Abstract
Multiple genome-wide scans involving sib-pairs or limited pedigrees have been extensively used for a wide number of complex genetic conditions. Comparing data from two or more scans, as well as combining data, require an understanding of the sources of genotyping errors and data discrepancies. We have conducted two genome-wide scans for age-related maculopathy using the Center for Inherited Disease Research (CIDR) and the Mammalian Genotyping Service (MGS). Thirty individuals were typed in common, in order to allow for the alignment of alleles and comparison of the data sets. The analysis of these 8914 genotypes distributed over 321 markers in common demonstrated excellent agreement between these two laboratories, which have low rates of internal errors. Under the assumption that within each genotype, the smaller MGS allele should correspond to the smaller CIDR allele, the alleles align well between the two centers, with only a small fraction (less than 0.65%) of the aligned alleles showing large differences in sizes. However, since called allele sizes are integer “labels” which may not directly reflect the true underlying allele sizes, it is important to carefully prepare in advance if one wishes to merge data from different laboratories. In particular, it would not suffice to attempt to align alleles by typing only one or two controls in common. Fortunately, for the purposes of linkage analysis, one can avoid merging difficulties by simply carrying out linkage analyses using laboratory-specific allele labels and allele frequencies for each laboratory-specific subset of the data.
As genotyping has become less expensive, it has become common to attempt to map disease genes via genome-wide scans (Weeks and Lathrop 1995). In fact, large-scale genotyping centers have been established to facilitate this process throughout the world. In the United States, the National Institutes of Health (NIH) have funded two prominent genotyping centers: the Mammalian Genotyping Service (MGS), led by Dr. James Weber and funded by the National Heart, Lung, and Blood Institute; and the Center for Inherited Disease Research (CIDR), led by Dr. David Valle and funded by 11 NIH Institutes. At these Centers, millions of genotypes are being generated per year: In 1999, MGS generated 5.54 million genotypes and CIDR generated approximately 1.2 million genotypes. These two centers, which both use multiallelic short tandem repeat (STR) markers, have extensive experience, and report very small internal error rates. MGS reports an average genotype error rate of 0.7%, based on blindly typing Centre d'Étude du Polymorphisme Humain (CEPH) family DNA samples in duplicate or triplicate on different gels (Weber and Broman 2001); note that the allele error rate is approximately 60% of the genotype error rate (as usually one of the two alleles is correct for most incorrect genotypes). CIDR reports an error rate of 0.18% based on more than 2 million genotypes; these error rates are based on four blind duplicates of investigator-supplied samples per 96-lane gel. These impressively low error rates indicate that allele calling is highly consistent within each of these genotyping centers. While it is implicitly recognized that marker genotypes are, like most experimentally derived data, an excellent but imperfect reflection of the underlying “true” genotypes, most published linkage studies fail to discuss genotype error rates in any detail, despite the fact that even a small genotyping error rate can have negative consequences, increasing the estimated recombination fraction (Terwilliger et al. 1990) and reducing the evidence for linkage (Göring and Terwilliger 2000; Abecasis et al. 2001). For example, Douglas et al (2000) found that a genotype error rate of 1% can cause a loss of 53%–58% of the linkage information for a trait locus of small effect; an error rate of 0.5% can cause a loss of 28%–30% of the linkage information. Similarly, genotype error rates as small as 3% can have serious effects on measures of linkage disequilibrium (Akey et al. 2001; Gordon et al. 2001).
As part of our large linkage study of age-related macular degeneration, we had our first set of 225 families genotyped at MGS (Weeks et al. 2000), and our second set of 196 families genotyped at CIDR, for much the same set of markers (Weeks et al. 2001). When we started the study, CIDR did not exist, and MGS was the only available center that was performing high-quality genome-wide scans. We were very fortunate that MGS accepted our application at a time when the demand for genome-wide scans allowed them to conduct studies that were not directly related to the mission of the National Heart Lung and Blood Institute. Once CIDR was established, we were strongly encouraged to use CIDR as our genotyping center in part because the National Eye Institute (NEI) (which funded our study) had announced its intention of being one of the supporting institutes for CIDR. Using the 30 samples from 15 families that had been genotyped by both centers, we hoped to be able to align the marker alleles and analyze both sets of families in one analysis. We report here on our comparisons of these genotypes and the implications of our results for combining data across genotyping centers.
