Abstract
The ribosome, as a catalyst for protein synthesis, is universal and essential for all organisms. Here we describe the structure of the genes encoding human ribosomal proteins (RPs) and compare this class of genes among several eukaryotes. Using genomic and full-length cDNA sequences, we characterized 73 RP genes and found that (1) transcription starts at a C residue within a characteristic oligopyrimidine tract; (2) the promoter region is GC rich, but often has a TATA box or similar sequence element; (3) the genes are small (4.4 kb), but have as many as 5.6 exons on average; (4) the initiator ATG is in the first or second exon and is within ± 5 bp of the first intron boundaries in about half of cases; and (5) 5′- and 3′-UTRs are significantly smaller (42 bp and 56 bp, respectively) than the genome average. Comparison of RP genes from humans, Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae revealed the coding sequences to be highly conserved (63% homology on average), although gene size and the number of exons vary. The positions of the introns are also conserved among these species as follows: 44% of human introns are present at the same position in either D. melanogaster or C. elegans, suggesting RP genes are highly suitable for studying the evolution of introns.
[The sequence data described in this paper have been submitted to the DDBJ/EMBL/GenBank databases under accession nos. AB055762–AB055780, AB056456, AB061820–AB061859, AB062066–AB062071, and AB070559.]
The ribosome is the cellular organelle responsible for protein synthesis in all cells. Recent analyses of the ribosome's structure using X-ray crystallography have enhanced our understanding of the structural basis of ribosome function (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000; Yusupov et al. 2001). In contrast, comparatively little is known about ribosome biogenesis, especially in higher eukaryotes. In mammalian cells, the biogenesis of cytoplasmic ribosomes requires assembly of 4 RNA molecules and 79 different proteins (Wool 1979). With the exception of two proteins, all of these components are present as single copies within the ribosome. Typically, mammalian cells contain ∼4 × 106 cytoplasmic ribosomes, which account for 80% of all cellular RNA and 5%–10% of cellular proteins.
Investigation of the mechanism that controls the coordinated expression of these components is a challenge. Three different RNA polymerases are involved in production of these RNAs and proteins, RNA polymerase I (POL I) is involved in production of the 28S, 18S, and 5.8S rRNAs, POL II in production of ribosomal proteins (RPs), and POL III in production of the 5S rRNA. The amino acid sequences of all rat and human RPs have been deduced (Wool et al. 1996), and the nucleotide sequences of thousands of eukaryotic rRNAs are now known (The Ribosome Database Project; Maidak et al. 2001). On the other hand, only a handful of mammalian RP genes have been studied in terms of their genomic structure. Unlike rRNAs, which are encoded by several hundred copies of genes, each mammalian RP is typically encoded by a single gene. Single functional genes generate large numbers of processed pseudogenes (Dudov and Perry 1984; Wagner and Perry 1985; Kuzumaki et al. 1987), which, however has hampered the cloning of the functional genes and, hence, analysis of their genomic structure. Even though some enhancer/promoter sites have been identified (Rhoads et al. 1986; Hariharan et al. 1989; Kenmochi et al. 1992; Toku and Tanaka 1996), we are far from understanding the basis of the coordinated expression of RP genes.
Despite the central role played by cytoplasmic ribosomes in organismal growth and development, the effects of their mutation have been largely ignored, particularly with respect to human disease. One might predict that genetic defects in the components of ribosomes would invariably result in early embryonic death. However, there is strong evidence in Drosophila that a quantitative deficiency of any one of the cytoplasmic RPs can yield the viable but abnormal Minute phenotype (Kongsuwan et al. 1985; Lambertsson 1998). Moreover, heterozygous mutations in the ribosomal protein S19 gene (RPS19) have been found in a subset of patients with Diamond-Blackfan anemia (Draptchinskaia et al. 1999; Willig et al. 1999), a rare form of chronic anemia characterized by the absence or low levels of erythroid precursors in the bone marrow (Diamond et al. 1976; Halperin et al. 1989). It has been suggested that RPS4, encoded by both the X and Y chromosomes, is an important factor for Turner syndrome (Fisher et al. 1990; Watanabe et al. 1993), a complex human phenotype associated with monosomy X (Zinn et al. 1994). Finally, RPL6 was mapped to a critical region for Noonan syndrome (Jamieson et al. 1994; Kenmochi et al. 2000), and because of similarities between the Noonan and Turner phenotypes (Noonan 1968; Allanson 1987), the gene is considered an attractive candidate for the disease. Although involvement of the RP genes in the pathogenesis of the aforementioned diseases has yet to be proved, we are intrigued by the possibility that defects in other RP genes might also underlie certain pathological conditions.
To explore this possibility, we mapped all human RP genes to the chromosomes and then compared the assigned positions with candidate regions for Mendelian disorders (Kenmochi et al. 1998; Uechi et al. 2001). The results emphasize the need to conduct systematic analysis of the genomic sequences of these genes to screen for mutations that could disturb ribosomal function. In the present study, we determined the genomic sequences of human RP genes, as well as the full-length cDNA sequences. Together with the previously determined sequences, we analyzed the characteristics of 73 RP genes with respect to intron/exon structure, transcription start site, promoter region, and the 5′ and 3′ noncoding regions. Comparative analysis of these genes among several eukaryotes was also carried out. Finally, we evaluated the currently available draft genome sequence using our data set of RP gene sequences.
RESULTS
Gene Structure
The human RP genes were cloned from the Keio BAC library by PCR using sequence-tagged sites (STSs) originally developed from partial genomic sequences of the genes (Kenmochi et al. 1998; Uechi et al. 2001). These STSs enabled us to distinguish the intron-containing functional genes from the processed pseudogenes, which, in turn, enabled us to clone 73 of the 80 human RP genes. Of these, 44 were newly sequenced in the present study by use of the shotgun method. The full-length cDNA sequences were also determined to analyze the transcription start sites. Together with the previously determined sequences, we analyzed 70 complete (including at least 400 bp of the 5′-flanking region) and 3 partial human RP gene sequences. The accession numbers for these genes, including both newly and previously determined sequences and the 5′-UTR sequences of the full-length cDNAs are listed in Table 1.
Table 1.
