein gene encoding hypothetical protein FKSG25 |
| Gene containing damaging mutations in redundant motifs |
| gene encoding hypothetical protein FLJ20079 |
| Genes with sequencing errors mistaken for polymorphisms |
| PSMB7, encodes proteasome subunit β7 |
| MYL6, encodes myosin light polypeptide 6 |
| RPL11, encodes 60S ribosomal protein L11 |
| FAU, encodes 40S ribosomal protein S30 |
| PSMC5, encodes 26S protease regulatory subunit 8 |
| TJP2, encodes tight junction protein 2 |
| EFNA2, encodes ephrin-A2 |
| XDH, encodes xanthine dehydrogenase |
Substitutions Already Shown to Be Involved in Disease
For 5 of the 16 genes with an excessive number of nsSNPs predicted to be damaging, most of the nsSNPs came from patients with disease, and the gene in which the nsSNPs were detected had been shown or suspected to contribute to the disease. These genes had a high fraction of nsSNPs predicted to affect protein function because many of their variants in dbSNP contribute to disease. SIFT correctly predicted 18/22 of the nsSNPs found in disease patients to affect protein function and 9/10 nsSNPs found in control patients as functionally neutral (Table 3). This provides additional evidence that SIFT can distinguish between nsSNPs involved in disease and those that are functionally neutral. All predictions for the five genes are shown in Table 3; we highlight certain aspects of SIFT by discussing several predictions in detail.
Table 3.
nsSNPs from dbSNP Predicted to Be Damaging Were Shown to Be Involved in Disease
| Melanocyte stimulating hormone receptor | ||
| Predictions supported by evidence | ||
| R151C | Damaging | Increased risk of cutaneous malignant melanoma (CMM) (Palmer et al. 1999). |
| R160W | Damaging | Doubles risk of CMM (Palmer et al. 1999). |
| D294H | Damaging | Doubles risk of CMM (Palmer et al. 1999). |
| L60V | Tolerated | No association with CMM (Palmer et al. 1999, Box et al. 2001). |
| R163Q | Tolerated | Detected from EST (Irizarry et al. 2000) and contig comparisons (Sachidanandam et al. 2001). No association with CMM (Box et al. 2001). |
| Prediction not supported by evidence | ||
| D84E | Damaging | Not associated with CMM (Healy et al. 1999; Palmer et al. 1999). |
| Peroxisome proliferator activated receptor α | ||
| Prediction supported by evidence | ||
| R127Q | Damaging | Detected in diabetic (Au et al. 1998; Brookes et al. 2000). |
| L162V | Damaging | Detected in diabetics and controls (Au et al. 1998; Brookes et al. 2000), but increases cholesterol and apolipoprotein B levels in diabetics, and perhaps nondiabetics (Flavell et al. 2000; Lacquemant et al. 2000; Vohl et al. 2000). |
| D304N | Damaging | Detected in diabetic (Au et al. 1998; Brookes et al. 2000). |
| R409T | Damaging | Detected in diabetic (Au et al. 1998; Brookes et al. 2000) |
| V227A | Tolerated | Found in similar frequencies in diabetics and nondiabetics (Au et al. 1998; Brookes et al. 2000; Hara et al. 2001). |
| A268V | Tolerated | Detected in diabetic (Au et al. 1998; Brookes et al. 2000) and EST comparison (Irizarry et al. 2000). No association with diabetes or coronary heart disease (Lacquemant et al. 2000). |
| Methylenetetrahydrofolate reductase (under balancing selection) | ||
| Prediction supported by evidence | ||
| A222V | Damaging | Common variant with diminished enzyme activity and increased risk of vascular disease and neural tube defects (Frosst et al. 1995) but also reduces risk of adult acute leukemia (Skibola et al. 1999), childhood leukemia (Wiemels et al. 2001), and colon cancer (Ma et al. 1997). |
| E429A | Damaging | Common variant with diminished enzyme activity (Weisberg et al. 1998) but reduces risk of adult acute leukemia (Skibola et al. 1999). |
| Effect unknown | ||
| R68Q | Damaging | Detected by comparison of contig to reference gene. |
| Fibrillin | ||
| Prediction supported by evidence | ||
| D1113G | Damaging | In Marfan patient (Liu et al. 1997/1998). |
| C1153Y | Damaging | In Marfan patient (Bairoch and Apweiler 2000). |
| I2023T | Damaging | In Marfan patient (Liu et al. 1997/1998). |
| C2038Y | Damaging | In Marfan patient (P. Oefner, pers. comm.) |
| C2053F | Damaging | In Marfan patient (P. Oefner, pers. comm.). |
| C2500S | Damaging | In Marfan patient (P. Oefner, pers. comm.). |
| G2514R | Damaging | In Marfan patient (P. Oefner, pers. comm.). |
| C2110R | Damaging | In Marfan patient (P. Oefner, pers. comm.). |
| C2170F | Damaging | In Marfan patient (P. Oefner, pers. comm.). |
| P1148A | Tolerated | In control individual (Liu et al. 1997/1998). |
| V2018I | Tolerated | In control individual (P. Oefner, pers. comm.). |
| Y2113F | Tolerated | In control individual (P. Oefner, pers. comm.). |
| D2329E | Tolerated | In control individual (P. Oefner, pers. comm.). |
| P2278S | Tolerated | In control individual (P. Oefner, pers. comm.). |
| Prediction not supported by evidence | ||
| S1077P | Tolerated | In Marfan patient (Liu et al. 1997/1998). |
| V1667I | Tolerated | In Marfan patient (Liu et al. 1997/1998). |
| N1341S | Tolerated | In Marfan patient (Liu et al. 1997/1998). |
| N1282S | Tolerated | In Marfan patient (Liu et al. 1997/1998). |
| Apolipoprotein A-I | ||
| Effect unknown | ||
| D126H | Damaging | Detected in individuals from either the top or bottom 2.5th percentile of a normalized blood pressure distribution (Halushka et al. 1999). |
| R184P | Damaging | Detected in individuals from either the top or bottom 2.5th percentile of a normalized blood pressure distribution (Halushka et al. 1999). |
| K131N | Tolerated | Detected from comparison of ESTs (Garg et al. 1999) and in an electrophoretic screening of newborns (von Eckardstein et al. 1990). |
Five genes with a high fraction of nsSNPs from dbSNP predicted to be damaging; these nsSNPs have been shown to be involved in disease. The proteins that these genes code for are listed. Under each protein, the first column is the amino acid substitution caused by the nsSNP. For example, the first row under melanocyte stimulating hormone receptor gives the prediction for an nsSNP that corresponded to a change from the original amino acid R to amino acid C at position 151 in the melanocyte stimulating hormone receptor protein. The second column is the SIFT prediction for that particular amino acid substitution, and the third column is evidence supporting or conflicting with the prediction. Most predictions are supported by previously published evidence.