RESULTS
Our 30 samples were genotyped at 321 markers in common. Due to varying genotype success rates, the number of samples genotyped in common ranged from 17 to 30, with an average of 27.8 genotypes in common. The rate of missing genotypes differed between the two centers, with 6.0% of the genotypes missing in the CIDR data and 1.5% missing in the MGS data. In total, there were 8914 genotypes in common (Table 1).
Table 1.
Linear Regression Results
| di | tri | tetra | Total | |
|---|---|---|---|---|
| Markers | 34 | 38 | 249 | 321 |
| Genotypes | 937 | 1023 | 6954 | 8914 |
| Alleles | 1874 | 2046 | 13,908 | 17,828 |
| Large residuals | 34 | 8 | 74 | 116 |
| Fraction of alleles | 0.0181 | 0.0039 | 0.0053 | 0.0065 |
| Markers involved | 10 | 3 | 32 | 45 |
| Fraction of markers | 0.29 | 0.08 | 0.13 | 0.14 |
| Markers with a non–one-to-one mapping | 10 | 3 | 46 | 59 |
| Alleles with a non–one-to-one mappinga | 29 | 2 | 178 | |
| Percent involving adjacent alleles | 82.8 | 100.0 | 50.0 |
Total numbers of alleles in common, markers in common, number of alleles with extreme residuals (see text), and number of markers that had alleles with extreme residuals, broken down by base-pair repeat type of the marker.
Here we count the number of times an allele in one data set matched with more than one other allele in the other data set.
Using PedCheck (O'Connell and Weeks 1998) to carry out Mendelian error checks, we found only 18 Mendelian inconsistencies (10 from CIDR, eight from MGS). This low number of inconsistencies was expected because we are studying small families due to the late age of onset of age-related macular degeneration, so it is difficult or impossible to detect Mendelian inconsistencies for many of our families. Additionally, the genotyping centers themselves did some screening for Mendelian errors prior to sending us the genotypes.
We examined how many genotypes were scored as homozygous by CIDR but heterozygous by MGS—this occurred only 27 out of 8914 times. Similarly, there were only 29 genotypes that were scored as heterozygous by CIDR but homozygous by MGS. We also examined how often a one-to-one mapping could be established between the alleles under the assumption that, within each genotype, the smaller MGS allele should correspond to the smaller CIDR allele. That is, a one-to-one mapping holds if each allele as called by CIDR is consistently matched with only one other allele as called by MGS. For the majority of the markers, 262 of 321 (81.6%), a one-to-one mapping held; however, no one-to-one mapping existed for 59 (18.4%) of the markers.
We then examined more closely the 59 markers where alleles did not align one-to-one. Figure 1 displays, for the first 15 of these markers, plots of the CIDR alleles versus the MGS alleles under the assumption that, within each genotype, the smaller MGS allele should correspond to the smaller CIDR allele. Note that, even in this small subset of problematic markers, we see that the majority of the CIDR–MGS allele pairs do follow a one-to-one map, forming a straight diagonal line. Some of the discrepancies are due to large differences in called allele sizes; for example, for D1S1612, the point off the diagonal line represents a CIDR–MGS allele pair where CIDR called the allele as a ‘97’ and MGS called it as a ‘122.’ Other differences appear to be due to small differences in called allele sizes; for example, at D1S1679, we have a ‘173’-‘172’ CIDR–MGS allele pair and a ‘174’-‘172’ pair. Similar 1-bp disagreements are seen at D1S2141 and D3S2406. One should also note that each point in these graphs may represent several CIDR–MGS allele pairs; for example, the two filled points off the diagonal at D1S1588 actually represent a total of five CIDR–MGS allele pairs.
Figure 1.
Plots of the Center for Inherited Disease Research (CIDR) alleles versus the Mammalian Genotyping Service (MGS) alleles, for the first 15 non-one-to-one markers. Here we assume that within each genotype, the smaller MGS allele should pair with the smaller CIDR allele. The diagonal line is the linear regression line, and off-diagonal points are shaded if they had large residuals (as defined in the text).