Structure of 73 Human RP Genes
| Gene | Size bp | CDS bp | No. of exons | GC cont percenta | CT tract bp | Position of ATGb | Exon 1 bp | 5′ UTR bp | 3′ UTR bp | Accession no. | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | 5′ UTR | ||||||||||
| RPSA | 5739 | 888 | 7 | 61 | 8 | 36 | 45 | 78 | 66 | U43901 | D28372 |
| RPS2 | 2757 | 882 | 7 | 67 | 12 | 6 | 20 | 23 | 28 | AC005363 | AB055772d |
| RPS3 | 6172 | 732 | 7 | 60 | 14 | −27 | 60 | 30 | 78 | AB061838d | D28344 |
| RPS3A | 5012 | 795 | 6 | 62 | 7 | −59 | 87 | 25 | 38 | AB062070dZ83334 | D28374 |
| RPS4X | 4609 | 792 | 7 | 63 | 12 | 0 | 26 | 23 | 61 | AF041428 | D28359 |
| RPS4Y | 25349 | 792 | 7 | 49 | 9 | 0 | 26 | 23 | 56 | AF041427 | AB055773d |
| RPS5c | — | 615 | — | 59 | 9 | 4 | 71 | 72 | 54 | AB061852dAB061853d | D28455 |
| RPS6 | 3980 | 750 | 6 | 58 | 11 | −3 | 48 | 42 | 38 | X67309 | D28348 |
| RPS7 | 5571 | 585 | 7 | 64 | 9 | 21 | 26 | 44 | 21 | Z25749 | AB055774d |
| RPS8 | 3162 | 627 | 6 | 66 | 11 | −1 | 27 | 23 | 55 | AB061855dX67247 | D28361 |
| RPS9 | 6787 | 585 | 5 | 57 | 10 | 28 | 31 | 56 | 69 | AB061839d | AB055774d |
| RPS10 | 8616 | 498 | 6 | 63 | 13 | 3 | 38 | 38 | 52 | AL157372 | D28427 |
| RPS11 | 3258 | 477 | 5 | 60 | 16 | −12 | 42 | 27 | 52 | AB028893 | D28407 |
| RPS12 | 2985 | 399 | 6 | 59 | 10 | 40 | 46 | 83 | 14 | AB061840d | D28378 |
| RPS13 | 3269 | 456 | 6 | 62 | 10 | −20 | 49 | 26 | 39 | AB062067dD88010 | D28429 |
| RPS14 | 5398 | 456 | 5 | 62 | 12 | 5 | 37 | 39 | 45 | AB061858dM13934 | D28352 |
| RPS15 | 2084 | 438 | 4 | 67 | 8 | 0 | 22 | 19 | 31 | M32405 | AB055776d |
| RPS15A | 7369 | 393 | 5 | 61 | 10 | 8 | 25 | 30 | 42 | AC020716 | D28347 |
| RPS16 | 2737 | 441 | 5 | 66 | 12 | −45 | 102 | 54 | 60 | AB061841d | D28392 |
| RPS17 | 3670 | 408 | 5 | 68 | 11 | 0 | 27 | 24 | 45 | AB062068dM18000 | AB055777d |
| RPS18 | 4429 | 459 | 6 | 55 | 11 | 0 | 46 | 43 | 39 | AL031228 | AB055778d |
| RPS19 | 11158 | 438 | 6 | 72 | 13 | 3 | 33 | 33 | 39 | AC010616 | D28389 |
| RPS20 | 1448 | 360 | 4 | 67 | 13 | 0 | 126 | 123 | 31 | AB061842d | D28358 |
| RPS21 | 1416 | 252 | 6 | 73 | 15 | 21 | 33 | 51 | 53 | AB061843d | D28422 |
| RPS23 | 2290 | 432 | 4 | 62 | 12 | −1 | 35 | 31 | 43 | AC005406 | D28396 |
| RPS24 | 4459 | 402 | 5 | 60 | 16 | 0 | 42 | 39 | 75 | AB062069dU12202 | D28424 |
| RPS25 | 2634 | 378 | 5 | 60 | 10 | 0 | 66 | 63 | 48 | AB061844d | D28369 |
| RPS26 | 2029 | 348 | 4 | 55 | 6 | 0 | 30 | 27 | 66 | AB061856dU41448 | AB056456d |
| RPS27 | 1387 | 255 | 4 | 53 | 9 | −3 | 40 | 34 | 54 | AB061845d | D28454 |
| RPS27A | 2926 | 471 | 6 | 54 | 12 | 20 | 25 | 42 | 32 | AB062071dAB061854d | D28404 |
| RPS28 | 895 | 210 | 4 | 67 | 9 | −36 | 70 | 31 | 139 | AB061846d | AB055779d |
| RPS29 | 2799 | 171 | 3 | 57 | 12 | −59 | 93 | 31 | 88 | AB061847d | AB055780d |
| RPS30 | 1503 | 402 | 5 | 65 | 16 | 11 | 45 | 53 | 50 | AB061859dX65921 | D28403 |
| RPL3 | 6740 | 1212 | 10 | 62 | 5 | 0 | 30 | 27 | 55 | AL022326 | D28415 |
| RPL4 | 5517 | 1284 | 10 | 50 | 12 | 0 | 59 | 56 | 87 | AB061820d | D23660 |
| RPL5 | 9885 | 894 | 8 | 61 | 10 | 0 | 78 | 75 | 59 | AB061848dAL162740 | AB055762d |
| RPL6 | 4415 | 867 | 7 | 54 | 11 | 3 | 19 | 19 | 33 | AB042820 | D28388 |
| RPL7c | — | 762 | — | 48 | 14 | −11 | 38 | 24 | — | L16557 | AB055763d |
| RPL7A | 3225 | 801 | 8 | 62 | 25 | 0 | 25 | 22 | 59 | X52138 | D28405 |
| RPL8 | 2622 | 774 | 6 | 73 | 11 | 14 | 25 | 36 | 35 | AB061821d | D28421 |
| RPL9 | 4784 | 579 | 8 | 58 | 10 | 4 | 25 | 26 | 103 | U09954 | D28399 |
| RPL10 | 2540 | 645 | 7 | 61 | 13 | 26 | 18 | 41 | 63 | AB061857dM81806 | D28410 |
| RPL10A | 2381 | 654 | 6 | 67 | 10 | −2 | 36 | 31 | 34 | AL022721 | AB055764d |
| RPL12 | 3730 | 498 | 7 | 63 | 6 | −34 | 125 | 88 | 45 | AL445222 | D28443 |
| RPL13A | 4254 | 612 | 8 | 61 | 14 | −12 | 38 | 23 | 43 | AB028893 | D28409 |
| RPL14 | 4960 | 663 | 6 | 64 | 14 | 0 | 32 | 29 | 42 | AB061822d | AB055765d |
| RPL15 | 2360 | 615 | 4 | 65 | 13 | 13 | 26 | 36 | 6 | AB061823d | D28417 |
| RPL17 | 4003 | 555 | 7 | 54 | 16 | 16 | 26 | 39 | 35 | AB061824d | D28373 |
| RPL18 | 3867 | 567 | 7 | 64 | 12 | 0 | 36 | 33 | 41 | AB061825d | D28461 |
| RPL18A | 3394 | 531 | 5 | 67 | 9 | −15 | 53 | 35 | 52 | AC005796 | D28393 |