SIFT detects nsSNPs that are damaging to a protein, although loss of protein function may not cause an obvious phenotype. Although a protein may not play an important role in the organism, if the amino acid substitution resulting from an nsSNP occurs at a conserved position, it will be predicted to affect function. For example, some nsSNPs in the melanocyte stimulating hormone receptor (MSHR) gene are associated with a twofold risk for cutaneous malignant melanoma (Palmer et al. 2000). Although MSHR is not under strong selection outside of African populations (Harding et al. 2000) and has a minor role in overall health, SIFT correctly predicted the appropriate nsSNPs as damaging because the amino acid substitutions occurred at conserved positions in the protein alignment used for prediction.
Some nsSNPs might be damaging to the protein, but their effects on health are difficult to ascertain. For example, when a candidate gene for diabetes, the gene encoding peroxisome proliferator activated receptor α (PPARα), was screened for polymorphisms in diabetics and nondiabetics, the nsSNP causing a L162V substitution in PPARα was found at similar frequencies in both populations. SIFT predicted this substitution to affect protein function (Table 3). The prediction might appear incorrect based on the lack of association with diabetes, but carriers of this nsSNP have higher cholesterol levels and increased apolipoprotein B concentrations, thus it has been proposed to increase the risk of coronary artery disease (Vohl et al. 2000; Lacquemant et al. 2000). SIFT was sensitive to this mutation and predicted it to be damaging because the position of substitution is conserved among orthologous proteins and other nuclear hormone receptors present in the alignment used for prediction. Because mutations in proteins can have pleiotropic effects, a mutation that initially does not appear to have an effect but is predicted to affect function by SIFT may have an effect that has not yet been assayed for.
SIFT can detect overdominant nsSNPs in which the heterozygote has a selective advantage. Individuals severely deficient in methylenetetrahydrofolate reductase (MTHFR) activity develop mental retardation and cardiovascular disease (OMIM #236250). However, reduced MTHFR activity can also confer protection against child and adult acute leukemia and colon cancer. SIFT correctly predicted the two common variants of MTHFR with reduced enzymatic activity to affect protein function (Table 3). A lowered risk for some diseases has selected for these variants that reduce enzyme activity, despite other detrimental effects on health. Overdominant nsSNPs can become common in a population although they affect protein function. Common nsSNPs are often expected to be functionally neutral; their identification as damaging to the protein and perhaps maintained by overdominance may lead to the understanding of some common diseases.
nsSNPs Erroneously Mapped from Pseudogenes
SIFT detected two genes for which the changes from dbSNP were mistakenly mapped from pseudogenes (Table 2). Programs that identify SNPs by aligning ESTs (expressed sequence tags) or genomic sequences might detect base differences between the functional gene and a pseudogene and erroneously report these differences as SNPs in the functional gene. For example, AGP1, the gene encoding α1-acid glycoprotein, was annotated to contain six missense changes in dbSNP, but the source of the differences was ESTs from AGP2. Although AGP2 is expressed, the protein has been suggested to lack function because it has evolved at an unconstrained rate (Merritt et al. 1990).
Damaging Mutations in Redundant Motifs
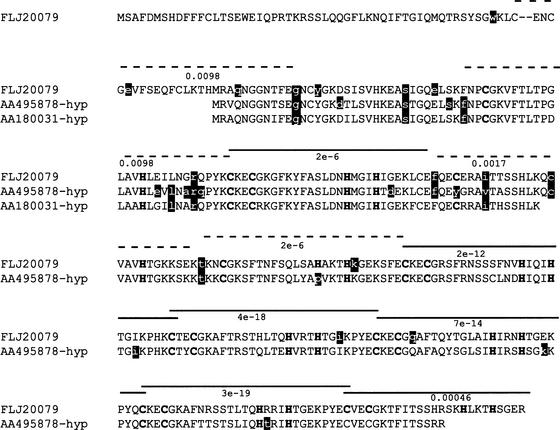
Like the pseudogene examples in the previous section, differences entered as nsSNPs into dbSNP for the gene encoding FLJ20079 actually matched other regions of the genome. However, this example is more complex because the other regions may code for functional genes (Fig. 1). After we inferred the hypothetical protein sequences from these regions, we observed that the amino acids predicted to affect protein function clustered in domains that were ancestrally derived from zinc-finger domains but could no longer function as zinc fingers (Fig. 1, dashed lines). Because these domains aligned to functional zinc-finger domains during prediction, the changes were predicted to affect protein function. These regions would have acquired a substitution that rendered the zinc finger nonfunctional; once the first deleterious substitution was acquired, other substitutions were allowed to accumulate in the nonfunctional domain. Thus, studying the location of amino acids predicted to be damaging in a protein might reveal regions that have lost their function when aligned to related sequences that have retained their function.
