Abstract
We performed a systematic maximum density subgraph (MDS) detection of conserved sequence regions to discover new, biologically relevant motifs from a set of 21,050 conceptually translated mouse cDNA (FANTOM1) sequences. A total of 3202 candidate sequences, which shared similar regions over >20 amino acid residues, were screened against known conserved regions listed in Pfam, ProDom, and InterPro. The filtering procedure resulted in 139 FANTOM1 sequences belonging to 49 new motif candidates. Using annotations and multiple sequence alignment information, we removed by visual inspection 42 candidates whose members were found to be false positives because of sequence redundancy, alternative splicing, low complexity, transcribed retroviral repeat elements contained in the region of the predicted open reading frame, and reports in the literature. The remaining seven motifs have been expanded by hidden Markov model (HMM) profile searches of SWISS-PROT/TrEMBL from 28 FANTOM1 sequences to 164 members and analyzed in detail on sequence and structure level to elucidate the possible functions of motifs and members. The novel and conserved motif MDS00105 is specific for the mammalian inhibitor of growth (ING) family. Three submotifs MDS00105.1–3 are specific for ING1/ING1L, ING1-homolog, and ING3 subfamilies. The motif MDS00105 together with a PHD finger domain constitutes a module for ING proteins. Structural motif MDS00113 represents a leucine zipper-like motif. Conserved motif MDS00145 is a novel 1-acyl-SN-glycerol-3-phosphate acyltransferase (AGPAT) submotif containing a transmembrane domain that distinguishes AGPAT3 and AGPAT4 from all other acyltransferase domain-containing proteins. Functional motif MDS00148 overlaps with the kazal-type serine protease inhibitor domain but has been detected only in an extracellular loop region of solute carrier 21 (SLC21) (organic anion transporters) family members, which may regulate the specificity of anion uptake. Our motif discovery not only aided in the functional characterization of new mouse orthologs for potential drug targets but also allowed us to predict that at least 16 other new motifs are waiting to be discovered from the current SWISS-PROT/TrEMBL database.
The growing number of complete genomes and estimates that the human proteome may contain up to 500,000 proteins (Banks et al. 2000) underline the importance of understanding protein sequences, motifs, and their functional units (modules) to derive potential functions and interactions. Motifs are conserved sequence patterns within a larger set of protein sequences that share common ancestry. Conserved motifs may be used to predict the functions of novel proteins if the relationship among the encoding genes is orthologous (Tatusov et al. 1996). However, the increasing number of paralogs and mosaic proteins evolved from gene duplications and genomic rearrangement mechanisms led to different interpretations of the term motif and the concept of modules as conserved building blocks of proteins that have a distinct function (Bork and Koonin 1996, Henikoff et al. 1997). A module can consist of one motif or multiple adjacent motifs. For example, C2H2 zinc fingers, leucine zippers, and POU domains are DNA-binding modules. However, structural motifs or active site conservation in a short stretch of sequences do not often reflect common ancestry. Therefore, biological interpretation of motif findings requires additional efforts, for example, literature, structural, and phylogenetic analysis.
To annotate and characterize new protein sequences or extend known protein families, motif searches are conducted by using regular expressions (PROSITE search), similarity search with matrices (BLASTP, profile search), or nonlinear pattern recognition (HMM or artificial neural network) with training sets comprising already known motifs. Strictly defined, new protein motifs are either conserved sequences of common ancestry or convergence (functional motifs) within several proteins that group together for the first time by similarity search and show statistical significance (Bork and Ouzounis 1995). This definition poses a problem for motif extension or submotifs. Therefore, we adopted a case-by-case approach using the functional importance as threshold for a novel motif. Here we have chosen a systematic linkage clustering based on sequence similarity, followed by visual inspection, sequence, topological, and literature analysis of the motif candidates to explore new motifs derived from 21,076 mouse cDNA clones (RIKEN Genome Exploration Research Group Phase II Team and FANTOM Consortium 2001).
RESULTS AND DISCUSSION
Computational Motif Detection
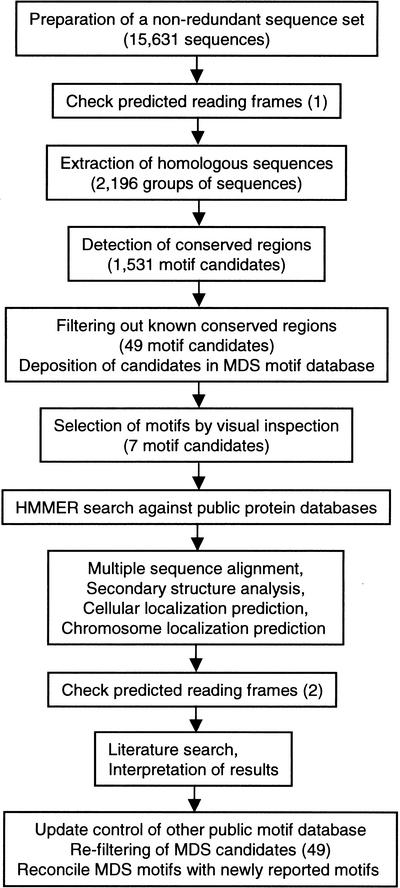
We used a linkage-clustering method with all-to-all sequence comparison (Matsuda et al. 1999) to extract 2196 homologous groups of sequences from a nonredundant set of RIKEN mouse cDNA sequences (Fig. 1). In 465 groups that contained four or more sequences, 1531 motif candidates were detected with a motif detection algorithm as described in the Methods section. The candidates, comprising 3202 sequences with 12,251 conserved regions, were screened against known conserved regions found in Pfam (Bateman et al. 2000), ProDom (Corpet et al. 2000), and InterPro (Apweiler et al. 2000) databases. The filtering procedure resulted in 49 novel motif candidates containing 139 sequences and 216 conserved regions. Subsequent inspection using annotations and multiple sequence alignments yielded seven possible new motifs (MDS00105, MDS00113, MDS00132, and MDS00145–MDS00148) comprising 28 sequences and 42 false positive motifs.
Figure 1.
Strategy of motif exploration.
HMMs were constructed from the motifs and applied in a HMMER search of the nonredundant SWISS-PROT/TrEMBL (SPTR) database (Bairoch et al. 2000). All motifs, including information on HMM scores, E-values, cutoff thresholds, motif alignments, and chromosomal localization can be accessed at the MDS motif database (URL http://motif.ics.es.osaka-u.ac.jp/MDS/). At present, the MDS motif database contains 164 sequences belonging to seven motifs. From 28 FANTOM1 sequences, seven sequences were derived from EST assemblies and therefore do not carry DDBJ accessions.
Estimation of Motif Coverage
The InterPro coverage of 21,050 FANTOM1 translated sequences is 27.9% (5873 sequences) (RIKEN Genome Exploration Research Group Phase II Team and FANTOM Consortium, 2001). The coverage of seven MDS motifs is 0.133% (28 sequences). If we extrapolate from the 136 hits of seven motifs to 707,571 sequences of the nonredundant SPTR database, excluding the 10,465 FANTOM1 sequences, the estimated number of new MDS motif-containing sequences would be 927 (0.133% of 697,106 sequences). Because the number of sequences per MDS motif varies from four (the minimum number of motif-containing sequences that our method detects) to 57 (MDS00148), the estimated number of not yet discovered MDS motifs in the current release of SPTR would range from 16 to 231. The low number of new motifs may reflect a constraint on the number of possible functions and interactions for a given protein in the proteome. In addition, some of the new motifs will be lineage-specific because of species-specific expansion of regulatory genes (Mortlock et al. 2000). If the effects of cDNA library normalization and insert size limitation are neglected, the FANTOM clones represent a random sample of the mouse transcriptome.