| RPL19 | 4411 | 591 | 6 | 57 | 12 | −2 | 33 | 28 | 79 | AC004408 | AB055766d |
| RPL21 | 5017 | 483 | 6 | 61 | 13 | 15 | 28 | 40 | 43 | AB061826d | D28406 |
| RPL22 | 12978 | 387 | 4 | 67 | 11 | −9 | 35 | 23 | 53 | AB061849dAL031847 | D28346 |
| RPL23 | 3645 | 423 | 5 | 54 | 19 | −10 | 39 | 26 | 41 | AB061827d | D28349 |
| RPL23A | 2968 | 471 | 5 | 56 | 8 | −22 | 37 | 12 | 54 | AF001689 | D28401 |
| RPL24 | 5606 | 474 | 6 | 55 | 16 | −2 | 47 | 42 | 43 | AB061828d | D28400 |
| RPL26 | 5664 | 438 | 4 | 58 | 14 | 8 | 37 | 42 | 41 | AB061829d | D28413 |
| RPL27c | — | 411 | — | 59 | 16 | 5 | 33 | 35 | 25 | AB061850dAB061851d | D28453 |
| RPL27A | 3052 | 447 | 5 | 65 | 12 | 0 | 25 | 22 | 39 | AB020236 | AB055767d |
| RPL30 | 3824 | 348 | 5 | 59 | 13 | 35 | 42 | 74 | 75 | AB070559d | D28438 |
| RPL31 | 4130 | 378 | 5 | 61 | 12 | 3 | 27 | 27 | 37 | AB061830d | D28386 |
| RPL32 | 5512 | 408 | 4 | 51 | 17 | 8 | 48 | 53 | 63 | AB061831d | D28385 |
| RPL34 | 4652 | 354 | 5 | 53 | 18 | 12 | 24 | 33 | 17 | AB061832d | D28420 |
| RPL35 | 4078 | 372 | 4 | 72 | 15 | 0 | 52 | 49 | 34 | AL354928 | D28448 |
| RPL36 | 1370 | 318 | 4 | 67 | 7 | 5 | 32 | 34 | 44 | AB061833d | AB055769d |
| RPL36A | 2881 | 321 | 5 | 58 | 13 | 0 | 36 | 33 | 50 | U78027 | D28414 |
| RPL37 | 2758 | 294 | 4 | 60 | 7 | 0 | 30 | 27 | 49 | AB061834d | AB055770d |
| RPL39 | 5137 | 156 | 3 | 60 | 7 | 0 | 70 | 67 | 178 | AB061835d | D28397 |
| RPL40 | 3381 | 387 | 5 | 70 | 14 | 11 | 31 | 39 | 90 | X56997 | D28425 |
| RPL41 | 1200 | 78 | 4 | 52 | 10 | 16 | 27 | 40 | 303 | AB062066dAB010874 | D28462 |
| RPP0 | 4405 | 954 | 8 | 63 | 10 | 51 | 29 | 77 | 66 | AC004263 | D28418 |
| RPP1 | 2727 | 345 | 4 | 62 | 11 | −69 | 202 | 130 | 38 | AB061836d | D28366 |
| RPP2 | 2906 | 348 | 5 | 71 | 17 | 4 | 73 | 74 | 38 | AB061837d | D28411 |
| Average | 4413 | 521 | 5.6 | 61 | 12 | 0.0 | 45 | 42 | 56 | ||
GC content between −250 and +250 bp.
Relative position of the initiator ATG with respect to the first intron boundaries; e.g., Ae’0'Åf means localization of the ATG at the 3′ end of exon 1; ‘3’ at the 5′ end of exon 2.
Partial sequence, not included in the calculation of the average values.
Sequence determined in this study.
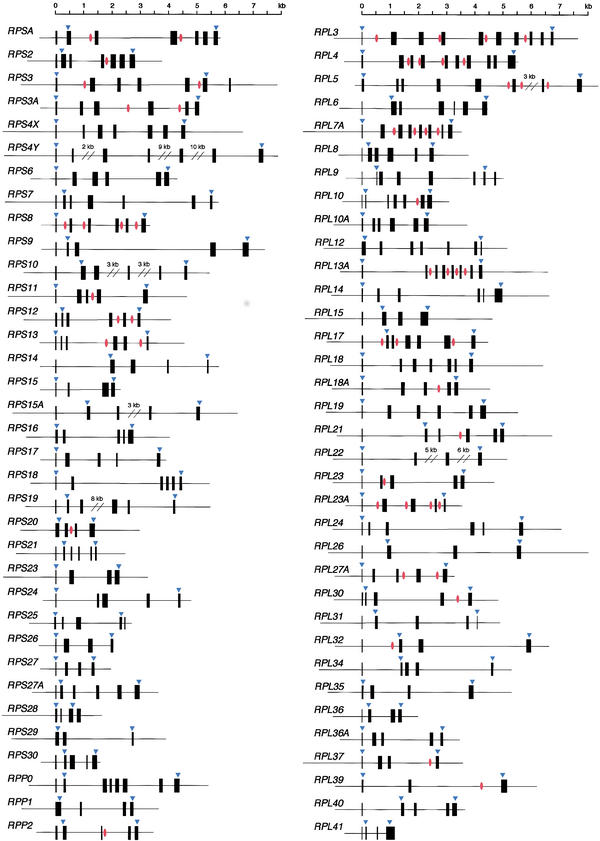
Figure 1 shows the intron/exon structures and the positions corresponding to the translation start and stop sites. The average size of the genes from the transcription start site was ∼4.4 kb; RPS4Y was the largest (25 kb), whereas RPS28 was the smallest (only 0.9 kb, Table 1). Each gene contained an average of 5.6 exons, ranging from 3 (RPS29 and RPL39) to 10 (RPL3 and RPL4). The translation initiator ATG was present either in the first or second exon, whereas the stop codon was in the last exon (all but RPS3, RPS25, RPS28, and RPL9). Interestingly, the ATG was always located near the splice sites of the first intron and, in 20 cases, was exactly at the 3′ end of the first exon (Table 1).
Figure 1.
Schematic representation of RP gene organization. Solid boxes indicate exons. Arrowheads show the position corresponding to the translation start and stop sites. Red circles represent the position of the snoRNA genes.
Summarized in Figure 2 are various features of these genes, including the sizes of the genes and coding sequences (CDSs), the sizes of the 5′- and 3′-noncoding regions, and the size and number of exons. According to the draft sequence of the human genome, the average sizes of genes, CDSs, exons, and the 5′- and 3′-noncoding regions are 27 kb, 1340 bp, 145 bp, and 300 bp and 770 bp, respectively (International Human Genome Sequencing Consortium 2001). RP genes, in contrast, were fairly small; with introns of only 760 bp on average, most were <5 kb in length. The first exons were also small, 45 bp on average, although the others were 124 bp, which is comparable with the genome average of 145 bp. The 5′- and 3′-noncoding regions were 42 and 56 bp, respectively, which is also significantly smaller (14 times smaller in the case of the 3′-noncoding region) than the genome averages (Table 1; Fig. 2). Similar features were reported in Xenopus laevis RP genes (Amaldi et al. 1995), suggesting they are common among vertebrate RP genes.