Figure 1.
Damaging mutations in redundant motifs. FLJ20079 is aligned with inferred proteins AA495878-hyp and AA180031-hyp. Protein sequences were inferred by obtaining the EST sequences AA495878 and AA180031 from which the dbSNP variants were derived. AA180031 and AA495878 were only 93% and 91% identical to the gene encoding FLJ20079, respectively, but 100% identical to other regions of the human genome. Genomic sequences surrounding the region that matched AA180031 and AA495878 were retrieved and translated. The protein sequences were aligned to FLJ20079 and the start of the proteins interpreted to be the first Met that aligned to FLJ20079. The inferred hypothetical sequences were named AA180031-hyp and AA495878-hyp, from the ESTs from which they were derived. The nonsynonymous/synonymous ratio (Yang 1997) for AA180031-hyp and AA495878-hyp with FLJ20079 is 0.50 and 0.55, respectively, indicating that these proteins are undergoing purifying selection and may be functional. The three proteins contain zinc-finger motifs. Each putative zinc finger is indicated by a dashed or solid line and the value beneath the line is the e-value score for the region from FLJ20079 with the C2H2 zinc-finger motif (IPB000822) (Henikoff and Henikoff 1994). Regions with solid lines on top have the Cys and His residues involved in binding the zinc atom conserved in the three sequences. Regions with dashed lines do not have at least one of the Cys and His amino acids and can no longer function as zinc-finger modules. Amino acids predicted to be damaging by SIFT are the white characters against black background; most occur in regions that no longer function as zinc-finger modules.
Sequencing Errors Mistaken for Polymorphisms
Most of the variation in the remaining eight genes with a high fraction of nsSNPs predicted to be damaging originated from comparison of sequences from ESTs and/or cDNA clones with the reference gene (Table 2). These sequences had multiple base changes with respect to the reference gene (http://blocks.fhcrc.org/∼pauline/SIFTing_databases.html). It is doubtful that the observed differences are real SNPs occurring together on a rare allele; it is more likely that errors occurred in the EST sequencing or SNP interpretation procedure.
Among these eight genes, there were six nonsynonymous changes detected from sequences that were identical to the reference gene except for the change causing the amino acid substitution. These could be real nsSNPs found in the population. For example, the nsSNP that causes a V39A substitution in proteasome subunit β7 was detected in five different individuals. Multiple independent observations support this as an nsSNP occurring in the human population and SIFT predicted the V39A substitution as tolerated. The other five substitutions were predicted to affect protein function by SIFT. These could be real nsSNPs rather than errors from SNP detection programs. As these were detected in single libraries, they may be rare mutations under negative selection.
DISCUSSION
Identifying Damaging nsSNPs
Currently, there are more than a million SNPs in dbSNP that can be screened for association with diseases. By predicting the nsSNPs most likely to be damaging, the number of SNPs screened for association with disease can be reduced to those that most likely alter gene function. SIFT returned predictions for 3084 of the 5780 nsSNPs in the dbSNP database (Table 1). Of these 3084 substitutions, SIFT identified 757 that are likely to affect protein function; these are of higher interest than nsSNPs predicted to be neutral because they are more likely to contribute to disease. Not all of these variants will be useful for screening for novel contribution to disease because some were already known to be involved in disease. Some mapped to pseudogenes and others were sequencing errors; these were mistakenly interpreted as polymorphisms but have no bearing on health.
If a marker is found to be associated with disease and the marker is an nsSNP, prediction tools such as SIFT can provide independent evidence as to whether the nsSNP itself contributes to disease. A major problem in association studies is the high false positive signal of markers that appears to be associated with disease when a large number of markers are tested (Emahazion et al. 2001). nsSNPs in PPARα, MTHFR, and MSHR have been shown to be associated with disease, but assays for reduction of protein function have only been conducted on a fraction of them. Because carrying out the appropriate assays may be time-consuming, SIFT can filter out nsSNPs that are unlikely to affect protein function before experimentation. Users can choose to minimize either false negative or false positive error, tailoring SIFT predictions to their needs.
How useful are prediction programs such as SIFT for detecting damaging nsSNPs in proteins with only subtle effects on health? A protein may play only a minor or redundant role in the organism, so that if its function is altered the organism is only mildly affected. Nevertheless, over the long periods of evolution represented in an alignment, natural selection will remove damaging substitutions from such proteins and their homologs. For this reason, it was possible for SIFT to predict nsSNPs in PPARα, MTHFR, and MSHR as damaging, although they have only minor effects on a carrier's health.
SIFT prediction accuracy for a particular protein will depend on the alignment obtained. The sequences in the alignment are restricted to those homologous sequences that are available in the protein database; therefore, the resulting alignment information is expected to vary from protein to protein. The protein alignments constructed by SIFT contain paralogs as well as orthologs; therefore, active-site residues specific to orthologs may not appear conserved. However, a random mutation is more likely to affect structure than activity because relatively few residues are involved at the active site of the protein and many more are necessary for maintaining structure. Thus, reasonable prediction accuracy was obtained on the datasets when paralogs were included in the alignment used for prediction, although the ideal alignment is one composed of a diverse set of orthologs. As protein databases grow with data from sequencing whole genomes, a larger number of orthologs will become available and SIFT prediction should become more accurate.