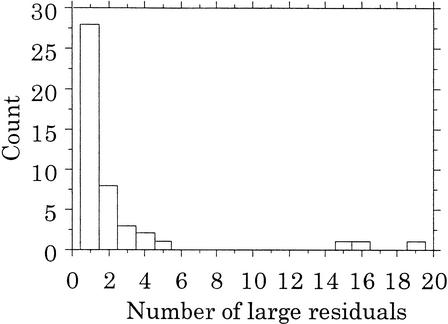
In order to generate an objective count of the number of off-diagonal points, we applied a linear regression approach across all 321 markers and identified 116 CIDR–MGS allele pairs with large residuals distributed across 45 markers (Table 1). Note that 14 of the 59 non-one-to-one markers do not have large residuals. These results indicate that the alleles align quite well between the two centers, with only a small fraction (less than 0.65%) of the aligned alleles showing large differences in sizes. Figure 2 indicates that 50 of these 116 discrepant CIDR–MGS allele pairs are due to just three markers; the other 42 markers have five or less discrepant CIDR–MGS pairs, and 28 of these markers have only one discrepant CIDR–MGS pair.
Figure 2.
Histogram of the number of large residuals per marker among 45 non-one-to-one markers. The height of each bar indicates the number of markers with the given number of large residuals (e.g., the short bar at the right indicates that there is one marker with 19 large residuals).
We then counted how many of the non-one-to-one matches, where one allele from one center aligns with more than one allele from the other center, involve adjacent alleles (from the other center). As Table 1 indicates, the majority (82.8%) of non-one-to-one mappings for the dinucleotide repeat markers are most likely due to differing decisions about placing an allele in adjacent bins. However, while it is intuitive that dinucleotide repeat markers would have a higher percentage of binning discrepancies, the tetranucleotide repeat markers still have a relatively large 50% binning discrepancy rate.
The majority of our families have only the offspring genotyped, as we were studying a late-onset disease and parents were not available for genotyping. Thus, in our data, it is difficult to determine identity by descent (IBD) sharing between siblings. Therefore, we instead examined how identity by state (IBS) sharing might change when the different sets of two genotypes are used. Out of 5247 relative pair-marker combinations (almost all sib pairs), we observed only 37 changes in IBS sharing. Thus, sharing status within pedigrees barely changes at all (0.71%) between the two sets of genotypes.
Linkage analyses of data sets that are not fully genotyped will usually be sensitive to the marker allele frequencies used in the analyses. Thus, it is of interest to compare the frequencies of the corresponding alleles in the two data sets to see how closely matched they are. For the 262 markers that have a one-to-one mapping, we examined the Euclidean distance (which can range from 0 to 1) between the vectors of matching allele frequencies, and the maximum absolute allele frequency difference. We found that 85.5% of the 262 markers have a maximum allele frequency difference of less than 0.050. Thus, when there is a one-to-one mapping, the allele frequency estimates usually agree fairly well between the two sets of genotypes. Similarly, the maximum distance seen was 0.131, with a maximum allele frequency difference of 0.111. Even at this marker, most of the allele frequencies agreed quite well. For the purposes of linkage analysis, it is reassuring that we have no occurrence where an allele is scored as very common in one data set and very rare in the other.
DISCUSSION
Multiple genome-wide scans involving sib-pairs or limited pedigrees have been extensively used for a wide number of complex genetic conditions. Comparing data from two or more scans, as well as combining data, require an understanding of the sources of genotyping errors and data discrepancies. These issues can only be partially avoided by having one's genotype scans done in the same center, as rapidly changing technologies can result in the generation of different allele labels in different scans, even from the same center. Additionally, the genotyping centers do not typically offer follow-up genotyping services, so, during subsequent fine-mapping studies, one must often deal with the issue of combining locally generated genotypes with genotypes generated at the genotyping center. Thus, the issues discussed here are of wide relevance to the research community.