Figure 2.
Distribution of RP gene features. Shown are size distributions of genes and CDSs (A), exons (B) and 5′-and 3′-UTRs (C), as well as the numbers of exons (D).
During our sequencing efforts, we found that many small nucleolar RNAs (snoRNAs) were encoded within the introns of the RP genes (Fig. 1). snoRNAs function as guide RNA, mostly in the modification of pre-ribosomal RNA — that is, site-specific ribose methylation and pseudouridylation through base pairing with the target RNA (Maxwell and Fournier 1995; Nicoloso et al. 1996; Smith and Steitz 1997; Huttenhofer et al. 2001). To date, 106 methylations and 91 pseudouridylations have been identified in human rRNA, and about one-half of these have been tentatively assigned to known snoRNAs. Together with the putative genes, 54 copies of 38 snoRNA genes were identified within introns of 26 RP genes, accounting for about one-third of the known snoRNAs.
Promoter Features
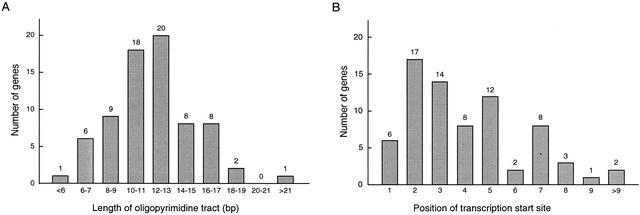
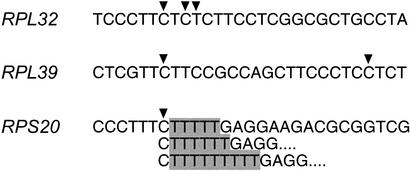
To determine the transcription start sites of the genes, we analyzed the 5′-UTR sequences of full-length cDNAs obtained using the oligo-capping method (Kato et al. 1994) and identified the start sites on the genomic sequences. As shown in Figure 3, transcription always started at a C residue within a characteristic oligopyrimidine tract that varied from 5 bp to 25 bp in length (12 bp on average). Most often, it was the second C residue that served as the transcription start site (Fig. 4). However, full-length cDNA analysis revealed that the position of the start site C residue can vary within a gene (Fig. 5); in some cases, transcription can begin at different C (or T) residues within a given oligopyrimidine tact (e.g., RPL32); transcription can also begin at different C residues within separate oligopyrimidine tracts (e.g., RPL39); finally, even when transcription always begins at the same C residue, its position may vary due to the presence of T stretches of variable length (e.g., RPS20). With respect to the last, the observed variation in the length of the T stretches does not appear to be an artifact of the oligo-capping method, as it was only present within a T stretch at the 5′ end of the gene and was also detected in cDNA prepared by a different method (Kato et al. 1994). These sequence variations will appear in the DDBJ/EMBL/GenBank DNA databases under accession numbers listed in Table 1 (5′ UTR).
Figure 3.
Characteristics of the promoter regions. Features including oligopyrimidine tracts (green), TATA boxes (pink), TATA-like sequences (yellow), and possible binding sites for Ets proteins (blue) are indicated. Arrowheads represent the position of the transcription start site. Upper- and lowercase letters denote exon and intron sequences, respectively.
Figure 4.
Features of the oligopyrimidine tract. (A) Size distribution: Min, 5 bp; Max, 25 bp; Mean, 11.6 bp. (B) Position of the transcription start site within a oligopyrimidine tract; Mean, 4.0.
Figure 5.
Variation of the transcription start sites. Three types of variations are detected: (1) transcription starts at a different C (or T) residue within an oligopyrimidine tract (e.g., RPL32); (2) transcription starts at a C residue in distinct oligopyrimidine tracts (e.g., RPL39); and (3) transcription start at the same C residue but the length of the T stretches vary (e.g., RPS20). These variations will appear in the DDBJ/EMBL/GenBank DNA database under accession nos. D28385, D28397, and D28358 (for a complete list, see Table 1). Arrowheads indicate the possible transcription start sites. T-stretches of variable length are shaded.
The average GC content in the 70 complete RP genes was 49%, that in the promoter regions (−250 to +250 bp) was 61% (Table 1), which is significantly higher than the genome-wide average of 41% (International Human Genome Sequencing Consortium 2001). The promoter region of RPS21 had the highest GC content, 73%. We found CpG islands in the promoter regions of all RP genes except RPL7 (data not shown), which is consistent with the characteristics of the housekeeping genes described by Gardiner-Garden and Frommer (1987).
To investigate the coordinated control of RP gene expression at the transcriptional level, the 5′-flanking regions were examined for sequence elements that might serve as transcription factor binding sites. We analyzed a region extending from the transcription start site up to the −400-bp position in all 73 RP genes using TFSEARCH (http://molsun1.cbrc.aist.go.jp/research/db/TFSEARCH.html). In general, the 5′-flanking region of housekeeping genes is GC rich and the promoter lacks TATA sequences. Likewise, this region of RP genes was highly GC rich, as described above; however, there were also many TATA or TATA-like sequences around at the −30-bp position. TATA box consensus sequences were seen in 7 cases, and TATA-like sequences were seen in 52 cases (Fig. 3). The presence of TATA boxes or related sequences has also been reported in other vertebrate RP genes (Hariharan et al. 1989; Nakasone et al. 1993; Higa et al. 1999), suggesting that they are a characteristic feature of RP genes.
Although none of the elements common to the 5′-flanking region was found in all 73 RP genes, possible transcription factor binding sites commonly seen included those for the GATA-binding protein family (45 cases), for CdxA (Chicken homeobox protein) (43 cases), for the Ets protein family (34 cases), and for Sp1 (20 cases). Among these, the key roles played by Ets and Sp1 in the transcription of RP genes have been reported previously (Hariharan and Perry 1989; Maeda et al. 1993; Genuario and Perry 1996; Higa et al. 1999). Possible Ets-binding sites in the upstream region (up to −50 bp) are shown in Figure 3.
Interspersed Repeats
We found 381 interspersed repeats in the sequences of 70 RP genes (partial sequences were excluded from this analysis). The sequences including 400 bp of the 5′- and 3′- flanking regions of the individual genes were searched for repeats using the RepeatMasker at the University of Washington (http://repeatmasker.genome.washington.edu). Alu elements were the most common; on average, they appeared 3.0 times in each gene, accounting for 13% of the entire sequence (211 copies in total). On the other hand, 23 Alu repeats were found in introns 2 and 3 of RPL22, which accounted for 46% of the entire gene, significantly more than the genome average of 10.6% (International Human Genome Sequencing Consortium 2001).