Hypothetical Protein Comprising Motifs
Three of seven new motifs have been found in hypothetical proteins. Because we lack experimental information on these proteins, we briefly summarize the predicted functions. MDS00132 members are encoded by mouse 2210414H16Rik and 330001H21Rik and human DKFZP586A0522 (SPTR accessions Q9H8H3, Q9H7R3, Q9Y422, and AAH08180) loci. The human proteins belong to the generic methyltransferase family (InterPro IPR001601) and contain adjacent to the N-terminal located MDS00132 a SAM (S-adenosylL-methionine) binding motif (IPR000051). Considering the 80% sequence identity and 90% similarity to DKFZP586A0522 >146 residues (data not shown), it is likely that hypothetical proteins 2210414H16RIK and 3300001H21Rik belong to the methyltransferase family. Motif MDS00146 comprises 21 members of hypothetical proteins or fragment derived from human, mouse, rat, fruitfly, and worm. Three members, mouse 1200017A24Rik (SPTR accession Q9DB92) and human BA165F24.1 (Q9H1Q3) and FLJ00026 (Q9H7P2), carry at their C-terminus an aminoacyl-transfer RNA synthetases class-II signature (IPR002106), indicating possible involvement in the protein synthesis. The homology with a rat TRG protein fragment (SPTR accession Q63603, not to be confused with TRG T-cell receptor gamma) did not reveal any functional information. Motif MDS00147 is located at the N-terminus of four mouse and two human hypothetical proteins. No other motifs have been detected in the sequences. The remaining motifs—MDS00105, MDS00145, MDS00148, and MDS00113—have been the subject of detailed analyses to explore potential functions of the motifs and their member sequences.
Inhibitor of Growth Motif
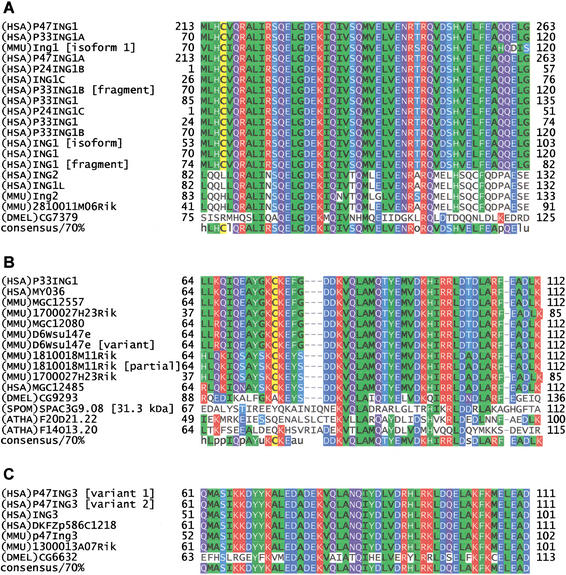
Motif MDS00105 is specific for the ING family, comprising three subfamilies: the ING1/ING1L subfamily (Zeremski et al. 1999, Gunduz et al. 2000, Saito et al. 2000), the ING3 subfamily, and the ING1-homolog subfamily including distant homologs in Drosophila melanogaster, Arabidopsis thaliana, and Schizosaccharomyces pombe (Fig. 2A,B,C). The putative translations of five RIKEN genes (21810011M06Rik, D6Wsu147e, 1700027H23Rik, 1810018M11Rik, and 1300013A07Rik), in addition to another 35 sequences derived from the nonredundant SPTR, share the conserved 51 amino acid MDS00105 region near the N-terminus. Previous research on ING1 has focused predominantly on the PHD finger region because it is also conserved in the yeast homolog YNG2 (Loewith et al. 2000). However, the homology breaks down toward the N-terminus. It was shown that the PHD finger is not required for histone acetyltransferase (HAT) activity association of YNG2 but for human P33ING1 (Loewith et al. 2000). Recently, P33ING1 has been associated with SIN3/HDAC1 complex-mediated transcriptional repression (Skowyra et al. 2001). P24ING1 is believed to be a p53-mediated growth suppressor that acts through transcriptional activation of CDKN1A.
Figure 2.
Multiple sequence alignments of the MDS00105 motif region. The proteins are designated by the species abbreviation and the name. The positions of motif MDS00105 are indicated by “+”. Submotif positions based on regular expressions are designated by “#&”. Multiple sequence alignments of representative complete sequences are shown in Supplement Figure 1A,B,C. (A) Alignment ING1 and ING1L MDS00105.1 motif region. The SWISS-PROT/TrEMBL accessions of the sequences are as follows: (HSA)P47ING1, Q9UIJ4; (HSA)P33ING1A, O43658; (MMU)Ing1 [isoform 1], Q9QXV3 isoform 1; (HSA)P47ING1A, Q9UK53; (HSA)P24ING1B, Q9UBC6; (HSA)ING1C, Q9P0U6; (HSA)P33ING1B [fragment]; CAC38067; (HSA)P33ING1, O00532; (HSA)P24ING1C, Q9NS8; (HSA)P33ING1, Q9UIJ2; (HSA)P33ING1B, Q9UK52; (HSA)ING1 [isoform], Q9H007; (HSA)ING1, Q9HD98; (HSA)ING1 [fragment], Q9HD99; (HSA)ING2, Q9H160; (HSA)ING1L, O95698; (MMU)Ing2, Q9ESK4; (MMU)2810011M06Rik, Q9CZD8; and (DMEL)CG7379, Q9VEF5. (B) Alignment of ING1 homolog MDS00105.2 motif region. The SWISS-PROT/TrEMBL accessions of the ING1 homolog sequences are as follows: (HSA)LOC51147 [P33ING1 homolog], Q9UNL4; (HSA)MY036, Q9H3J0; (HSA)MGC12557, AAH07781; (HSA)MGC4757 [similar to (MMU)1700027H23Rik], AAH13038; (HSA)MGC12080, AAH09127; (MMU)D6Wsu147e, Q9D7F9; (MMU)D6Wsu147e [variant]; (MMU)1810018M11Rik, Q9D8Y8; (MMU)1810018M11Rik [partial]; (MMU)1700027H23Rik, Q9D9V8; (HSA)MGC12485 [similar to (MMU)1819918M11Rik], Q9BS30; (DMEL)CG9293, Q9VJY8; (SPOM)C3G9.08, O42871; (ATHA)14013, Q9LIQ6; and (ATHA)F20D21.22, Q9SLJ4. (C) Alignment of the ING3 MDS00105.3 motif region. The SWISS-PROT/TrEMBL accessions of the other ING3 subfamily sequences are as follows: (HSA)P47ING3 [variant 1], Q9NXR8; (HSA)P47ING3 [variant 2], Q9HC99; (HSA)ING3, O60394; (MMU)p47Ing3, Q9ERB2; (MMU)13000 13A07Rik, Q9DBG0; (HSA)DKFZp586C1218, CAC48260; and (DMEL)CG6632, Q9VWS0.
The PHD finger contains seven conserved Cys and one His residues with metal-chelating potential. These residues are conserved throughout all subfamily members of the ING family. In contrast, the MDS00105 sequence region shows moderate sequence similarity among all ING family members but is conserved between human, mouse, and fruitfly within each subfamily. We therefore reconstructed HMM profiles of MDS00105 for each of the three subfamilies and derived the three submotifs MDS00105.1–MDS00105.3 that distinguish ING1/ING1L, ING1-homolog, and ING3 subfamily members from each other. The E-values of each submotif HMM search result significantly increased toward random values when it hit a sequence of another submotif member. For example, within the ING1/ING1L submotif HMM the E-values ranged from 4.1E-33 to 1E-27 for the ING1/ING1L members, whereas they reached 1E-2 and 3.3E-1 for the ING1-homolog and ING3 members, respectively. The E-value based discrimination was also supported by regular expression submotifs Q-E-L-G-D-E-K-[IM]-Q, K-E-[FY]-[SG]-D-D-K-V-Q and [LM]-E-D-A-D-E-K-V-[AQ] that are specific for ING1/ING1L, ING1-homolog, and ING3 subfamilies, respectively (Fig. 2A,B,C). The A. thaliana and yeast sequences that were aligned with the ING1-homolog sequences (Suppl. Fig. 1B) do not bear the K-E-[FY]-[SG]-D-D-K-V-Q signature, but they do share similar residues [DTN]-E-K-V-[LTQ].