Our results indicate that, as might be expected from the high quality of the intralaboratory genotype calls, interlaboratory genotype calls agree quite well between CIDR and MGS. However, we note that it would not suffice to attempt to align alleles by typing only one or two controls in common. Our results indicate that the size difference between alleles within a given genotype differ quite a bit across laboratories. This is expected, not only because the allele calls are based on gel mobility, but also because the genotyping was done on different instruments using different analysis software at the two centers. Similarly, Ghosh et al. (1997) found systematic differences in allele sizing of up to nearly two bp on a 373 ABI sequencer as compared to a 377 sequencer. However, even if large alleles migrate more slowly in one of the laboratories than in the other, we have shown that it is possible to establish a one-to-one mapping between the alleles across the two centers for the majority (81.6%) of the markers examined here. Note that our criterion for deciding whether a marker has a one-to-one mapping is rather strict, as all but a single pair of alleles of a given marker may match one-to-one, and that marker would still be scored as ‘non-one-to-one’ (see Fig. 2).
We would emphasize that allele calling for STRs is based purely on gel mobility, not on actual sequences of the amplified fragment (as discussed by Weber and Broman 2001). When one uses mobility of an amplified fragment relative to a ladder of standard fragments, then differences in fragment mobilities that are dependent upon sequence and not on absolute numbers of nucleotides can dramatically confound results. If allele labels represent true allele sizes, one could get the alleles to agree by computing the best bp shift which, when applied to both CIDR alleles in each genotype at once, minimizes the number of differences in genotypes between CIDR and MGS. However, after applying the best possible bp shift, 16.8% (1495/8914) of the genotypes differ between MGS and CIDR. Furthermore, even if one were to apply different bp shifts to different genotypes within a given marker (e.g., used a shift of two bp for both alleles within some genotypes and a shift of one bp for both alleles within the rest of the genotypes), 13.7% (1220/8914) of the genotypes would disagree; therefore, this approach is not recommended.
A few previous studies examined rates of errors when genotypes were done in duplicate. Brzustowicz et al. (1993) examined 21 individuals within the CEPH collection who were members of more than one CEPH family and so were genotyped more than once; they found an error rate of about 3%. Ghosh et al. (1997) genotyped 50 individuals in duplicate for 105 markers, and, using a variety of quality control measures, obtained an allele-specific error rate of 0.08%. More recently, Ewen et al. (2000) found, based on duplicate genotyping, a discordance rate of 0.76% when testing within gels and discordance rates of 2.36% (with a fine mapping set of nonoptimized markers) and 0.16% (with a commercially optimized set of markers) when testing across gels. These studies examined the rate of discordance when the markers are redone within the same laboratory. Here, it is encouraging and gratifying that we observed similarly low discordance rates between two different genotyping laboratories.
As discussed above, the majority of the disagreements appear to be due to different binning decisions into adjacent bins. For example, D1S3728 was scored by MGS as having alleles that all followed perfectly the expected 4-bp ladder, while the CIDR allele size histograms clearly show two groups of ‘noninteger’ alleles that do not fit into the expected 4-bp ladder. Similarly, MGS and CIDR fail to agree on several genotypes for D12S391, but this is not surprising since both MGS and CIDR scored this marker as having some noninteger alleles that differed by only one bp from their more common counterpart in the expected allele ladder. Clearly, differing decisions about ‘lumping’ or ‘splitting’ could lead to many of the disagreements we observed. Indeed, it is well recognized that binning of alleles is a difficult and challenging problem, with binning errors accounting for 21% to 40% of allele calling errors in a recent study (Ewen et al. 2000). Several groups have proposed improved protocols for binning alleles (Ghosh et al. 1997; Idury and Cardon 1997; Li et al. 2001).
Those involved in large, multicenter consortiums have recognized that it can be challenging to obtain consistent scoring of alleles across centers. Solutions to this problem have included using arbitrary allele frequencies for each marker (Easton et al. 1993), matching and renaming alleles by frequency across studies (Dorr et al. 1997), and using center-specific marker allele frequencies (Xu 2000). We initially thought that we could simply align the genotypes by a simple bp shift, but our results indicate that this was too simplistic. Similarly, many laboratories use a few CEPH genotypes to align genotyping calls across gels. However, our results indicate that while this might work fine for intralaboratory alignment, it is unlikely to work well for interlaboratory alignments. It seems clear that the best way to combine marker data from different laboratories is not to try to adjust allele labels to match, but rather to use center-specific marker alleles with center-specific allele frequencies in the statistical analyses. Unfortunately, this approach has been a bit cumbersome to implement in practice. For example, Daly et al. (1997) accomplished this by creating duplicate fictitious markers located on top of each other, and, at each marker, specifying everyone as untyped except for pedigrees from a particular population. Fortunately, it is fairly straightforward to modify existing software to allow for center-specific marker alleles, and already the authors of VITESSE (O'Connell and Weeks 1995) and Allegro (Gudbjartsson et al. 1999, 2000) have made useful improvements in this regard.