Comparative Analysis
The structures of human RP genes were compared with those from the fruitfly D. melanogaster, the nematode worm C. elegans, and the budding yeast S. cerevisiae, all of which are eukaryotes whose entire genome has been sequenced. Although the CDSs were comparable in both size and sequence and showed 59% to 69% homology between any two of these species, the genomic structures were varied and showed significant changes to have occurred during evolution (Tables 2 and 3). The human RP genes were 4–5 times larger than those from the other species because of increases in the size and number of introns. In contrast, the exons were somewhat smaller (Table 2). All human RP genes had at least two introns, whereas 36% of the yeast genes had no introns. Interestingly, nematode worm RP genes had more introns than fruitfly genes and a single worm gene and 13 fly genes had no introns.
Table 2.
Comparison of Gene Structures
| Human | Fly | Worm | Yeast | |
|---|---|---|---|---|
| Gene length(bp) | 4316 | 922 | 742 | 764 |
| CDS length (bp) | 541 | 524 | 520 | 498 |
| Number of exons | 5.3 | 2.5 | 3.0 | 1.6 |
| Exon length (bp) | 103 | 206 | 172 | 303 |
| Intron length (bp) | 888 | 258 | 110 | 413 |
Genomic sequences (n = 60) corresponding to the region between the translation start and stop sites were analyzed.
Table 3.
CDS Homology Between Any Two Species in Eukaryotes
| Human | Fly | Worm | Yeast | |
|---|---|---|---|---|
| Human | 100% | 69% | 64% | 61% |
| Fly | 100% | 67% | 59% | |
| Worm | 100% | 59% | ||
| Yeast | 100% |
n = 60.
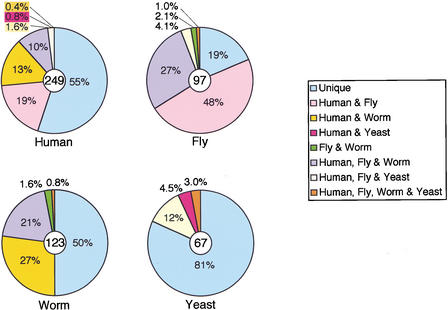
We also compared the positions of the introns among these species. In humans, we found 249 introns within the coding regions of the genes. Among them, the insertion sites of 136 were unique to the human genes, 77 were the same in humans and flies, and 60 were the same in humans and worms. Of these, 26 introns (10% of the total) were common to all three species (Fig. 6). In contrast, only 7 introns shared the same insertion sites in humans and yeast, and the position of only one, the second intron of RPL14, was conserved among all four species. About 80% of fruitfly introns were present in human RP genes, but only 30% of these introns appeared in worms. A comparison of the intron insertion sites in eukaryotic RPL8 genes is summarized in Figure 7.
Figure 6.
Comparison of intron positions among human, fruitfly (D. melanogaster), nematode worm (C. elegans), and yeast (S. cerevisiae) RP genes. Intron positions for 60 genes were compared. 'Unique' represents the ratio of introns that are specific to that particular species. The number of analyzed introns was 249 in Human, 97 in Fly, 123 in Worm, and 67 in Yeast.
Figure 7.
Comparison of the intron/exon structures and the CDS homology of RPL8. (A) Yellow and red lines indicate the corresponding positions of the human splice sites with those in other species. Yeast has two copies of the RPL8 gene, designated as Yeast-A and Yeast-B. Solid boxes represent exons, and arrowheads represent the position corresponding to the translation start and stop. (B) CDS homology of RPL8 between any two of the four species.
DISCUSSION
Evaluation of the Draft Genome Sequence
To evaluate the publicly available draft sequence of the human genome, our data set of RP gene sequences was compared with those appearing in the draft sequence. We found 32 RP genes in the finished sequence and 43 genes in the unfinished sequence, which together accounted for 94% of the RP genes; the same value as the claimed coverage of the human genome (International Human Genome Sequencing Consortium 2001). Although sequences that appeared in the finished sequence are accurate, those in the unfinished sequences still have some minor problems (as of July 10, 2001), including misassembled sequences and/or sequence gaps in five cases (data not shown). This suggests that, for the time being, the draft sequence should be carefully interpreted. Moreover, even if the sequence is accurate, we need to know the transcription start sites to determine the complete gene structures and identify the promoters. Generation of full-length cDNA sequences, as was done in the present study, should facilitate this analysis.
Promoter Structure and Gene Expression
In prokaryotes, the RP genes are organized into a small number of operons, each containing genes for up to 11 RPs under the control of a single promoter (Nomura et al. 1984). In contrast, in humans, RP genes are scattered over the genome (Kenmochi et al. 1998; Uechi et al. 2001). But, although encoded at widely dispersed genomic sites, RPs are assembled into the ribosome with stoichiometric precision; thus, clustering of RP genes into operons, as in bacteria, is not an important means of regulated coproduction of RPs in humans. The situation is similar in other eukaryotes, such as D. melanogaster, C. elegans, and S. cerevisiae. It has been argued that the translational control of RP gene expression is the most prevalent regulatory mechanism operating in higher eukaryotes (Amaldi et al. 1995; Meyuhas et al. 1996). Nevertheless, in yeast, regulation at the transcriptional level seems to dominate RP production (Warner 1999). Recent experiments using DNA array technology have shown that expression of RP mRNAs in yeast is strictly regulated in a manner responsive to changes in growth conditions (Brown and Botstein 1999). Systematic analysis of the human transcriptome also suggests that transcriptional regulation plays an essential part in the expression of this class of genes (Kawamoto et al. 2000; N. Kenmochi and K. Okubo, unpubl.). Although we found possible binding sites for various transcription factors in the 5′-flanking regions, common regulatory factors such as Rap1, which controls most yeast RP gene expression (Lascaris et al. 1999), have not yet been identified elsewhere. The only sequence element that emerged from our studies so far is the oligopyrimidine tract, which is located at the transcription start site of the genes. Searches for additional regulatory elements, combined with analyses of the expression profiles under various conditions, will need to be carried out if a better understanding of the coordinated control of RP production in humans is to be achieved.
Evolution of Introns
By comparing the positions of introns in RP genes from various species (Fig. 6), we found that about one-half of nematode worm introns (60 of 123) are represented at the same position in the corresponding human gene, but 33 of these introns are not present in fruitflies. Moreover, 26 of these introns apparently disappeared from the fruitfly genome, resulting in a reduced number of introns in the corresponding gene (e.g., RPL8; see Fig. 7). It would be interesting to know whether, during evolution, these introns were deleted from the fruitfly genome after the three species had separated, or whether they were inserted into the same positions in the human and worm genomes.