The ING1-homolog (Suppl. Fig. 1B) and ING3 (Suppl. Fig. 1C) subfamily members, which are moderately similar to ING1 and ING1L, are still uncharacterized. The ING1 and ING1L proteins are highly similar except for the N-terminus and a short region within the MDS00105 motif (Suppl. Fig. 1A). The variation in the N-terminal region is the result of alternative transcripts (Zeremski et al. 1999). The ING3 subfamily contains up to 14 consecutive serine residues between the MDS00105.3 submotif and the PHD finger. All ING members were predicted to be nuclear proteins that share a C-terminal PHD (plant homeo domain) finger domain (Aasland et al. 1995, Pascual et al. 2000), indicating involvement in DNA binding and transcriptional regulation.
21810011M06Rik or Ing2 (Fig. 2A, Suppl. Fig. 1A) is the mouse ortholog of human ING1L (Chr 4q35.1), which appears to be the paralog of ING1 (Chr 13q34). D6WSU147E, 1700027H23Rik, and 1810018M11Rik are members of the ING1-homolog subfamily (Fig. 2B, Suppl. Fig. 1B). D6Wsu147e appears to be the ortholog of the human ING1-homolog and was mapped to mouse Chr 6 (59.3 cM). The human ING1-homolog (LocusLink accession LOC51147) has been mapped to the syntenic region of Chr 12pter-12q14.3. ING1-homologs, 1700027H23RIK and 1810018M11RIK, are identical to each other except for the N-terminal region. They are 64% identical to D6WSU147E and human ING1-homolog (p33 variant) and may therefore represent paralogs of ING1-homolog. 1300013A07Rik seems to be the ortholog of human ING3 (Chr 7q31) and a variant of mouse ING3 (Fig. 2C, Suppl. Fig1C). We identified an unspliced intron between potential donor splice site 799 (ACAG|GTAA) and position 1277, which may lead to premature termination and render the translation product nonfunctional.
Considering the earlier described experimental findings, we hypothesize that MDS00105 and its submotifs represent binding sites for distinct subfamily specific protein–protein interaction with HAT, HDAC, MYC (Helbing et al. 1997) and other cell cycle-related proteins, whereas the unique regions of each subfamily member may modulate interactions. For example, ING1-homolog subfamily members share a tyrosine kinase phosphorylation site in the immediate vicinity of the motif, whereas ING3 and ING1/ING1L subfamily members lack this site. The conservation of the submotifs in human and Drosophila coincides also with the presence of TP53 and its Drosophila homolog p53 (Ollmann et al. 2000). Yeast and plants lack TP53 homologs and do not share the conserved submotifs. Therefore, the functions of yeast and plant ING homologs in transcriptional and cell cycle regulation may have been differently conserved.
1-Acyl-SN-Glycerol-3-Phosphate Acyltransferase Subfamily Motif
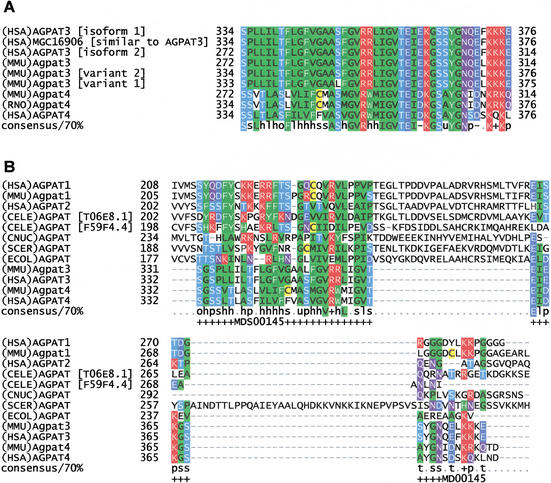
Motif MDS00145 is specific for mammalian 1-acyl-SN-glycerol-3-phosphate acyltransferases (AGPAT) AGPAT3 and AGPAT4 (Fig. 3A). RIKEN clones 4930526L14 and 2210417G15 represent Agpat3 (Chr 10 41.8 cM), which is the ortholog of human AGPAT3 on Chr 21q22.3. The FANTOM1 mapping of RIKEN clones 4930526L14 and 2210417G15 to Chr 16 69.90–71.20 appears to be caused by an Agpat3 related sequence on Chr 16. Agpat4 (clone 1500003P24) has been mapped to mouse Chr 17, 7.3–8.2 cM and a syntenic region on human Chr 6 that contains AGPAT4 and is close to MAP3K3. AGPAT4 is a paralog of AGPAT1 (Chr 6p21) located in major histocompatibility complex class III region.
Figure 3.
(A) Multiple sequence alignment of AGPAT3 and AGPAT4 specific motif MDS00145. The SWISS-PROT/TrEMBL accessions of the sequences are as follows: (HSA)AGPAT3 [isoform 1], Q9NRZ7; (HSA)MGC16906, AAH11971; (HSA)AGPAT3 [isoform 2], Q9NRZ701; (MMU)Agpat3, Q9D517; (MMU)Agpat4, Q9DB84; (RNO)Agpat4, BAB62290; and (HSA)AGPAT4, Q9NRZ5. The putative translations of Agpat3 variant 1 (clone ID 2210417G15) and variant 2 (clone ID 4831410M06) are derived from DDBJ entry AK008965 and FANTOM1 database because variant 2 has no DDBJ accession number. (B) Alignment of AGPAT3 and AGPAT4 C-terminal region with other AGPAT family members. The positions of the AGPAT3 and AGPAT4 specific motif MDS00145 are indicated by “+”. The multiple sequence alignment of the complete sequences is shown in Suppl. Fig. 2.
PSORT (Nakai et al. 1999) program results indicated that AGPAT3 and AGPAT4 are endoplasmic reticulum (ER) membrane proteins. The latter is in concordance with previous findings for human AGPAT1 (Aguado and Campbell 1998). If the signal peptide is not cleaved at predicted positions 19 or 73—as it is often observed for ER membrane proteins—AGPATs carry three transmembrane helices. MDS00145 spans the C-terminal region facing the ER lumen (Suppl. Fig. 2). AGPAT3 and AGPAT4 lack a 25 amino acid region at the C-terminus that causes a split of MDS00145 when AGPAT1 and AGPAT2 in the alignment (Fig. 3B). The Pfam defined acyltransferase domain is located between the second and third transmembrane helices and shared by all AGPAT members.
AGPATs (EC2.3.1.51) are involved in phospholipid metabolism by converting lysophosphatidic acid (LPA) into phosphatidic acid (PA). The AGPAT substrate is 1-acylglycerol 3-phospate. Neither the catalytic nor the substrate binding sites of AGPAT are known. Because AGPAT1 and AGPAT2 overexpression experiments by West et al. (1997) showed enhanced IL6 and TNF-alpha transcription and synthesis on IL1B stimulation, the AGPAT family members are potential targets to modulate inflammatory responses. The transmembrane domain-based alignment of mouse and human AGPAT sequences with distant homologs revealed two conserved blocks in the ER and cytoplasmic loop region. The latter was also described in an experimental E. coli AGPAT study (Lewin et al. 1999). The presence of potential active sites on both sites of the ER is corroborated by the recently discovered reverse, ATP-independent and CoA-dependent LPA synthesis from PA (Yamashita et al. 2001). The signature E-G-T-R-X(4–8)-[SD]X(1–18)-L-P is conserved among all AGPAT family members. We propose that these residues are required for substrate binding on the cytoplasmic site. Possible catalytic sites may involve the N-H-X4-D and AGPAT3 and four specific H-K-L-Y-Q-E and G-N-X-[DE] signatures (Suppl. Fig. 2) because His or Gln and Glu or Asp can facilitate a nucleophilic attack on thioester groups of the glycerol phosphate group of fatty acid CoAs. The divergence of AGPATs in the ER-sided region around MDS00145 motif of AGPAT3 and AGPAT4 indicates a regulatory function MDS00145 for transacylation specificity or activity.