For a linkage study to be successful, one needs consistent allele identity within families, and an ability to assign proper allele frequencies. Thus, one could use different sets of alleles (and allele frequencies) in different subsets of families. However, although maintaining allele identity across all families is not crucial for the success of a linkage analysis project, it is vital if one wishes to test for association. Thus, based on the results presented here, we would strongly advocate that, when exploring a narrow region of interest for association using multiallelic STR markers, all genotyping be carried out in a single laboratory, preferably on the same instrument. The forensic community uses marker-specific allelic ladders to improve allele calls (Puers et al. 1993; Griffiths et al. 1998), but it is not clear if such a costly approach is justified based on the low levels of error noted here. Even within an allelic ladder, one may not necessarily obtain correct relative allele sizes (Haberl and Tautz 1999).
One could probably reduce the genotyping error to nearly zero by duplicate typing, but that would cut in half the number of projects that could be genotyped. Instead, we would recommend explicitly allowing for genotype error in the analyses. This can be done by altering the penetrances at each marker to permit the observed genotype to correspond to several alternative genotypes (Ott 1985; Lincoln and Lander 1992), or via error filtration with multiple pairwise analyses (Shields et al. 1991), or by using hypercomplex-valued recombination fractions (Göring and Terwilliger 2000). Alternatively, one can attempt to identify those genotypes with a high posterior probability of being in error, given all the marker data (Douglas et al. 2000; Papp et al. 2000; Sobel and Lange 2000), as we have done in our recent genome-wide scan for genes influencing age-related macular degeneration (Weeks et al. 2000).
METHODS
Family Selection and Genotyping
As part of our large linkage study of age-related macular degeneration, we had our first set of 225 families genotyped at MGS (Weeks et al. 2000), and our second set of 196 families genotyped at CIDR. The DNA was extracted from leukocytes using a salting-out procedure (Miller et al. 1988), diluted to 40ng/μL and shipped out to the genotyping services. Although both centers utilized much of the same set of markers, different experimental methods were used by the two genotyping centers: the markers at MGS were genotyped on a custom-built scanning fluorescence detector, which is wider and shorter (14 cm long) than the ABI 377XL sequencers (36 cm long) used at CIDR. Genotyping controls were used by each center: MGS used CEPH samples in duplicate to align gels, while CIDR used four samples supplied by us as quality controls along with CEPH samples to align gels. Regarding the families that we sent to CIDR, we included 30 samples from 15 families (12 of these have more than one member genotyped) who had been previously genotyped by MGS. These individuals were selected because they were members of families that had been expanded since the first genome-wide scan by MGS.
Linear Regression
To generate an objective count of the number of off-diagonal points, we applied a linear regression approach across all 321 markers. Under the assumption that, within each genotype, the smaller MGS allele should correspond to the smaller CIDR allele, we used linear regression to derive the best straight line through the CIDR versus MGS plot, and then looked for points with large deviations from that line. A point was counted if the point's residual differed from the median of the residuals by more than a constant; we defined this constant as the base-pair repeat size of the marker minus one. This should identify, in an objective manner, those disagreements that generate points that deviate from the diagonal line; however, it will not count discrepancies involving small differences in called allele sizes, such as the discrepancy at D1S1679 (Fig. 1).
SUPPLEMENTARY INFORMATION
Additional supplementary information about our results are at our web page at http://watson.hgen.pitt.edu/compare/ and Supplementary Table 1 is available online at http://www.genome.org.
WEB SITE REFERENCES
http://research.marshfieldclinic.org/genetics/, the NHLBI Mammalian Genotyping Service.
http://watson.hgen.pitt.edu/compare/, authors' web page.