Despite variation among protein families attributable to different evolutionary pressures and the heterogeneous set of sequence alignments used, our results show that SIFT works sufficiently well on a large scale so that it can be used as a first-pass filter to identify the substitutions worth pursuing. SIFT performance is similar to that of tools that require structure, as described below, so a more refined approach may not necessarily improve performance given the complexity of protein evolution.
Comparison of SIFT with Other Prediction Tools
Approximately 30% of the proteins encoded by the human genome are likely to be homologous to proteins with known structures (Guex et al. 1999). Therefore, the prediction tools of Sunyaev et al. (2001) and Chasman and Adams (2001), which require structural information, are restricted to these proteins. SIFT needs only homologous sequences for prediction and was able to predict on 60% of the protein sequences that contained dbSNP nonsynonymous variants (Table 1), providing twice the coverage of other tools.
Although SIFT does not use structural information, all three tools appear to perform similarly (Table 4). Sixty-nine percent of amino acid substitutions annotated to be involved in disease were predicted to be damaging by SIFT and by Sunyaev et al. (2001). SIFT (Ng and Henikoff 2001) and Chasman and Adams (2001) predicted similarly for neutral substitutions that did not alter LacI function; each had a false positive error of ∼30%. It is possible that SIFT performs similarly to tools that use structural information because constraints inferred from protein sequence alignments are based ultimately on structural constraints.
Table 4.
Summary of Results for Prediction Tools
| Ng and Henikoff | Chasman and Adams (2001) | Sunyaev et al. (2001) | ||||
|---|---|---|---|---|---|---|
| Number of substitutions predicted on (number of proteins predicted on) | % predicted to have effect | Number of substitutions predicted on (number of proteins predicted on) | % predicted to have effect | Number of substitutions predicted on (number of proteins predicted on) | % predicted to have effect | |
| Predictions on substitutions that affect function | ||||||
| Substitutions annotated to be involved in disease from databases | 5218 (462) | 69% (3626/5218)a | — | — | 1551 (N/A) | 69% (1071/1551)b |
| Substitutions in LacI that affect function | 1764 (1) | 63% (1113/1764)cd | 733 (1) | 75% (551/733)e | — | — |
| Prediction on substitutions that do not affect function | ||||||
| Substitutions in LacI that do not give an altered phenotype | 2240 (1) | 28% (627/2240)c | 1131 (1) | 31% (345/1131)f | — | — |
| Substitutions between species | — | — | — | — | 440 (N/A) | 9% (41/440)b |
| Nonsynonymous polymorphisms | ||||||
| WI-nsSNPs | 115 (53) | 19% (22/115)a | 53 (N/A) | 15% (8/53)g | — | — |
| Validated polymorphisms from databases | — | — | — | — | 245 (N/A) | 32% (79/245)b |
| dbSNP | 3084 (1789) | 25% (757/3084)a | — | — | — | — |
From Table 1.
From Table 1 of Sunyaev et al. (2001).
The SIFT prediction accuracies for LacI were originally published in Ng and Henikoff (2001). The numbers shown here are slightly different from the original reference because they are based on SIFT version 2.
Because paralogues were included in the LacI alignment used for SIFT prediction, many of the wrongly predicted residues recognition residues not conserved among paralogues (Ng and Henikoff 2001). This tended to lower prediction accuracy.
(prediction = effect & actual = effect)/actual = effect. Values are taken from the leftmost column in Table 5 from Chasman and Adams (2001), the test set with the highest total prediction accuracy which used the structure of LacI for predictions. When the structure of the Pur repressor, a LacI paralogue, was used for prediction on the LacI dataset, only 19% (69/368) [rightmost column of Table 5 from Chasman and Adams (2001)] of the substitutions that have an effect were correctly predicted as damaging.
(prediction = effect & actual = no effect)/actual = no effect. Values are taken from the leftmost column in Table 5 from Chasman and Adams (2001), the dataset with the highest total prediction accuracy.
From Table 7b of Chasman and Adams (2001).
Estimating the Number of Damaging nsSNPs in an Individual
By extrapolating their results to the human genome, Sunyaev et al. (2001) and Chasman and Adams (2001) have estimated that an individual would have on average 2000 and 9500 damaging nsSNPs, respectively. Our results do not support these estimates; the percentage of nsSNPs predicted to be damaging in dbSNP (25%) was close to the false positive error expected (19%) if all variants in dbSNP are functionally neutral (Table 1). Moreover, we found that some of the 6% difference between these two estimates can be accounted for by database contamination.
To calculate the percentage of nsSNPs that are damaging, ideally one should use an unbiased set of nsSNPs, estimate the percentage of nsSNPs predicted to be damaging, and then subtract the false positive error for functionally neutral substitutions. The WI-nsSNPs dataset is an unbiased set of nsSNPs, but because the genes screened were few in number and are candidates for disease, one still should be cautious in extrapolating from this dataset to the entire human genome. When SIFT was applied to WI-nsSNPs, there was no significant difference between the percentage predicted to be damaging for these SNPs and the false positive error (19% vs. 20%, respectively), indicating that the number of damaging nsSNPs per individual falls within our prediction error (Table 5).
Table 5.