Solute Carrier 21 Family Motif
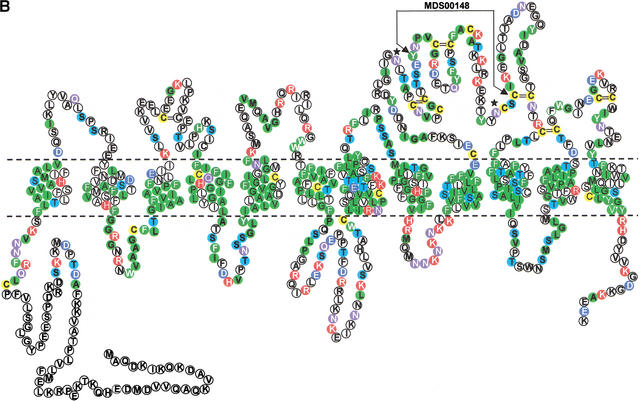
Motif MDS00148 (Fig. 4A) spans a 35–37 amino acids long extracellular oriented loop region between two transmembrane domains that is conserved among members of the mammalian SLC21 family (organic anion transporters), organic anion transporter polypeptide-related (OATPRP), related Drosophila and Caenorhabditis elegans organic anion transporters (Tsuji et al. 1999). All member sequences have seven to 12 transmembrane regions depending on the transmembrane helix prediction algorithm used. We assume 12 transmembrane regions. Sequences of rat Slc21a10 (Lst-1), rat Oat-k2, and 1700022M03Rik are shorter because of exon deletion or truncation (Suppl. Fig. 3A). SLC21 family proteins catalyze the Na+-independent transport of organic anions (e.g., estrogen and prostaglandin) as well as conjugated and unconjugated bile acids (taurocholate and cholate). SLC21 members are tissue specific, for example, SLC21A6 (Abe et al. 1998, Noe et al. 1997) is expressed in brain and liver and kidney but not in testis, heart, spleen, lung, and muscle. SLC21A6 can uptake taurocholate, estrogen conjugates, digoxin, and cardiac glycosides.
Figure 4.
(A) Multiple sequence alignment of extracellular domain containing the OATP family motif MDS00148. The proteins are designated by the species abbreviation and the name. The positions of motif MDS00148 are designated by “+”. Potential N-glycosylation sites N-x-[ST] are indicated. A multiple sequence alignment of MDS00148 representatives covering the entire sequence length and a phylogenetic tree are shown in Suppl. Fig. 3A and B. The SWISS-PROT/TrEMBL accessions of the sequences are as follows: (MMU)4921511I05Rik, Q9D5W6; (MMU)4933404A18Rik, Q9D4B7; (MMU)1700022M03Rik, Q9DA23; (RNO)Tst1, AAK63015; (MFA)hypo. protein [61.1 kD], BAB69708; (DMEL)CG11332, Q9I7P1; (HSA)SLC21A12, Q9H4T8; (HSA)SLC21A12 [POATP], Q9UI35; (HSA)MGC22943 [similar to SLC21A12], AAH15727; (HSA)SLC21A12 [OATP-E], Q9UIG7; (HSA)LOC115191 [SLC21A12-related], Q9H8P2; (RNO)Oatp-e, Q99N01; (MMU)Slc21a2, Q9EPT5; (RNO)Slc21a2, Q00910; (HSA)SLC21A2, Q92959; (MMU)5830414C08Rik, Q9JKV0; (HSA)OATPRP3, Q9GZV2; (HSA)SLC21A11, Q9UIG8; (HSA)MGC659 [similar to SLC21A11], Q9BW73, (RNO)Pgt2, Q99N02; (RNO)Oat-k2, Q9WTM0; (RNO)Slc21a4, P70502; (MMU)Slc21a1, Q9QXZ6; (RNO)Oatp2, O35913; (RNO)Oatp3, O88397; (RNO)Slc21a7, Q9EQR8; (RNO)Slc21a3, P46720; (MMU)MGC6720, AAH13594; (MMU)Slc21a7, AAK39416; (MMU)Slc21a5, Q9EP96; (MMU)Slc21a13, Q99J94; (RNO)Oatp5, Q9QYE2; (HSA)OATP, AAG30037; (HSA)SLC21A3 [OATP-A], CAB97006; (HSA)SLC21A3 [OATP], P46721; (HSA)SLC21A3 [OATP1B], Q9UL38; (MMU)Slc21a14, Q9ERB5; (RNO)LOC845111, Q9EPZ7, (HSA)SLC21A14, Q9NYB5; (MFA)hypo. protein [66.7 kD], Q9GMU6; (HSA)SLC21A8, Q9NPD5; (HSA)SLC21A6, Q9Y6L6; (MMU)Slc21a10(MLST-1), Q9JJJ1; (MMU)Slc21a10(Lst-1), Q9JJL3; (MMU)MG1351894, Q9JI79; (RNO)Slc21a10(Lst-1a), Q9JHF6; (RNO)Slc21a10(Lst-1c), Q9JIM2; (RNO)Slc21a6, Q9QZX8; (HSA)OATRPR4, Q9H2Y9; (DMEL)CG3811, Q9VLB3; (DMEL)CG3380, Q9W269; (DMEL)CG3380 [variant], AAK77236; (DMEL)CG3387, Q9W271; (HSA)OATPRP2, Q9H2Z0; (HSA)SLC21A9, O94956; (HSA)DKFZP586I0322, Q9UFU1; (RNO)moatp1 [splice variant], Q9JHI3; (CELE)Y70G10A.3, Q9XWC5; (DMEL)CG7571, Q9VVH9; and (CELE)F21G4.1, Q93550. Motif sequences that contain an hmmpfam-predicted kazal-type serine protease inhibitor domain are labeled with a “k”. (B) Proposed topology of an organic anion transporter. The primary structure is shown for 4921511I05Rik, assuming 12 transmembrane helices. The intra- and extracellular domains are separated by horizontal dashed lines, which symbolize the cell membrane. The motif region MDS00148 is indicated by arrows. Potential extracellular N-glycosylation sites are symbolized by asterisks. Potential disulfide bonds of Cys residues are shown as double lines.
The motif MDS00148 is located in the extracellular region between transmembrane helices 9 and 10. Interestingly, 33 of 60 proteins shown in Fig. 4A were predicted to carry in this region a kazal-type serine protease inhibitor domain as defined by InterPro (IPR002350, Release 3.2). The 3D structure of kazal-type domains typically includes two short α-helices and a three-stranded antiparallel ß-sheet. Secondary structure prediction using the DSC program (King et al. 1997) and the PredictProtein server at http://maple.bioc.columbia.edu/predictprotein/ (Rost 1996) confirmed the antiparallel ß-sheet but not the presence of α-helices. Moderate structural similarities of the kazal-type domain to the follistatin and osteonectin domains were noted earlier, but closer analysis showed that they differ in the cysteine bonding pattern and N-terminal regions (Hohenester et al. 1997). Our HMM profile detected none of the serine protease inhibitor members, including membrane-associated agrins. The hmmpfam search using the Pfam profile domains detected a kazal-type domains with E-values ranging from 0.0057 to 0.041 in 11 sequences (Fig. 4A). When applying the threshold of the Pfam gathering method to build Pfam full alignments for the kazal-type domain in an hmmpfam search against the Pfam Release 6.6, only two sequences, (HSA)SLC21A3 [OATP] and (HSA)SLC21A3 [OATP1B] (SPTR accessions P46721 and Q9UL38), showed a bit score >3.0, which qualifies them as kazal-type domain members. In the absence of crystal structure data of an SLC21 family protein, we conclude that MDS00148 represents a novel module with some structural similarities to the kazal-type domain. MDS00148 might have evolved from a protease inhibitor domain but acquired different functionality when transferred into an ancestral transmembrane SLC21 family protein.