In that regard, RP genes are well suited for studying the evolution of introns. Advantages they offer include a large number of family members (79 proteins), a large number of introns per length of CDS (8 introns/kb), and highly conserved CDS sequences (e.g., human and fruitfly CDSs share 69% homology; see Table 3). The size and sequence of CDSs are very similar among eukaryotes; consequently, they are highly homologous. Furthermore, the amino acid sequences are nearly identical in mammals, and one can find a yeast homolog for all but one of the human RPs. Therefore, it is fairly easy to compare the intron positions within RP gene sequences. In fact, we identified 26 introns located at the same position in human, fruitfly, and nematode worm RP genes, although a large fraction of the introns are unique to the individual species. In addition, many snoRNAs are encoded within the RP gene introns (Maxwell and Fournier 1995), and transcriptional control elements are also found there (Chung and Perry 1989), perhaps indicative of new roles for introns in eukaryotic gene expression. RP genes thus provide a large data set useful for investigating the evolution of introns and their function.
Implications for Human Disease
Evolutionary and genetic considerations allow us to predict the roles of RP genes in human disease. Among multicellular animals, the consequences of mutations in RP genes have been studied most thoroughly in Drosophila. Here, mutations resulting in reduced expression of individual RPs yield the Minute phenotype characterized by short and thin bristles, reduced body size, diminished fertility, and recessive lethality (Schultz 1929; Lambertsson 1998). Because a full complement of RPs is required to assemble a functional ribosome, Minute cells are thought to contain fewer ribosomes and thus have less capacity for protein synthesis (Kay and Jacobs-Lorena 1987). As RPs are highly conserved between Drosophila and humans, it is likely that defects in human RPs will also result in ribosomal dysfunction leading to pathological conditions. In fact, as mentioned earlier, RPS4 and RPL6 are postulated to be candidate genes for Turner and Noonan syndromes, respectively (Fisher et al. 1990; Jamieson et al. 1994; Kenmochi et al. 2000). Moreover, RPS19 is mutated in patients with Diamond-Blackfan anemia, so far the only reported case in which RP gene mutation is associated with human disease (Draptchinskaia et al. 1999; Willig et al. 1999). Nevertheless, it remains unclear how phenotypes arise from RP defects. It would be of great interest to us to know the mechanism by which specific RP mutations disturb normal cell function and lead to abnormal phenotypes.
Meanwhile, transcriptome analysis of Ts65Dn, a segmental trisomy mouse and a model of Down syndrome, has shown that expression patterns of 14 RP genes in the brains of 30-day-old mice are significantly different from those of the normal mice, nine are underexpressed, and the others are overexpressed (Chrast et al. 2000). This implicates abnormal ribosome biogenesis in the development and maintenance of Down syndrome phenotypes. Although no RP genes are present on chromosome 21 (Uechi et al. 2001), we found that these 14 RP genes have potential recognition sites for the GA-binding protein (GABP) in the promoter region and/or the first intron (data not shown). Because the gene encoding a subunit of GABP is located in the Down syndrome locus, near the APP gene in 21q21–q22.1 (Baxter et al. 2000), and because GABP is thought to act as both an activator and repressor of RP gene transcription (Genuario and Perry 1996), this protein might be involved in the pathogenesis of Down syndrome through abnormal ribosome biogenesis.
Recent reports indicate that RPL38 is essential for early embryogenesis and skeletal development, as shown in studies using mouse skeletal mutations, Tail-short (Ts), Tail-short shionogi (Tss), and Rabo torcido (Rbt). The phenotypes of these mice are similar and are characterized by a shortened kinky tail, neural tube defects, and various skeletal abnormalities including homeotic transformation of the axial skeleton (Hustert et al. 1996; Ishijima et al. 1998; Tsukahara et al. 2000). Heterozygous mutations in the Rpl38 gene were detected in all of these mice, and a wild-type Rpl38 transgene rescued the Ts phenotype, confirming the direct involvement of RPL38 deficiency in abnormal mouse skeletal development (T. Shiroishi, pers. comm.). In addition, Volarevic et al. (2000) further implicated RP defects in abnormal phenotypes in mice when they conditionally deleted the gene encoding RPS6 in mouse liver and found that cell cycle progression was blocked in hepatocytes after partial hepatectomy.
We recently completed chromosomal mapping of the human RP genes and found certain genes that might be involved in disease by comparing their assigned positions with candidate regions for Mendelian disorders (Uechi et al. 2001). The sequence data presented here allow us now to screen for mutations in patients. Although RPS19 is the only case with mutations in patients at present, more mutations in other RP genes may be identified from such screening. Thus, together with the mapping data, our sequence data should serve as a powerful tool for studying ribosomapathy, a new class of human disease.
METHODS
Cloning
cDNA clones were isolated from the full-length cDNA libraries prepared from mRNAs of human tissues and cell lines using the DNA–RNA chimeric oligo-capping method described by Kato et al. (1994). BAC clones were isolated from the Keio BAC library by the PCR screening method (Asakawa et al. 1997) using STSs specific to the human RP genes (Kenmochi et al. 1998; Uechi et al. 2001). BAC DNAs were sheared by the shotgun method using a nebulizer (Kawasaki et al. 1997), and the 3–5-kb fragments were subcloned into XL1-Blue Escherichia coli cells using the pUC19 plasmid vector. Subclones containing the RP genes were selected by colony PCR and then sequenced.
Sequencing
Nucleotide sequences were determined by use of the shotgun sequencing method as described previously (Kawasaki et al. 1997). Plasmid DNAs from the isolated subclones were fragmented (1.1–1.3 kb) and inserted into the pHSG398 vector. After electroporation to XL1-Blue cells, DNAs from 48–96 clones were sequenced from both ends using ABI PRISM DNA sequencers. These sequencing conditions provide 2.0–9.6 times redundancy. Sequencing data were edited and assembled using the Staden or Phred/Phrap/Consed software packages (Bonfield et al. 1995; Ewing and Green 1998; Ewing et al. 1998). When necessary, sequencing primers were designed within the cDNA sequences and used for primer walking to determine ambiguous nucleotides and to fill unsequenced gaps. These sequences will appear in the DDBJ/EMBL/GenBank DNA databases under accession numbers AB055762–AB055780, AB056456, AB061820–AB061859, AB062066–AB062071 and AB070559, which are listed in Table 1.
Sequence Analysis
Intron/exon boundaries were determined by comparing the genomic sequences with the corresponding cDNA sequences. Transcription start sites were deduced from the 5′-UTR sequences of the full-length cDNAs. GC contents and sequence homologies were calculated using GENETYX version 11 (Software Development). We searched the 5′-flanking regions for possible binding sites of transcription factors using TFSEARCH at http://molsun1.cbrc.aist.go.jp/research/db/TFSEARCH.html. Regions up to the −400 bp were analyzed with a threshold score of 90. When searching for TATA-like sequences, however, the threshold score was reduced to 50.