Comparison of Amino Acid Prediction Tools on nsSNPs to Estimate the Percentage of nsSNPs that Affect Protein Function in an Individual
| Ng and Henikoff | Chasman and Adams (2001) | Sunyaev et al. (2001) | Sunyaev et al. (corrected) | |||||
|---|---|---|---|---|---|---|---|---|
| Dataset | Prediction | Dataset | Prediction | Dataset | Prediction | Dataset | Prediction | |
| % nsSNPs predicted to affect protein function − | WI-nsSNPs | 19% (22/115)a | WI-nsSNPs | 15% (8/53)b | nsSNPs from public databasesc | 32% (79/245)d | nsSNPs from public databases, biased mutations removed | 19% (41/207)e |
| False positive error (% neutral substitutions predicted to affect protein function) = | LacI | 20%a | LacI | 30% (345/1131)f | Substitutions between human proteins and their orthologues | 9% (41/399)d | Substitutions between human proteins and their orthologues | 9% (41/399)g |
| % nsSNPs that affect protein function | No difference | No extrapolation can be made | ∼20% | ∼10%, after removing biased nonsynonymous variantsb | ||||
For this estimation, an unbiased set of nsSNPs (detected from normal individuals) should be used and the false positive error subtracted from the percentage of nsSNPs predicted to be damaging.
From Table 1.
From Table 7b of Chasman and Adams (2001).
The databases from which the polymorphisms were obtained also contained disease-causing mutations so are not representative of random nsSNPs; see Discussion.
From Table 1 of Sunyaev et al. (2001).
See Discussion, section “Estimating the Number of Damaging nsSNPs in an Individual.”
Calculating the false positive error using the values from the leftmost column of Table 5 from Chasman and Adams (2001); this test set has the highest total prediction accuracy. Out of 1131 substitutions in LacI that have no effect, 345 substitutions were predicted to affect function (false positive error).
The value (41/399) used to calculate the false positive error was taken from Table 1 of Sunyaev et al. (2001). But since proteins with contaminating variants were removed, this should be readjusted.
See Discussion for why the 9% false positive error is a lower limit and therefore the 10% is an overestimate of the percentage of damaging nsSNPs that affect protein function.
What accounts for the difference in results? Chasman and Adams (2001) estimated 27% of nsSNPs are damaging based on the WI-nsSNPs but did not take into account their false positive prediction error. Their tool calculates the probability that a substitution affects function, and if this is below 0.5, the substitution is predicted to be functionally neutral. The 27% estimate was obtained by averaging the probabilities for all WI-nsSNPs. This type of analysis will fail to get a 0% estimate of damaging nsSNPs even if all substitutions are functionally neutral. On a set of neutral substitutions, low probabilities will correctly predict these substitutions as neutral, but when the probabilities are averaged, a nonzero value will be obtained. Because their approach cannot be used to estimate the percentage of damaging nsSNPs, we instead examine the number of WI-nsSNPs that Chasman and Adams (2001) predicted to be damaging and compare it with their false positive error for functionally neutral substitutions. They predicted 15% of the WI-nsSNPs as damaging (Table 5). This is lower than their 31% false positive error observed for functionally neutral substitutions (Table 4); therefore, no extrapolation for the number of damaging nsSNPs in a human genome can be made.
In the case of Sunyaev et. al. (2001), we examined the origin of the 79 nsSNPs they predicted to affect protein function and found that some are biased; therefore, they should not be included in the estimate of damaging nsSNPs per individual. Eighteen of the 79 nsSNPs are found in the HLA class I protein, most mapping to the peptide-binding region that is favored by diversifying selection (Janeway and Travers 1996). An additional 17 polymorphisms predicted to affect protein function were first discovered in an individual or population afflicted with disease in a gene known or suspected to contribute to the disease. These are far more likely to be involved in disease, and thus predicted as damaging, than random nsSNPs. Three substitutions from in vitro mutagenesis studies were also in the dataset. We were unable to account for the origin of all 79 nsSNPs, but we concluded that at least 38 mutations were biased in the manner discussed above and are not representative of random nsSNPs (http://blocks.fhcrc.org/∼pauline/SIFTing_databases.html). After removing these mutations, the percentage of polymorphisms predicted to be damaging decreased to 19% (Table 5). After subtracting the 9% false positive error they reported, this reduces the proportion of damaging nsSNPs to 10%.
The 9% false positive error reported by Sunyaev et al. (2001) was based on applying their tool to substitutions that have occurred between human proteins and their orthologs. These substitutions have undergone millions of years of selection and must have had selection coefficients very near zero to become fixed (with the exception of substitutions that have been driven by positive selection). Conditional mutations, those that affect protein function conditional on an environment that may no longer exist (Fay et al. 2001), are excluded from Sunyaev et al.'s control set. Such substitutions will exist as SNPs that will eventually be culled out over time, but they have undetectable effects on an individual's health. Thus, Sunyaev et al.'s control set is the easiest set of substitutions to predict on because even long evolutionary periods are insufficient for them to be culled out. Hence, the 9% false positive error is a lower limit for their prediction method. The 10% difference between Sunyaev et al.'s 9% false positive error and 19% nsSNPs predicted to be damaging (after correcting for biased nsSNPs) is an estimate of damaging nsSNPs that severely affect protein function, as well as the slightly deleterious nsSNPs that might eventually be removed by natural selection. This latter class may be irrelevant to human disease. Another study has estimated that ∼20% of nsSNPs are selected against by comparing the frequencies of common and rare nsSNPs (Fay et al. 2001). This estimate, like Sunyaev et al.'s, includes damaging, as well as slightly deleterious, mutations. The discrepancy between the two values may result from differences in the datasets and their small sample sizes.