Acknowledgments
Our first genome-wide scan was genotyped by the NHLBI Mammalian Genotyping Service (http://research.marshfieldclinic.org/genetics/), which is led by Dr. James Weber. The Mammalian Genotyping Service is funded by NHLBI under contract number N01-HV-48141 to the Marshfield Medical Research Foundation. Genotyping services for our second genome-wide scan were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, Contract Number N01-HG-65403. Our work was supported by NEI grant EY09859 (MBG) and Research to Prevent Blindness, Inc., NY, NY. Clinical support was provided by The Eye & Ear Foundation of Pittsburgh, Pittsburgh, PA, Smith Kettlewell Research Foundation, San Francisco, CA (T. Otis Paul), and V. Kann Rasmussen Foundation (supporting the efforts of Dr. Philip Rosenfeld). A full listing of the members of our clinical consortium and contributing clinical practices can be found in our recent paper describing the linkage results (Weeks et al. 2001). We are indebted to the invaluable discussions and suggestions made by James Weber, Francis Collins, Elizabeth Pugh, Kim Doheny, Michael Boehnke, and Michael Barmada during the preparation of this manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Present address: Dept. of Human Genetics, University of Pittsburgh, Crabtree Hall, Room A302A, 130 DeSoto St., Pittsburgh, PA 15261, USA.
E-MAIL dweeks@watson.hgen.pitt.edu; FAX (412) 624-3020.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.211502.
References
- Abecasis GR, Cherny SS, Cardon LR. The impact of genotyping error on linkage and association analysis of quantitative traits. Eur J Hum Genet. 2001;9:130–134. doi: 10.1038/sj.ejhg.5200594. [DOI] [PubMed] [Google Scholar]
- Akey JM, Zhang K, Xiong M, Doris P, Jin L. The effect that genotyping errors have on the robustness of common linkage-disequilibrium measures. Am J Hum Genet. 2001;68:1447–1456. doi: 10.1086/320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzustowicz LM, Merette C, Xie X, Townsend L, Gilliam TC, Ott J. Molecular and statistical approaches to the detection and correction of errors in genotype databases. Am J Hum Genet. 1993;53:1137–1145. [PMC free article] [PubMed] [Google Scholar]
- Daly M, Kirby A, Kruglyak L. Do multiple data sets provide support for a bipolar illness susceptibility locus on chromosome 18? Genet Epidemiol. 1997;14:599–604. doi: 10.1002/(SICI)1098-2272(1997)14:6<599::AID-GEPI8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Dorr DA, Rice JP, Armstrong C, Reich T, Blehar M. A meta-analysis of chromosome 18 linkage data for bipolar illness. Genet Epidemiol. 1997;14:617–622. doi: 10.1002/(SICI)1098-2272(1997)14:6<617::AID-GEPI11>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Douglas JA, Boehnke M, Lange K. A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data. Am J Hum Genet. 2000;66:1287–1297. doi: 10.1086/302861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Bishop DT, Ford D, Crockford GP. Genetic linkage analysis in familial breast and ovarian cancer: Results from 214 families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- Ewen KR, Bahlo M, Treloar SA, Levinson DF, Mowry B, Barlow JW, Foote SJ. Identification and analysis of error types in high-throughput genotyping. Am J Hum Genet. 2000;67:727–736. doi: 10.1086/303048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Karanjawala ZE, Hauser ER, Ally D, Knapp JI, Rayman JB, Musick A, Tannenbaum J, Te C, Shapiro S, et al. Methods for precise sizing, automated binning of alleles, and reduction of error rates in large-scale genotyping using fluorescently labeled dinucleotide markers. FUSION (Finland-U.S. Investigation of NIDDM Genetics) Study Group. Genome Res. 1997;7:165–178. doi: 10.1101/gr.7.2.165. [DOI] [PubMed] [Google Scholar]