The RP gene sequences appearing in the draft sequence of the human genome were obtained by BLASTN search at NCBI (as of July 10, 2001) using the human cDNA sequences as the query. Sequences of the nematode worm C. elegans were obtained by BLASTP search from the C. elegans Genome Project at the Sanger Center (http://www.sanger.ac.uk/Projects/C_elegans/), and sequences of the fruitfly D. melanogaster were from the Berkeley Drosophila Genome Project (BDGP, http://www.fruitfly.org/). Sequences of the yeast S. cerevisiae were obtained by keyword search from SGD at Stanford University (http://genome-www.stanford.edu/Saccharomyces/).
WEB SITE REFERENCES
http://genome-www.stanford.edu/Saccharomyces/; sequences of the yeast S. cerevisiae were obtained by keyword search from SGD at Stanford University.
http://molsun1.cbrc.aist.go.jp/research/db/TFSEARCH.html; TFSEARCH.
http://repeatmasker.genome.washington.edu; Repeat Masker.
http://www.fruitfly.org/; the Berkeley Drosophila Genome Project (BDGP).
http://www.sanger.ac.uk/Projects/C_elegans/; the C. elegans Genome Project at the Sanger Center.
Acknowledgments
We thank Dr. Atsushi Shimizu and the genome sequencing team at Keio University School of Medicine. This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and Fund for “Research for the Future” Program from the Japan Society for the Promotion of Science.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kenmochi@post.miyazaki-med.ac.jp; FAX 81-985-85-1514.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.214202.
REFERENCES
- Allanson JE. Noonan syndrome. J Med Genet. 1987;24:9–13. doi: 10.1136/jmg.24.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaldi F, Camacho-Vanegas O, Cardinall B, Cecconi F, Crosio C, Loreni F, Mariottini P, Pellizzoni L, Pierandrei-Amaldi P. Structure and expression of ribosomal protein genes in Xenopus laevis. Biochem Cell Biol. 1995;73:969–977. doi: 10.1139/o95-104. [DOI] [PubMed] [Google Scholar]
- Asakawa S, Abe I, Kudoh Y, Kishi N, Wang Y, Kubota R, Kudoh J, Kawasaki K, Minoshima S, Shimizu N. Human BAC library: Construction and rapid screening. Gene. 1997;191:69–79. doi: 10.1016/s0378-1119(97)00044-9. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Baxter LL, Moran TH, Richtsmeier JT, Troncoso J, Reeves RH. Discovery and genetic localization of Down syndrome cerebellar phenotypes using the Ts65Dn mouse. Hum Mol Genet. 2000;9:195–202. doi: 10.1093/hmg/9.2.195. [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Smith KF, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33s–37s. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- Chrast R, Scott HS, Papasavvas MP, Rossier C, Antonarakis ES, Barras C, Davisson MT, Schmidt C, Estivill X, Dierssen M, et al. The mouse brain transcriptome by SAGE: Differences in gene expression between P30 brains of the partial trisomy 16 mouse model of Down syndrome (Ts65Dn) and normals. Genome Res. 2000;10:2006–2021. doi: 10.1101/gr.10.12.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Perry RP. Importance of introns for expression of mouse ribosomal protein gene rpL32. Mol Cell Biol. 1989;9:2075–2082. doi: 10.1128/mcb.9.5.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond LK, Wang WC, Alter BP. Congenital hypoplastic anemia. Adv Pediatr. 1976;22:349–378. [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Dudov KP, Perry RP. The gene family encoding the mouse ribosomal protein L32 contains a uniquely expressed intron-containing gene and an unmutated processed gene. Cell. 1984;37:457–468. doi: 10.1016/0092-8674(84)90376-3. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fisher EM, Beer-Romero P, Brown LG, Ridley A, McNeil JA, Lawrence JB, Willard HF, Bieber FR, Page DC. Homologous ribosomal protein genes on the human X and Y chromosomes: Escape from X inactivation and possible implications for Turner syndrome. Cell. 1990;63:1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Genuario RR, Perry RP. The GA-binding protein can serve as both an activator and repressor of ribosomal protein gene transcription. J Biol Chem. 1996;271:4388–4395. doi: 10.1074/jbc.271.8.4388. [DOI] [PubMed] [Google Scholar]
- Halperin DS, Estrov Z, Freedman MH. Diamond-Blackfan anemia: Promotion of marrow erythropoiesis in vitro by recombinant interleukin-3. Blood. 1989;73:1168–1174. [PubMed] [Google Scholar]
- Hariharan N, Perry RP. A characterization of the elements comprising the promoter of the mouse ribosomal protein gene RPS16. Nucleic Acids Res. 1989;17:5323–5337. doi: 10.1093/nar/17.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan N, Kelley DE, Perry RP. Equipotent mouse ribosomal protein promoters have a similar architecture that includes internal sequence elements. Genes & Dev. 1989;3:1789–1800. doi: 10.1101/gad.3.11.1789. [DOI] [PubMed] [Google Scholar]
- Higa S, Yoshihama M, Tanaka T, Kenmochi N. Gene organization and sequence of the region containing the ribosomal protein genes RPL13A and RPS11 in the human genome and conserved features in the mouse genome. Gene. 1999;240:371–377. doi: 10.1016/s0378-1119(99)00429-1. [DOI] [PubMed] [Google Scholar]
- Hustert E, Scherer G, Olowson M, Guenet JL, Balling R. Rbt (Rabo torcido), a new mouse skeletal mutation involved in anteroposterior patterning of the axial skeleton, maps close to the Ts (tail-short) locus and distal to the Sox9 locus on chromosome 11. Mamm Genome. 1996;7:881–885. doi: 10.1007/s003359900261. [DOI] [PubMed] [Google Scholar]
- Huttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: An experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Ishijima J, Yasui H, Morishima M, Shiroishi T. Dominant lethality of the mouse skeletal mutation tail-short (Ts) is determined by the Ts allele from mating partners. Genomics. 1998;49:341–350. doi: 10.1006/geno.1998.5277. [DOI] [PubMed] [Google Scholar]
- Jamieson CR, van der Burgt I, Brady AF, van Reen M, Elsawi MM, Hol F, Jeffery S, Patton MA, Mariman E. Mapping a gene for Noonan syndrome to the long arm of chromosome 12. Nat Genet. 1994;8:357–360. doi: 10.1038/ng1294-357. [DOI] [PubMed] [Google Scholar]
- Kato S, Sekine S, Oh SW, Kim NS, Umezawa Y, Abe N, Yokoyama-Kobayashi M, Aoki T. Construction of a human full-length cDNA bank. Gene. 1994;150:243–250. doi: 10.1016/0378-1119(94)90433-2. [DOI] [PubMed] [Google Scholar]