The presence of 11 Cys residues in the pattern CX(22–23,27–28)-C-X(3,5)-C-X-C-X(7,8)-C-X(10–11)-C-X3-C-X(7,14–16)-C-X-C-X(13–37)-C-X(3–5)-C could support four loops (Fig. 4B) that are stabilized by disulfide bonds between Cys residues. The other extracellular regions between transmembrane helices form shorter loops. The loops of the extracellular regions may interact with each other through a network of hydrogen and disulfide bonds to form a lid-like structure. Because the extracellular region between transmembrane helices 9 and 10 is conserved between mammals, Drosophila and C. elegans, we propose that substrate specificity is determined by conserved cysteines, positively charged residues located between the conserved cysteine residues, and the loop length of extracellular regions. Cysteines have been shown to be involved in the transport activity of dopamine and serotonin transporters (Chen et al. 2000, Chen et al. 1997). Based on the similarities in the extracellular loop regions and phylogenetic analysis (Suppl. Fig. 3B), we predict that the novel mouse SLC21 members may share anion uptake specificities of SLC21A12 [OATP-E]. SLC21A12 can transport estradiol-17ß-glucuronide, ß-lactam antibiotic benzylpenicillin, and prostaglandin E2 (Tamai et al. 2000). SLC21A12 is expressed in various cancer cells but not in normal blood cells.
While revising this paper we noticed that the Pfam Release 6.6 (August 2001) includes two domains of the SLC21 family (OATP_N, PF03132; OATP_C, PF03137). In an hmmpfam search against the Pfam Release 6.6, 58 and 47 of the 60 proteins are predicted to have (according to the Pfam gathering method) the OATP_N domain and the OATP_C domain, respectively. The graphical view of MDS00148 in the MDS motif database visualizes the multidomain information. The OATP_N domain overlaps with the C-terminus of the MDS00148 motif in 1, 3, 4, 6, and 12 residues for 25, 1, 16, 2, and 1 sequences, respectively. The Pfam OATP_N motif appears to be the N-terminal extension of the MDS00148 motif that was originally detected on September 28, 2000. No overlaps have been detected for OATP_C. Because the filtering of motif candidates against known conserved regions was conducted before the release of Pfam 6.6, the partially overlapping MDS00148 motif candidates were originally not removed. To counter the problems arising from updates of other motif databases, the motif discovery pipeline (Fig. 1) has been retrofitted with an “update screening” step.
Leucine Zipper-Like Motif
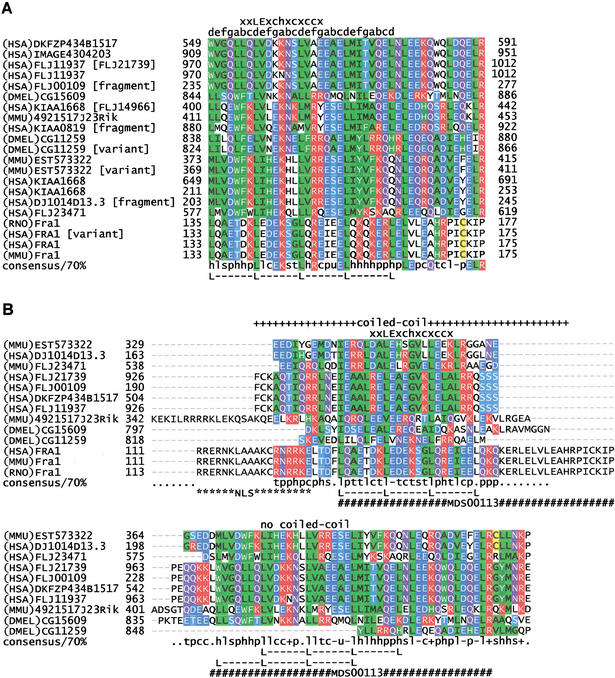
Motif MDS00113 includes 20 members (Fig. 5A) with conserved sequences of 43 amino acids length that carry either a leucine zipper signature characteristic for Fos related antigen 1 (FRA1) or a leucine zipper-like motif (16 members). We analyzed 13 representative members in detail (Fig. 5B). Ten of 13 members contain a leucine heptad repeats. The program “pattern search” of ANTHEPROT V5.0 rel.1.0.5 (Geourjon et al. 1995) calculated the theoretical frequency of the heptad repeat L-x6-L-x6-L-x6-L as 1.07E-5 using PROSITE Rel.16.0 (Hofmann et al. 1999). Because the leucine repeat could have occurred by chance, it would be risky to infer the leucine zipper function of DNA binding. Therefore, we reanalyzed the sequences for features of known and characterized leucine zippers occurring in transcription factors. Sequences were evaluated for (1) alpha helical coiled-coil region (O'Shea et al. 1989) with mostly 3,4-hydrophobic repeat of apolar amino acids at positions a and d of the helix, (2) an overlap of the coiled-coil region with the leucine heptad repeat, (3) coiled-coil trigger sequences [IVL]-X-[DE]-I-X-[RK]-X and [IVL]-[DE]-X-I-X-[RK]-X (Frank et al. 2000) or the 13-residue trigger motif xxLExchxcxccx (Kammerer et al. 1998, Wu et al. 2000), (4) a basic DNA binding region preceding the heptad repeat, and (5) nuclear localization signals (Hicks 1995). Applying these criteria, only the FRA1 sequences qualified as basic leucine zipper with DNA binding function (Landschulz et al. 1988 and 1989). Eight sequences contain a nuclear localization signal. Three proteins (FLJ21739, FLJ11937, and DKFZP434B1517) were predicted by PSORT as cytoplasmic proteins, whereas all have a “COILS” (Lupas et al. 1991) predicted coiled-coil region. Eleven sequences bear a coiled-coil trigger sequence or trigger motif. Only the three FRA1 sequences have a coiled-coil region and a trigger motif overlapping with the heptad repeat, whereas EST573322, 4921517J23Rik, DJ1014D13.3, CG15609, FLJ00109, and FLJ11937 contain a potential tandem coiled-coil region that is separated by the heptad repeat (Fig. 5C). Drosophila protein CG11259 has a tandem coiled-coil region with a leucine heptad repeat located in the first coiled-coil. The basic region of these seven sequences is either nonexistent or does not immediately precede the heptad repeat (Fig. 5B).
Figure 5.
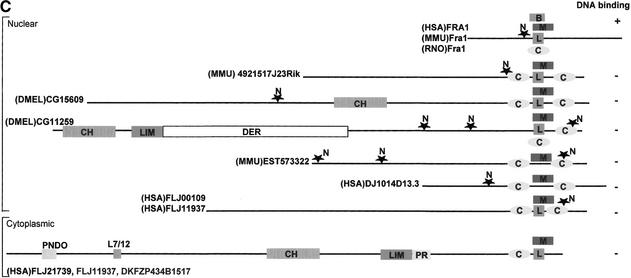
(A) ClustalW sequence alignment of motif MDS00113. (The SWISS-PROT/TrEMBL accessions of the sequences are as follows: (HSA)DKFZP434B1517, Q9UFF7; (HSA)IMAGE4304203, AAH09972; (HSA)FLJ11937 [FLJ21739], Q9H6X6; (HSA)FLJ11937, Q9HAA1; (HSA)FLJ00109, Q9H7I0; DMEL)CG15609, Q9V7X1; (HSA)KIAA1668 [FLJ14966], BAB55422; (MMU)4921517J23Rik, Q9D5U9; (HSA)KIAA0819 [fragment], O94909; (DMEL)CG11259 [variant], AAK93415; DMEL)CG11259, Q9VU34; (MMU)EST57332, 4930438G05; (MMU)EST57332 [variant], 4933434L05; (HSA)KIAA1668, Q9BY92; (HSA)KIAA1668 [Unknown], Q9BVL9; (HSA)DJ1014D13.3, Q9UH43; (HSA)FLJ23471, Q9H5F9; (RNO)Fra1, P10158 (HAS)FRA1 [variant], CAC50237; (HSA)FRA1, P15407; and (MMU)Fra1, P48755. The putative translations of segment EST573322 and its variant (clone ID 4930438G05 and 4933434L05) were taken directly from the FANTOM1 database because the sequence has no DDBJ accession number. (B) Topology-based multiple sequence alignment of 13 MDS00113 representatives. Regions that were predicted as coiled coil were aligned in the upper part. The lower part of the alignment contains the noncoiled regions. The positions of motif MDS00113 and leucine heptad repeats, the predicted coiled coil regions, and the coiled coil trigger sequence (xxLExchxcxccx) are indicated. Letters a to g indicate helix positions. NLS symbolizes the nuclear localization signal. (C) Domain map of MDS00113 containing putative proteins. The abbreviations and InterPro accessions are as follows: M, MDS00113; L, leucine heptad repeat; N, nuclear localization signal; C, coiled coil; CH, calponin-homology domain, IPR001715; LIM, LIM domain, IPR001718; DER, delayed-early response, IPR002259; B, basic-leucine zipper, IPR001871; PNDO, pyridine nucleotide-disulfide oxidoreductase; PRR, proline-rich region, IPR000694; and L7/12, ribosomal L7/L12 C-terminal domain, IPR000206.