- Gordon D, Heath SC, Liu X, Ott J. A transmission/disequilibrium test that allows for genotyping errors in the analysis of single-nucleotide polymorphism data. Am J Hum Genet. 2001;69:371–380. doi: 10.1086/321981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD. Linkage analysis in the presence of errors II: Marker-locus genotyping errors modeled with hypercomplex recombination fractions [published erratum appears in Am. J. Hum. Genet. 2000 Apr;66(4):1472] Am J Hum Genet. 2000;66:1107–1118. doi: 10.1086/302798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RA, Barber MD, Johnson PE, Gillbard SM, Haywood MD, Smith CD, Arnold J, Burke T, Urquhart AJ, Gill P. New reference allelic ladders to improve allelic designation in a multiplex STR system. Int J Legal Med. 1998;111:267–272. doi: 10.1007/s004140050167. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Kong CA. Fast multipoint linkage calculation with Allegro. Am J Hum Genet. 1999;65:A60. [Google Scholar]
- Haberl M, Tautz D. Comparative allele sizing can produce inaccurate allele size differences for microsatellites. Mol Ecol. 1999;8:1347–1349. doi: 10.1046/j.1365-294x.1999.00692_1.x. [DOI] [PubMed] [Google Scholar]
- Idury RM, Cardon LR. A simple method for automated allele binning in microsatellite markers. Genome Res. 1997;7:1104–1109. doi: 10.1101/gr.7.11.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Deng H, Lai DB, Xu F, Chen J, Gao G, Recker RR, Deng HW. Toward high-throughput genotyping: Dynamic and automatic software for manipulating large-scale genotype data using fluorescently labeled dinucleotide markers. Genome Res. 2001;11:1304–1314. doi: 10.1101/gr.159701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln SE, Lander ES. Systematic detection of errors in genetic linkage data. Genomics. 1992;14:604–610. doi: 10.1016/s0888-7543(05)80158-2. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet/ITL. 1995;11:402–408. doi: 10.1038/ng1295-402. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: A program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J. Analysis of human genetic linkage. Baltimore: Johns Hopkins University Press; 1999. p. 249. [Google Scholar]
- Papp JC, Kearsey G, Lange K. Improving the quality of genotyping data. Am J Hum Genet (Suppl.) 2000;67:301. [Google Scholar]
- Puers C, Hammond HA, Jin L, Caskey CT, Schumm JW. Identification of repeat sequence heterogeneity at the polymorphic short tandem repeat locus HUMTH01[AATG]n and reassignment of alleles in population analysis by using a locus-specific allelic ladder. Am J Hum Genet. 1993;53:953–958. [PMC free article] [PubMed] [Google Scholar]
- Shields DC, Collins A, Buetow KH, Morton NE. Error filtration, interference, and the human linkage map. Proc Natl Acad Sci. 1991;88:6501–6505. doi: 10.1073/pnas.88.15.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Lange K. Mistyping probabilities from a multipoint analysis of general pedigrees. Am J Hum Genet (Suppl.) 2000;67:35. [Google Scholar]
- Terwilliger JD, Weeks DE, Ott J. Laboratory errors in the reading of marker alleles cause massive reductions in lod score and lead to gross overestimates of the recombination fraction. Am J Hum Genet (Suppl.) 1990;47:A201. [Google Scholar]
- Weber JL, Broman KW. Genotyping for human whole-genome scans: Past, present, and future. Adv Genet. 2001;42:77–96. doi: 10.1016/s0065-2660(01)42016-5. [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Mah TS, Paul TO, Morse L, Ngo-Chang J, Dailey JP, Ferrell RE, Gorin MB. A full genome scan for age-related maculopathy. Hum Mol Genet. 2000;9:1329–1349. doi: 10.1093/hmg/9.9.1329. [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Tsai H-J, Mah TS, Rosenfeld PJ, Paul TO, Eller AW, Morse LS, Dailey JP, Ferrell RE, et al. Age-related maculopathy: An expanded genome-wide scan with evidence of susceptibility loci within the 1q31 and 17q25 regions. Am J Ophthalmol. 2001;132:682–692. doi: 10.1016/s0002-9394(01)01214-4. [DOI] [PubMed] [Google Scholar]
- Weeks DE, Lathrop GM. Polygenic disease: Methods for mapping complex disease traits. Trends Genet. 1995;11:513–519. doi: 10.1016/s0168-9525(00)89163-5. [DOI] [PubMed] [Google Scholar]
- Xu J. Combined analysis of hereditary prostate cancer linkage to 1q24-25: Results from 772 hereditary prostate cancer families from the International Consortium for Prostate Cancer Genetics. Am J Hum Genet. 2000;66:945–957. doi: 10.1086/302807. [DOI] [PMC free article] [PubMed] [Google Scholar]