- Kawamoto S, Yoshii J, Mizuno K, Ito K, Miyamoto Y, Ohnishi T, Matoba R, Hori N, Matsumoto Y, Okumura T, et al. BodyMap: A collection of 3′ ESTs for analysis of human gene expression information. Genome Res. 2000;10:1817–1827. doi: 10.1101/gr.151500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits JL, Wang J, Shimizu N. One-megabase sequence analysis of the human immunoglobulin λ gene locus. Genome Res. 1997;7:250–261. doi: 10.1101/gr.7.3.250. [DOI] [PubMed] [Google Scholar]
- Kay MA, Jacobs-Lorena M. Developmental genetics of ribosome synthesis in Drosophila. Trends Genet. 1987;3:347–351. [Google Scholar]
- Kenmochi N, Maeda N, Tanaka T. The structure and complete sequence of the gene encoding chicken ribosomal protein L5. Gene. 1992;119:215–219. doi: 10.1016/0378-1119(92)90274-s. [DOI] [PubMed] [Google Scholar]
- Kenmochi N, Kawaguchi T, Rozen S, Davis E, Goodman N, Hudson TJ, Tanaka T, Page DC. A map of 75 human ribosomal protein genes. Genome Res. 1998;8:509–523. doi: 10.1101/gr.8.5.509. [DOI] [PubMed] [Google Scholar]
- Kenmochi N, Yoshihama M, Higa S, Tanaka T. The human ribosomal protein L6 gene in a critical region for Noonan syndrome. J Hum Genet. 2000;45:290–293. doi: 10.1007/s100380070018. [DOI] [PubMed] [Google Scholar]
- Kongsuwan K, Yu Q, Vincent A, Frisardi MC, Rosbash M, Lengyel JA, Merriam J. A Drosophila Minute gene encodes a ribosomal protein. Nature. 1985;317:555–558. doi: 10.1038/317555a0. [DOI] [PubMed] [Google Scholar]
- Kuzumaki T, Tanaka T, Ishikawa K, Ogata K. Rat ribosomal protein L35a multigene family: Molecular structure and characterization of three L35a-related pseudogenes. Biochim Biophys Acta. 1987;909:99–106. doi: 10.1016/0167-4781(87)90031-5. [DOI] [PubMed] [Google Scholar]
- Lambertsson A. The Minute genes in Drosophila and their molecular functions. Adv Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- Lascaris RF, Mager WH, Planta RJ. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics. 1999;15:267–277. doi: 10.1093/bioinformatics/15.4.267. [DOI] [PubMed] [Google Scholar]
- Maeda N, Kenmochi N, Tanaka T. The complete nucleotide sequence of chicken ribosomal protein L7a gene and the multiple factor binding sites in its 5′-flanking region. Biochimie. 1993;75:785–790. doi: 10.1016/0300-9084(93)90128-f. [DOI] [PubMed] [Google Scholar]
- Maidak BL, Cole JR, Lilburn TG, Parker CT, Jr, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM. The RDP-II (Ribosomal Database Project) Nucleic Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Meyuhas O, Avni D, Shama S. Translational control of ribosomal protein mRNAs in eukaryotes. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 363–388. [Google Scholar]
- Nakasone K, Kenmochi N, Toku S, Tanaka T. The structure of the gene encoding chicken ribosomal protein L30. Biochim Biophys Acta. 1993;1174:75–78. doi: 10.1016/0167-4781(93)90094-t. [DOI] [PubMed] [Google Scholar]
- Nicoloso M, Qu LH, Michot B, Bachellerie JP. Intron-encoded, antisense small nucleolar RNAs: The characterization of nine novel species points to their direct role as guides for the 2‘-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Noonan JA. Hypertelorism with Turner phenotype. A new syndrome with associated congenital heart disease. Am J Dis Child. 1968;116:373–380. doi: 10.1001/archpedi.1968.02100020377005. [DOI] [PubMed] [Google Scholar]
- Rhoads DD, Dixit A, Roufa DJ. Primary structure of human ribosomal protein S14 and the gene that encodes it. Mol Cell Biol. 1986;6:2774–2783. doi: 10.1128/mcb.6.8.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Schultz J. The Minute reaction in the development of Drosophila melanogaster. Genetics. 1929;14:366–419. doi: 10.1093/genetics/14.4.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Sno storm in the nucleolus: New roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Toku S, Tanaka T. A characterization of transcriptional regulatory elements in chicken ribosomal protein L37a gene. Eur J Biochem. 1996;238:136–142. doi: 10.1111/j.1432-1033.1996.0136q.x. [DOI] [PubMed] [Google Scholar]
- Tsukahara K, Hirasawa T, Makino S. Tss (Tail-short Shionogi), a new short tail mutation found in the BALB/cMs strain, maps quite closely to the Tail-short (Ts) locus on mouse chromosome 11. Exp Anim. 2000;49:131–135. doi: 10.1538/expanim.49.131. [DOI] [PubMed] [Google Scholar]
- Uechi T, Tanaka T, Kenmochi N. A complete map of the human ribosomal protein genes: Assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics. 2001;72:223–230. doi: 10.1006/geno.2000.6470. [DOI] [PubMed] [Google Scholar]
- Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- Wagner M, Perry RP. Characterization of the multigene family encoding the mouse S16 ribosomal protein: Strategy for distinguishing an expressed gene from its processed pseudogene counterparts by an analysis of total genomic DNA. Mol Cell Biol. 1985;5:3560–3576. doi: 10.1128/mcb.5.12.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Zinn AR, Page DC, Nishimoto T. Functional equivalence of human X- and Y-encoded isoforms of ribosomal protein S4 consistent with a role in Turner syndrome. Nat Genet. 1993;4:268–271. doi: 10.1038/ng0793-268. [DOI] [PubMed] [Google Scholar]
- Willig TN, Draptchinskaia N, Dianzani I, Ball S, Niemeyer C, Ramenghi U, Orfali K, Gustavsson P, Garelli E, Brusco A, et al. Mutations in ribosomal protein S19 gene and diamond blackfan anemia: Wide variations in phenotypic expression. Blood. 1999;94:4294–4306. [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wool IG. The structure and function of eukaryotic ribosomes. Annu Rev Biochem. 1979;48:719–754. doi: 10.1146/annurev.bi.48.070179.003443. [DOI] [PubMed] [Google Scholar]
- Wool IG, Chan YL, Glük A. Mammalian ribosomes: The structure and the evolution of the proteins. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 685–732. [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zinn AR, Alagappan RK, Brown LG, Wool I, Page DC. Structure and function of ribosomal protein S4 genes on the human and mouse sex chromosomes. Mol Cell Biol. 1994;14:2485–2492. doi: 10.1128/mcb.14.4.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]