The conservation of structural features among evolutionary unrelated sequences indicates domain shuffling. Given the sequence conservation with FRA1, it is conceivable that an ancestral functional basic leucine zipper region was subjected to recombination and mutation events that degenerated the basic leucine zipper. Domain mapping using the InterPro Scan program provided further evidence for domain shuffling. We detected a calponin homology, LIM, delayed early response, pyridine nucleotide-disulfide oxidoreductase, ribosomal L7/L12, and Pro-rich domains (Fig. 5C). The dispersed location of LIM and calponin homology domains among FLJ21739, FLJ11937, DKFZP434B1517, CG15609, and CG11259 does not indicate any ancestral relationship.
Combining the results, we propose that the tandem coiled-coil containing proteins bind to proteins, rather than DNA, in a similar fashion as the group D basic helix-loop-helix (HLH) proteins (Atchley and Fitch 1997). Group D HLH proteins, which lack the basic region in the first helix, can form protein–protein dimers that act, for example, as a negative dominant regulator of transcription factor MyoD on DNA binding. The effect of the other domains on the proposed protein binding function of these multidomain proteins remains unclear.
False Positive Motifs
From 42 false positives we describe seven cases that were not obvious from visual inspection and required detailed analysis. Motif MDS00118 contains 4931406F04Rik, testis expressed gene 11 (Tex21-pending, 4931412D23RIK, 4931421K24RIK), and amyotrophic lateral sclerosis 2 (juvenile) chromosome region candidate 3 (ALS2CR3). 4931406F04Rik and Tex21-pending show 95% identity over the entire sequence length. There is no sequence similarity between Tex21-pending and ALS2CR3 except for 51 amino acid region of MDS00118. Closer inspection of the region revealed that only 10(21) of 26 residues within MDS00118 (10–35) were identical (similar). The identity (similarity) was limited to short stretches of [IVL]-D-L or [KR]-E-E-[LI] that are believed to be a low complexity region that was not masked by the SEG filter.
Motif candidate MDS00131 is identical to a 3 × 37 residues tandem repeat reported in mouse nucleolar RNA helicase II DDX21 (Valdez and Wang 2000). The motif candidate is a false positive because each repeat unit contains the known signature sequence of a mononuclear localization signal. The second false positive, MDS00102, represents the immunoglobulin variable-like N-domains reported in sequences of the carcinoembryonic antigens (Keck et al. 1995).
Secondary structure analysis and fold predictions of sequences containing motifs MDS00121, MDS00129, MDS00137, and MDS00139 (Suppl. Fig. 4) did not produce any conclusive results. We therefore went back to DNA level and checked the ORF prediction and for repeat elements. A RepeatMasker (Smit and Green 1997) search revealed that the DECODER (Fukunishi and Hayashizaki 2001) predicted ORFs contain B1 (MDS00122), B2 (MDS00121), intracisternal A-particle (IAP) LTR (MDS00137), and mammalian apparent LTR-retrotransposon (MDS00139) repeat elements, respectively. Therefore, the sequences constitute transcribed repeats or repeat containing transcripts. We report the finding as a warning regarding data processing and demonstration of the limits of semiautomated annotation approaches.
RIKEN clone 2610016M14 is of biological interest because it carries a potential in-frame inserted IAP LTR in the DNA mismatch repair gene Msh3 (Watanabe et al. 1996). IAPs are endogenous proviral elements that originated from defective retroviruses. IAPs are highly expressed in transformed cell lines and during normal mouse embryogenesis. There are about 1000 IAP-related elements per haploid mouse genome, but only a few IAPs are transcribed (Mietz et al. 1992). The IAP-LTR insertion is located seven base pairs downstream of exon 5, relative to the mouse genomic Msh3 sequence, and may have resulted in exon skipping (Wandersee et al. 2001). Loss of Msh3 function causes a partial mismatch repair defect and may enhance tumorigenesis but does not result in cancer predisposition (de Wind et al. 1999).
CONCLUSION
Motif discovery can be performed with relative ease on genome scale. However, the exploration of new motifs or submotifs for potential biological functions is a time-consuming process depending on human expert knowledge. This case study enabled us to derive valuable process information and rules for semiautomation that will not only aid the next round of functional annotation of mouse cDNA clones (FANTOM) but also the initial steps of protein–protein interaction, regulatory, and active site target selections in a drug discovery process.
METHODS
Preparation of a Nonredundant Sequence Set
The FANTOM cDNA collection contains 21,076 clones. We have used DECODER to predict the ORF of each cDNA sequence, which yielded 21,050 potential coding sequences. As the clone set showed some redundancies that could lead to false positive motifs, we clustered the putative translations using DDS (Huang et al. 1997) and ClustalW (Thompson et al. 1994). From each cluster, we selected the longest sequence as representative of the cluster. As a result, we obtained 15,631 nonredundant sequences.
Extraction of Homologous Sequences and Clustering
Sequences were compared against each other with BLASTP of the NCBI-Toolkit (Altschul et al. 1997) applying an E-value of 0.1 and using the SEG filter option (Wootton 1993) to remove low-complexity regions. The BLASTP results of each comparison were then analyzed with a clustering algorithm (Matsuda et al. 1999) to extract homologous groups of sequences. The clustering algorithm is based on graph theory. Briefly, each sequence is considered as a vertex. If the similarity between any pair of sequences exceeds a user-defined threshold (E-value 0.1, in our case), an arc is drawn between the sequence pair or the two vertices.
The resulting graph is also covered by subgraphs whose vertices are connected with at least a fraction P (a user-defined ratio; here P = 40%) of the other vertices. The groups of subgraphs may overlap with each other if some sequences share two or more homologous regions with a different set of sequences, for example, multidomain-containing sequences. The method is equivalent to complete-linkage clustering if P is set to 100%. In contrast, single-linkage clustering requires only one arc to any member in a group and P becomes virtually 0% when the number of members is large.
Detection of Homologous Regions in Subgraph Groups
Homologous regions in groups were detected by another graph-theoretic method. We extracted all fixed-length subsequences (blocks) and performed ungapped pairwise alignments among all block pairs. The length of the initial block was 20 amino acids. The alignment score was measured using the BLOSUM 50 score matrix. A block graph was constructed by regarding blocks as vertices. The vertices were connected by weighted arcs if the blocks showed at least the user-defined similarity (BLOSUM score 50). Highly connected components in the block graph were detected with a maximum-density subgraph (MDS) algorithm (Matsuda 2000). Here, density is a graph-theoretic term that is defined as the ratio of the sum of the similarity scores between blocks to the number of blocks. Homologous regions longer than 20 amino acids were obtained by combining overlapping blocks that were detected by this method.