Based on the foregoing analysis, we were unable to conclude that the percentage of damaging nsSNPs that can affect human health is as high as 20% to 30%. We suggest there is a low number of nsSNPs that affect protein function in each individual because estimates lie within false positive error. This low number is supported by a study that examined the prereproductive mortality in the children of first-cousin marriages and estimated the average human is heterozygote for 1.4 lethal equivalents, or ∼0.002% of human genes (Bittles and Neel 1994).We conclude that there are very few damaging nsSNPs in an individual's genome that could impact health.
METHODS
Predicting Damaging Amino Acid Substitutions
SIFT uses sequence homology to predict whether an amino acid substitution predicts protein function and has two major steps (Ng and Henikoff 2001). In the first step, sequences closely related to the protein are chosen and the alignment of these sequences is what prediction is based on. In the second step, a scaled probability for the substitution of interest is calculated based on the amino acids observed at the position of substitution in the alignment generated from the first step. The substitution is predicted to affect protein function if its scaled probability falls below a cutoff. In SIFT version 2 (available at http://blocks.fhcrc.org/∼pauline/SIFT.html), the method by which sequences are chosen for the alignment has been changed. The user can opt for either a low false negative error, which predicts most of the substitutions that affect protein function, or a low false positive error, which predicts fewer substitutions that affect function but with a higher level of certainty.
SIFT version 2 first obtains related sequences, which are assumed to be functional, by searching SWISS-PROT/TrEMBL (Bairoch and Apweiler 2000) with PSI-BLAST (Altschul et al. 1997) for two iterations (-e 0.0001, -h 0.002). The sequences found by PSI-BLAST that are more than 90% identical to each other are clumped together and a consensus sequence is obtained for each clump by choosing the most frequently occurring amino acid for each position in the sequence. An iterative procedure is then used to choose the related sequences. The procedure starts by giving the query sequence to PSI-BLAST to search among the consensus sequences. The top hit is added and aligned to the query sequence. Conservation, as measured by information content (Schneider et al. 1986), is calculated for each position in the alignment, and the median of these values is obtained. The median conservation can range from 4.3 (sequences nearly 100% identical to each other) to 0 (all 20 amino acids are represented at the majority of positions in the sequence alignment). If the median conservation over all positions does not fall below a user-defined cutoff, the hit is retained in the alignment and a PSI-BLAST checkpoint file is built from the alignment. The checkpoint file is used as a query for PSI-BLAST to search among the remaining consensus sequences and the highest-scoring hit is added to the alignment only if the median conservation does not fall below the cutoff. The process repeats and sequences are continually added to the growing alignment until the median conservation cutoff is reached.
For efficiency, a new PSI-BLAST search is conducted after five sequences have been added. Once the process stops and the consensus sequences to be included determined, the protein sequences corresponding to these consensus sequences are obtained and their PSI-BLAST alignment used. To prevent the alignment from being contaminated by pseudogenes or protein sequences containing the polymorphism, sequences >90% identical to the query sequence are removed. SIFT allows a range of cutoffs, and similar results are obtained when sequences 95% and 99% identical to the query are removed (http://blocks.fhcrc.org/∼pauline/SIFTing_databases.html). The alignment is used for the second step of SIFT prediction as described previously with the gap option turned off (Ng and Henikoff 2001).
The user sets the median conservation cutoff to minimize either false negative error or false positive error. We used the mutation dataset from Escherichia coli LacI (Pace et al. 1997) to decide the range of median conservation values that work best. When the median conservation ranges from 2.25 to 3.25, the total prediction accuracy (number correctly predicted/number total substitutions assayed) on LacI remains the same (68%). Therefore, for prediction on the databases described here, we used 2.75 as the median conservation cutoff. If the sequences represented at the position of substitution had median conservation >3.25, this indicated that there were not enough homologous sequences in the database; therefore, no prediction was made.
When SIFT returns the prediction for an amino acid substitution, it also returns the median conservation for the sequences used in the alignment. A lower value provides greater confidence that the prediction for a substitution has a low false positive error because a low median conservation value reflects that very diverse sequences were used in the alignment. Then a substitution predicted to be damaging has occurred at a position that has been well conserved among the diverse set of proteins despite the diversity of amino acid compositions at other positions. This indicates that the position of substitution is constantly under negative selection; therefore, it is likely that the change is damaging.
Databases
To identify amino acid substitutions involved in disease, we searched SWISS-PROT 39.11 and TrEMBL 15.11 (http://www.expasy.ch/sprot, Bairoch and Apweiler 2000) with the keywords disease and mutation. We found 7397 disease-causing substitutions from 606 proteins after removing any substitution annotated as polymorphism or probable polymorphism.
nsSNPs in normal individuals were detected by the Whitehead Institute (Cargill et al. 1999). This dataset, downloaded from http://www.genome.wi.mit.edu/cvar_snps, is referred to as WI-nsSNPs.
Amino acid variants from dbSNP (build #95) (http://www.ncbi.nlm.nih.gov/SNP, Sherry et al. 2001) were found by searching dbSNP for variants with FXN-“coding nonsynonymous” in the organism Homo sapiens. Entries that listed the amino acid position affected were retrieved. For a given substitution, the reference amino acid was checked to match the amino acid in the protein sequence corresponding to the accession number referred to in the refSNP file. If the substitution did not match, it was discarded. If a substitution was referenced to more than one protein, such as in isoforms, the duplicated substitutions were removed so that the substitution was represented only once. Only one substitution per position was predicted on. After applying this filter, 5780 substitutions from 3005 protein sequences remained.