Filtering Out Known Conserved Regions
The detected blocks were screened for known conserved regions detected by HMMER 2.1.1 (Washington University, http://hmmer.wustl.edu/) in Pfam (Release 5.5), BLASTP in ProDom (Release 2000.1), and InterPro Scan in InterPro (Release 2.0) databases. Blocks that overlapped with at least one residue of known motifs or domains were discarded. The remaining blocks were labeled with the original discovery date and subjected to further overlap screening with the updates of ProDom, InterPro and Pfam. If a previously novel motif candidate overlapped with a newly reported motif of the updated motif databases, the motif candidate was reinspected manually to evaluate the significance of the overlap regarding motif extension or submotif.
HMM Search against RIKEN and SWISS-PROT/TrEMBL Sequences
Of 21,076 sequences, 1908 are EST assemblies that were not submitted to DDBJ (see also http://fantom.gsc.riken.go.jp/doc/faq.html), and 8703 of 19,168 submitted sequences do not have a coding sequence (CDS) assignment in GenBank Release 125. The nonredundant SWISS-PROT/TrEMBL database (SPTR) (Bairoch et al. 2000) composed of SWISS-PROT 40.0, TrEMBL 18.0, and TrEMBL_new of October 23, 2001 contains 10,465 translated sequences of the FANTOM1 set. A total of 10,603 translated FANTOM1 sequences are not in SPTR. To expand the number of motif members, we constructed (HMMs) from the conserved blocks and searched with HMMER the SPTR database, 10,603 DECODER-predicted FANTOM1 translations, and 1908 DECODER-predicted translations of the EST assemblies.
MDS Motif Database
The results of the HMM searches and accessory information such as HMM scores, E-values, protein names, motif alignments, and chromosomal mapping information were stored in flat files. A graphical user interface and Perl/CGI scripts facilitate the search and display of motifs and false positives. The MDS motif database is accessible at URL http://motif.ics.es.osaka-u.ac.jp/MDS/.
Extended Sequence Analyses
Secondary structure analyses of sequences were performed with the locally installed ANTHEPROT V5.0 rel.1.0.5 software, DSC package (King et al. 1997), and on the external PredictProtein server (Rost 1996). Functional sites, for example, phosphorylation and N-glycosylation sites were predicted with ANTHEPROT from the PROSITE database (Hofmann et al. 1999). The locally installed PSORT2 program was used to predict the cellular localization of proteins. Chromosomal localization information was retrieved from the FANTOM map of RIKEN clones on human genome and mouse chromosomes and LocusLink (Pruitt et. al. 2001). Alignments were performed with the locally installed ClustalW 1.8 program, postprocessed with the coloring software MView 1.41 (Brown et al. 1998) and edited by hand to improve the alignment quality. For the analysis of SLC21 family sequences, we constructed a phylogenetic tree by the maximum-likelihood method using MOLPHY ProtML (Adachi and Hasegawa 1996). The tree was obtained by the “Quick Add Search” using the Jones-Taylor-Thornton model (Jones et al. 1992) of amino acid substitution and retaining the 300 top ranking trees (options -jf -q -n 300). Bootstrap values of the tree were calculated by analyzing 1000 replicates using the resampling of the estimated log-likelihood (RELL) method (Kishino et al. 1990).
Acknowledgments
We thank Wolfgang Fleischmann (European Bioinformatics Institute), Richard Baldarelli (MGI, The Jackson Laboratory) for valuable comments, and Tadashi Ogawa (Computer/Informatics Facilities, RIKEN GSC) for technical assistance. This work was supported in part by ACT-JST (Research and Development for Applying Advanced Computational Science and Technology) of Japan Science and Technology Corporation (JST) to H.M. and Y.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL matsuda@ics.es.osaka-u.ac.jp; FAX 81-6-6850-6602.
E-MAIL schoen@gsc.riken.go.jp; FAX 81-45-503-9158.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.193702. Article published online before print in February 2002.
REFERENCES
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Abe T, Kakyo M, Sakagami H, Tokui T, Nishio T, Tanemoto M, Nomura H, Hebert SC, Matsuno S, Kondo H, et al. Molecular characterization and tissue distribution of a new organic anion transporter subtype (oatp3) that transports thyroid hormones and taurocholate and comparison with oatp2. J Biol Chem. 1998;273:22395–22401. doi: 10.1074/jbc.273.35.22395. [DOI] [PubMed] [Google Scholar]
- Adachi J, Hasegawa M. MOLPHY version 2.3: Programs for molecular phylogenetics based on maximum likelihood, Computer Science Monographs, no. 28. Tokyo: The Institute of Statistical Mathematics; 1996. ftp://ftp.ism.ac.jp/pub/ISMLIB/MOLPHY . ( ftp://ftp.ism.ac.jp/pub/ISMLIB/MOLPHY). ). [Google Scholar]
- Aguado B, Campbell RD. Characterization of a human lysophosphatidic acid acyltransferase that is encoded by a gene located in the class III region of the human major histocompatibility complex. J Biol Chem. 1998;273:4096–4105. doi: 10.1074/jbc.273.7.4096. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. InterPro—an integrated documentation resource for protein families, domains, and functional sites. Bioinformatics. 2000;16:1145–1150. doi: 10.1093/bioinformatics/16.12.1145. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks RE, Dunn MJ, Hochstrasser DF, Sanchez JC, Blackstock W, Pappin DJ, Selby PJ. Proteomics: New perspectives, new biomedical opportunities. Lancet. 2000;356:1749–1756. doi: 10.1016/S0140-6736(00)03214-1. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Koonin E. Protein sequence motifs. Curr Opin Struct Biol. 1996;6:366–376. doi: 10.1016/s0959-440x(96)80057-1. [DOI] [PubMed] [Google Scholar]
- Bork P, Ouzounis C. Ready for a motif submission? A proposed checklist.Trends Biochem. Sci. 1995;20:104. doi: 10.1016/s0968-0004(00)88974-4. [DOI] [PubMed] [Google Scholar]
- Brown NP, Leroy C, Sander C. MView: A web-compatible database search or multiple alignment viewer. Bioinformatics. 1998;14:380–381. doi: 10.1093/bioinformatics/14.4.380. [DOI] [PubMed] [Google Scholar]
- Chen N, Ferrer JV, Javitch JA, Justice JB. Transport-dependent accessibility of a cytoplasmic loop cysteine in the human dopamine transporter. J Biol Chem. 2000;275:1608–1614. doi: 10.1074/jbc.275.3.1608. [DOI] [PubMed] [Google Scholar]
- Chen JG, Liu-Chen S, Rudnick G. External cysteine residues in the serotonin transporter. Biochemistry. 1997;36:1479–1486. doi: 10.1021/bi962256g. [DOI] [PubMed] [Google Scholar]
- Corpet F, Servant F, Gouzy J, Kahn D. ProDom and ProDom-CG: Tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res. 2000;28:267–269. doi: 10.1093/nar/28.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wind N, Dekker M, Claij N, Jansen L, van Klink Y, Radman M, Riggins G, van der Valk M, van't Wout K, te Riele H. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat Genet. 1999;23:359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- Frank S, Lustig A, Schulthess T, Engel J, Kammerer RA. A distinct seven-residue trigger sequence is indispensable for proper coiled-coil formation of the human macrophage scavenger receptor oligomerization domain. J Biol Chem. 2000;275:11672–11671. doi: 10.1074/jbc.275.16.11672. [DOI] [PubMed] [Google Scholar]
- Fukunishi Y, Hayashizaki Y. Amino-acid translation program for full-length cDNA sequences with frame-shift errors. Physiol Genomics. 2001;5:81–87. doi: 10.1152/physiolgenomics.2001.5.2.81. [DOI] [PubMed] [Google Scholar]
- Geourjon C, Deleage G. ANTHEPROT 2.0: A three-dimensional module fully coupled with protein sequence analysis methods. J Mol Graph. 1995;13:209–212. doi: 10.1016/0263-7855(95)00035-5. , 199–200. [DOI] [PubMed] [Google Scholar]
- Gunduz M, Ouchida M, Fukushima K, Hanafusa H, Etani T, Nishioka S, Nishizaki K, Shimizu K. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 2000;60:3143–3146. [PubMed] [Google Scholar]
- Helbing C, Veilette C, Riabowol K, Johnston RN, Garkavetsev I. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res. 1997;57:1255–1258. [PubMed] [Google Scholar]
- Henikoff S, Greene EA, Pietrokovski S, Bork P, Attwood TK, Hood L. Gene families: The taxonomy of protein paralogs and chimeras. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- Hicks GR, Raikhel NV. Protein import into the nucleus: An integrated view. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenester E, Maurer P, Timpl R. Crystal structure of a pair of follistatin-like and EF-hand calcium-binding domains in BM-40. EMBO J. 1997;16:3778–3786. doi: 10.1093/emboj/16.13.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Adams MD, Zhou H, Kerlavage AR. A tool for analyzing and annotating genomic sequences. Genomics. 1997;46:37–45. doi: 10.1006/geno.1997.4984. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences, Comput. Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kammerer RA, Schulthess T, Landwehr R, Lustig A, Engel J, Aebi U, Steinmetz MO. An autonomous folding unit mediates the assembly of two-stranded coiled coils. Proc Natl Acad Sci. 1998;95:13419–13424. doi: 10.1073/pnas.95.23.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck U, Nedellec P, Beauchemin N, Thompson J, Zimmermann W. The cea10 gene encodes a secreted member of the murine carcinoembryonic antigen family and is expressed in the placenta, gastrointestinal tract and bone marrow. Eur J Biochem. 1995;229:455–464. doi: 10.1111/j.1432-1033.1995.0455k.x. [DOI] [PubMed] [Google Scholar]
- King RD, Saqi M, Sayle R, Sternberg MJ. DSC: Public domain protein secondary structure predication. Comput Appl Biosci. 1997;13:473–474. doi: 10.1093/bioinformatics/13.4.473. [DOI] [PubMed] [Google Scholar]
- Kishino H, Miyata T, Hasegawa M. Maximum likelihood inference of protein phylogeny, and the origin of chloroplasts, J. Mol Evol. 1990;17:368–376. [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- ————— The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;243:1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- Lewin TM, Wang P, Coleman RA. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33ING1 are associated with histone acetyltransferase activities. Mol Cell Biol. 2000;20:3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Matsuda H, Ishihara T, Hashimoto A. Classifying molecular sequences using a linkage graph with their pairwise similarities. Theoretical Computer Science. 1999;210:305–325. [Google Scholar]
- Matsuda H. Detection of conserved domains in protein sequences using a maximum-density subgraph algorithm. IEICE Trans Fundamentals Electron Commun Comput Sci. 2000;E83-A:713–721. [Google Scholar]
- Mietz JA, Fewell JW, Kuff EL. Selective activation of a discrete family of endogenous proviral elements in normal BALB/c lymphocytes. Mol Cell Biol. 1992;12:220–228. doi: 10.1128/mcb.12.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortlock DP, Sateesh P, Innis JW. Evolution of N-terminal sequences of the vertebrate HOXA13 protein. Mamm Genome. 2000;11:151–158. doi: 10.1007/s003350010029. [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. 1999. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Noe B, Hagenbuch B, Stieger B, Meier PJ. Isolation of a multispecific organic anion and cardiac glycoside transporter from rat brain. Proc Natl Acad Sci. 1997;94:10346–10350. doi: 10.1073/pnas.94.19.10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, et al. Drosophilap53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- O'Shea EK, Rutkowski R, Kim PS. Evidence that the leucine zipper is a coiled coil. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J Mol Biol. 2000;304:723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIKEN Genome Exploration Research Group Phase II Team and FANTOM Consortium. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- Rost B. PHD: Predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- Saito A, Furukawa T, Fukushige S, Koyama S, Hoshi M, Hayashi Y, Horii AJ. p24/ING1-ALT1 and p47/ING1-ALT2, distinct alternative transcripts of p33/ING1. Hum Genet. 2000;45:177–181. doi: 10.1007/s100380050206. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Zeremski M, Neznanov N, Li M, Choi Y, Uesugi M, Hauser CA, Gu W, Gudkov AV, Qin J. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J Biol Chem. 2001;276:8734–8739. doi: 10.1074/jbc.M007664200. [DOI] [PubMed] [Google Scholar]
- Smit, A.F.A and Green, P. 1997. RepeatMasker at http://ftp.genome.washington.edu/RM/RepeatMasker.html
- Tamai I, Nezu J, Uchino H, Sai Y, Oku A, Shimane M, Tsuji A. Molecular identification and characterization of novel members of the human organic anion transporter (OATP) family. Biochem Biophys Res Commun. 2000;273:251–260. doi: 10.1006/bbrc.2000.2922. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Mushegian AR, Bork P, Brown NP, Hayes WS, Borodovski M, Rudd KE, Koonin EV. Metabolism and evolution of Haemophilus influenzae deduced from a whole genome comparison with Escherichia coli. Current Biology. 1996;6:279–291. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji A, Tamai I. Organic anion transporters. Pharm Biotechnol. 1999;12:471–491. doi: 10.1007/0-306-46812-3_16. [DOI] [PubMed] [Google Scholar]
- Valdez BC, Wang W. Mouse RNA helicase II/Gu: cDNA and genomic sequences, chromosomal localization, and regulation of expression. Genomics. 2000;66:184–194. doi: 10.1006/geno.2000.6209. [DOI] [PubMed] [Google Scholar]
- Wandersee NJ, Roesch AN, Hamblen NR, de Moes J, van Der Valk MA, Bronson RT, Gimm JA, Mohandas N, Demant P, Barker JE. Defective spectrin integrity and neonatal thrombosis in the first mouse model for severe hereditary elliptocytosis. Blood. 2001;97:543–550. doi: 10.1182/blood.v97.2.543. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Ikejima M, Suzuki N, Shimada T. Genomic organization and expression of the human MSH3 gene. Genomics. 1996;31:311–318. doi: 10.1006/geno.1996.0053. [DOI] [PubMed] [Google Scholar]
- West J, Tompkins CK, Balantac N, Nudelmann E, Mengs B, White T, Bursten S, Coleman J, Kumar A, Singer JW, et al. Cloning and expression of two human lysophosphatidic acid acyltransferase cDNAs that enhance cytokine-induced signaling responses in cells. DNA Cell Biol. 1997;16:691–701. doi: 10.1089/dna.1997.16.691. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Statistics of local complexity in amino acid sequences and sequence databases. Comput Chem. 1993;17:149–163. [Google Scholar]
- Wu KC, Bryan JT, Morasso MI, Jang SI, Lee JH, Yang JM, Marekov LN, Parry DA, Steinert PM. Coiled-coil trigger motifs in the 1B and 2B rod domain segments are required for the stability of keratin intermediate filaments. Mol Biol Cell. 2000;11:3539–3558. doi: 10.1091/mbc.11.10.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Kawagishi N, Miyashita T, Nagatsuka T, Sugiura T, Kume K, Shimizu T, Waku K. ATP-independent fatty acyl-coenzyme A synthesis from phospholipid: Coenzyme A-dependent transacylation activity toward lysophosphatidic acid catalyzed by acyl-coenzyme A:lysophosphatidic acid acyltransferase. J Biol Chem. 2001;276:26745–26752. doi: 10.1074/jbc.M101795200. [DOI] [PubMed] [Google Scholar]
- Zeremski M, Hill JE, Kwek SS, Grigorian IA, Gurova KV, Garkavtsev IV, Diatchenko L, Koonin EV, Gudkov AV. Structure and regulation of the mouse ING1 gene. Three alternative transcripts encode two PHD finger proteins that have opposite effects on p53 function. J Biol Chem. 1999;274:32172–32181. doi: 10.1074/jbc.274.45.32172. [DOI] [PubMed] [Google Scholar]