Database predictions are available at http://blocks.fhcrc.org/∼pauline/SIFTing_databases.html
Estimation of False Positive Error
To test the hypothesis that all substitutions from a database are neutral, the percentage predicted to be damaging on the test set was compared with the percentage predicted to be damaging on a set of substitutions known to be neutral. More than 4000 single amino acid substitutions had been introduced into LacI and both neutral and negative phenotypes were assayed (Pace et al. 1997). In our previous study, this dataset was used to measure SIFT performance (Ng and Henikoff 2001). Because the effects of substitutions are known in this protein, we used this dataset as a standard to calibrate the expected prediction accuracy. SIFT's prediction accuracy for LacI is 68% for all substitutions with a median conservation cutoff of 2.75. However, the mutation data for LacI was generated from assaying 12 or 13 amino acid substitutions at each position, and some of the amino acid substitutions tested could not have occurred from a single base change, which is presumed for substitutions in the polymorphism test set. Because performance on amino acid substitutions that require multiple base changes has no relevance for the substitutions assayed on the databases, and some types of substitutions will occur more often than others, prediction accuracy must be calibrated for the composition of the test set being predicted on. The tolerated prediction accuracy weighted by composition of the test set was calculated as:
 |
1 |
For example, 107/3084 variants from dbSNP that were predicted on were substitutions from Ala to Thr. SIFT accurately predicts 75% of the Ala→Thr and Thr→Ala neutral substitutions as tolerated in the LacI dataset. This is the left term in Equation 1 for i = Ala and j = Thr. Rather than the right term simply being 107/3084, the denominator is reduced because not all combinations of substitutions were assayed in the LacI dataset. Tolerated prediction accuracy based on the LacI data is available for 2499 of the substitutions from dbSNP; thus, the contribution of the Ala→Thr substitution to the weighted tolerated accuracy is 0.75 *107/2499. The weighted tolerated prediction accuracy is the sum over all substitutions aai and aaj for which LacI tolerated prediction accuracy can be calculated and is weighted by the proportion of substitutions of aai and aaj occurring in the polymorphism database. The weighted false positive error is the weighted tolerated prediction accuracy subtracted from 100.
Genes with a High Fraction of nsSNPs Predicted to Affect Protein Function
We approximated the predictions for 217 genes with at least three nsSNP entries from dbSNP according to a binomial distribution. SIFT, with median conservation 2.75, has a false positive error of 0.30 for the entire LacI dataset. If x is the number of substitutions predicted to be damaging by SIFT and n is the total number of substitutions predicted on for the protein, the probability that at least x variants predicted to affect function is:
 |
2 |
Genes with probability <0.1 were considered to have a high fraction of nsSNPs predicted to affect protein function and were further investigated.
WEB SITE REFERENCES
http://blocks.fhcrc.org/∼pauline/SIFT.html); site at which SIFT version 2 is available.
http://blocks.fhcrc.org/∼pauline/SIFTing_databases.html; site at which database predictions are available.
http://www.expasy.ch/sprot; mutations annotated to be involved in disease were retrieved from SWISS-PROT/TrEMBL
http://www.genome.wi.mit.edu/cvar_snps; dataset of nsSNPs in normal individuals as detected by the Whitehead Institute and referred to as WI-nsSNPs.aa
http://www.ncbi.nlm.nih.gov/SNP; dbSNP, a public SNP database.
Acknowledgments
We thank Harmit Malik and Jorja Henikoff for their support. Kami Ahmad, Leonid Kruglyak, and Wendy Thomas gave thoughtful comments on the manuscript. P. Ng is a Department of Energy Computational Science Graduate Fellow. This work was supported by a grant from NIH (GM20009).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL steveh@fhcrc.org; FAX (206) 667-5889.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.212802.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au KG, Zhang J, Purdy GD, Fraser DJ, Lee D, Noren NK, Cronin MT, Chen J. Polymorphism screening of the human peroxisome proliferator activated receptor α gene in diabetic patients by ABI sequencing and high density oligonucleotide array technology. Am J Hum Genet. 1998;63:abs997. (data on poster, noted by A.J. Brookes and entered into HGBASE). [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box NF, Duff DL, Irving RE, Russell A, Chen W, Griffyths LR, Parsons PG, Green AC, Sturm RA. Melanocortin-1 receptor genotype is a risk factor for basal and squamous cell carcinoma. J Invest Dermatol. 2001;116:224–229. doi: 10.1046/j.1523-1747.2001.01224.x. [DOI] [PubMed] [Google Scholar]
- Brookes AJ, Lehvaslaiho H, Siegfried M, Boehm JG, Yuan YP, Sarkar CM, Bork P, Ortigao F. HGBASE: A database of SNPs and other variations in and around human genes. Nucleic Acids Res. 2000;28:356–360. doi: 10.1093/nar/28.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles AH, Neel JV. The costs of human inbreeding and their implications for variations at the DNA level. Nat Genet. 1994;8:117–121. doi: 10.1038/ng1094-117. [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Chasman D, Adams RM. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: Structure-based assessment of amino acid variation. J Mol Biol. 2001;307:683–706. doi: 10.1006/jmbi.2001.4510. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Ball EV, Krawczak M. The human gene mutation database. Nucleic Acids Res. 1998;26:285–287. doi: 10.1093/nar/26.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emahazion T, Feuk L, Jobs M, Sawyer SL, Fredman D, St. Clair D, Prince JA, Brookes AJ. SNP association studies in Alzheimer's disease highlight problems for complex disease analysis. Trends Genet. 2001;17:407–413. doi: 10.1016/s0168-9525(01)02342-3. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu C-I. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell DM, Pineda Torra I, Jamshidi Y, Evans D, Diamon JR, Elkeles RS, Bujac SR, Miller G, Talmud PJ, Staels B, et al. Variation in the PPARα gene is associated with altered function in vitro and plasma lipid concentrations in Type II diabetic subjects. Diabetologia. 2000;43:673–680. doi: 10.1007/s001250051357. [DOI] [PubMed] [Google Scholar]
- Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP, et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- Garg K, Green P, Nickerson DA. Identification of candidate coding region single nucleotide polymorphisms in 165 human genes using assembled expressed sequence tags. Genome Res. 1999;9:1087–1092. doi: 10.1101/gr.9.11.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Diemand A, Peitsch MC. Protein modelling for all. Trends Biochem Sci. 1999;24:364–367. doi: 10.1016/s0968-0004(99)01427-9. [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan J, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Hara M, Wang X, Paz VP, Iwasaki N, Honda M, Iwamoto Y, Bell GI. Identification of three missense mutations in the peroxisome proliferator-activated receptor α gene in Japanese subjects with mature-onset diabetes of the young. J Hum Genet. 2001;46:285–288. doi: 10.1007/s100380170080. [DOI] [PubMed] [Google Scholar]
- Harding RM, Healy E, Ray AJ, Ellis NS, Flanagan N, Todd C, Dixon C, Sajantila A, Jackson IJ, Birch-Machin MA, et al. Evidence for variable selective pressures at MC1R. Am J Hum Genet. 2000;66:1351–1361. doi: 10.1086/302863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy E, Todd C, Jackson IJ, Birch-Machin M, Rees JL. Skin type, melanoma, and melanocortin 1 receptor variants. J Invest Dermatol. 1999;112:512–513. doi: 10.1046/j.1523-1747.1999.00554.x. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Protein family classification based on searching a database of blocks. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- Irizarry K, Kustanovich V, Li C, Brown N, Nelson S, Wong W, Lee CJ. Genome-wide analysis of single-nucleotide polymorphisms in human expressed sequences. Nat Genet. 2000;26:233–236. doi: 10.1038/79981. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Jr, Travers P. Immunobiology: The immune system in health and disease. 2nd ed. London, UK: Current Biology Ltd.; 1996. The major histocompatibility complex of genes: Organization and polymorphism; pp. 4:20–4:31. [Google Scholar]
- Lacquemant C, Lepretre F, Pineda Torra I, Manraj M, Charpentier G, Ruiz J, Staels B, Froguel PH. Mutation screening of the PPARα gene in type 2 diabetes associated with coronary heart disease. Diabetes Metab. 2000;26:393–401. [PubMed] [Google Scholar]
- Liu W, Oefner PJ, Qian C, Odom RS, Francke U. Denaturing HPLC-identified novel FBN1 mutations, polymorphisms, and sequence variants in Marfan syndrome and related connective tissue disorder. Genet Testing. 1997/1998;1:237–242. doi: 10.1089/gte.1997.1.237. [DOI] [PubMed] [Google Scholar]
- Ma J, Stampfer MJ, Giovannucci E, Artigas C, Hunter DJ, Fuchs C, Willet WC, Selhub J, Hennekens CH, Rozen R. Methylenetetrahydrofolate reductase polymorphism, dietary interacations, and risk of colorectal cancer. Cancer Res. 1997;57:1098–1102. [PubMed] [Google Scholar]
- Merritt CM, Easteal S, Board PG. Evolution of human α1-acid glycoprotein genes and surrounding Alu repeats. Genomics. 1990;6:659–665. doi: 10.1016/0888-7543(90)90501-k. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace HC, Kercher MA, Lu P, Markiewicz P, Miller JH, Chang G, Lewis M. Lac repressor genetic map in real space. Trends Biochem Sci. 1997;22:334–339. doi: 10.1016/s0968-0004(97)01104-3. [DOI] [PubMed] [Google Scholar]
- Palmer JS, Duffy DL, Box NF, Aitken JF, O'Gorman LE, Green AC, Hayward NK, Martin NG, Sturm RA. Melanocortin-1 receptor polymorphisms and risk of melanoma: Is the association explained solely by pigmentation phenotype? Am J Hum Genet. 2000;66:176–186. doi: 10.1086/302711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stormo GD, Gold L, Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibola CF, Smith MT, Kane E, Roman E, Rollinson S, Cartwright RA, Morgan G. Polymorphisms in the methylenetetrahydrofolate reductase gene are associated with susceptibility to acute leukemia in adults. Proc Natl Acad Sci. 1999;96:12810–12815. doi: 10.1073/pnas.96.22.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, III, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Vohl M, Lepage P, Gaudet D, Brewer CG, Betard C, Perron P, Houde G, Cellier C, Faith JM, Despres JP, et al. Molecular scanning of the human PPARα gene: Association of the L162V mutation with hyperapobetalipoproteinemia. J Lipid Res. 2000;41:945–952. [PubMed] [Google Scholar]
- von Eckardstein A, Funke H, Walter M, Altland K, Benninghoven A, Assmann G. Structural analysis of human apolipoprotein A-I variants. J Biol Chem. 1990;265:8610–8617. [PubMed] [Google Scholar]
- Weisberg I, Tran P, Christensen B, Sibani S, Rozen R. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab. 1998;64:169–172. doi: 10.1006/mgme.1998.2714. [DOI] [PubMed] [Google Scholar]
- Wiemels JL, Smith RN, Taylor GM, Eden OB, Alexander FE, Greaves MF United Kingdom Childhood Cancer Study Investigators. Methylenetetrahydrofolate reductase (MTHFR) polymorphisms and risk of molecularly defined subtypes of childhood acute leukemia. Proc Natl Acad Sci. 2001;98:4004–4009. doi: 10.1073/pnas.061408298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]