Abstract
Deciphering the human genome includes locating the promoters that initiate transcription and identifying the exons of genes. Many promoter prediction programs have been proposed, but when they are applied to extended regions of the genome, most of their predictions are false-positives. The extensive collection of gene transcript sequences is an important new source of information, which has not been used previously in promoter predictions. Our approach is to enhance the specificity of predictions by restricting the genomic regions that are searched using gene transcript alignments as anchors in the genome for gene modeling. We developed a consensus promoter prediction method combining previously developed algorithms with the GENSCAN gene modeling program. Our method, CONPRO (CONsensus PROmoter), identifies promoters with very high confidence, and the predicted promoters are guaranteed to be associated with genes. On our test data set, the method correctly detects promoters for approximately half of all human genes (37%–71%), and most predictions are true promoters (85%–90%). Applying our method to the human genome and human genes from the Unigene data set, we find the promoters for 13,744 genes. Of these, 6440 are genes with a functionally cloned mRNA, and 7304 are novel genes for which only expressed sequence tags (ESTs) are available. Candidate promoters for many novel genes will be a useful resource in elucidating complex biological response mechanisms. CONPRO is available for searching promoters in the human genome (http://stl.bioinformatics.med.umich.edu/conpro).
With the publication of the human genome sequence (International Human Genome Sequencing Consortium 2001; Venter et al. 2001), we now face the daunting task of understanding how the genome functions. Considerable progress has been made in detecting genes by using de novo prediction with programs such as GENSCAN (Burge and Karlin 1997) and genomic sequence alignment of expressed sequences using programs such as BLASTX (Gish and States 1993). The reliability of gene coding sequence identification has been improving, but prediction and characterization of regulatory sequences remain challenging problems. Here, we focus on detecting promoters, which are in the class of regulatory sequences.
A promoter is the region of genomic sequence proximal to the transcription start site (TSS) that is responsible for the initiation of transcription. Promoters are integral components of genes and mediate important transcriptional regulation of genes. A small collection of experimentally defined and carefully curated human promoters is available in the Eukaryotic Promoter Database (EPD; Périer et al. 1998). EPD promoters are sequence fragments of length 500 bp located upstream of TSSs. Recently, 5′ cap cloning techniques have been used to generate libraries enriched in full-length transcripts (Suzuki et al. 2000), but the coverage of these libraries is incomplete and experimental definition of the 5′ untranslated region (UTR) remains challenging. For most human genes, promoters have not been defined or studied, but understanding the regulation of gene expression is an important aspect of understanding the gene function. Reliable recognition and characterization of promoters therefore is a high priority in studying the human genome. The knowledge of promoters will be useful in elucidating regulation and expression mechanisms of genes and may even shed light on the function of novel and uncharacterized genes.
A well-established measure for promoter prediction accuracy scores a prediction of TSS as positive if it is within the range of 200 bp upstream to 100 bp downstream of the true TSS (Fickett et al. 1997). Several groups have developed methods for in silico promoter prediction (Fickett and Hatzigeorgiou 1997; Scherf et al. 2000), such as algorithms considering statistical models for promoters, Markov model audic (Audic and Claverie 1997); neural network learning NNPP (Reese and Eeckman 1995), promoter2.0 (Knudsen 1999); individual residue or oligomer composition, PromFD (Chen et al. 1997); PromFind (Hutchinson 1996); PromoterInspector (Scherf et al. 2000); and the density of transcription factor binding sites in promoters, TSSG and TSSW (Solovyev and Salamov 1997), PromFD, and PROSCAN (Prestridge 1995). There are two major limitations to the practical application of these methods in the whole genome annotation; they produce many false predictions and the predicted promoters are not associated with genes. For most methods, the false-positive rate is estimated at approximately one per kilobase (Fickett and Hatzigeorgiou 1997). In another study, the ratio of true predictions to false predictions is a few percent, with the exception of one method, PromoterInspector, which show predicted accuracy of 43% (Scherf et al. 2000). Still, most predictions using existing methods are false predictions. Because the human genome is >3 billion base pairs in length, it is not practical to apply any one of the promoter prediction methods to the human genome.
Several aspects of genome structure remain to be fully exploited in promoter identification. One possible new feature might be the shape and flexibility of the DNA conformation in promoter regions. Although the contribution of DNA structure to transcription factor binding site recognition has been studied (Liu et al. 1998, 2001), there is still not enough information for characterizing the DNA structure for the whole promoter regions. The ease of helix opening has also been proposed as a possible indicator of a promoter or TSS (Benham 1993). Nevertheless, with the effort that has already been applied to de novo promoter prediction, we believe that creation of yet another novel method is unlikely to lead to dramatic improvements in algorithmic performance. This naturally raises the possibility that combining previously developed methods might provide a superior strategy for promoter identification.
Computational identification of genes and computational identification of promoters have been viewed as distinct problems, but they, in fact, are tightly interwoven. For biological studies, predicted promoters need to be associated with genes. Here, we make use of experimental evidence (gene transcripts) for the location of actively transcribed genes to improve the ability to recognize promoters. The approach is to first align gene transcripts (expressed sequence tags [ESTs] or mRNAs) with genomic sequences to anchor the position of the genes. Because many transcript sequences, especially ESTs, are less than full length (Suzuki et al. 2000), we use GENSCAN to identify the missing 5′ exons of the genes and build a gene model extending upstream of this aligned transcript anchor. The region upstream of the 5′ most predicted exon or mRNA is searched for candidate promoters using TSSG, TSSW, PROSCAN, PromFD, and NNPP. Finally, the predictions of these five different methods are compared, and we generate consensus predictions. TSSG, TSSW, and PROSCAN detect promoters by the presence of transcription factor binding sites. PromFD uses both presence of transcription factor binding sites and base composition bias as indicators of promoters. NNPP uses neural learning to build an overall model for promoters. These five methods predict promoters from different aspects and they complement each other. If several of these complementary methods predict the same promoter, it is unlikely the prediction is false.
The consensus promoter predictor, CONPRO (CONsensus PROmoter), correctly detects promoters for nearly half of human genes in our test data set (71% of genes with known mRNA; 37% of genes with only ESTs known). Among the promoters we identified, most of them are true promoters (90% for genes with mRNA; 85% for genes with only ESTs known). Applying our method to the human genome, considering only the transcripts aligning with large genomic contigs, we find promoters for 13,744 human genes, 7304 of which are novel genes with only ESTs known, and 6440 of them have mRNA known.
RESULTS
In the Unigene build 125, 17,624 of 86,213 clusters contain a known gene, which means a functionally cloned mRNA or complete coding region. The remaining 68,589 clusters contain only ESTs. Most Unigene clusters have ETSs, but many do not have 5′ ESTs. For our analysis, we treat genes as Unigene clusters and divide them into three major categories: clusters with a functionally cloned mRNA, clusters including 5′ ESTs, and clusters with only 3′ ESTs. Our goal is to identify the promoters in the recently published human genome (International Human Genome Sequencing Consortium 2001; Venter et al. 2001) for human genes in these three major categories.
Analysis of Genes for Which a Functionally Cloned mRNA Is Available
We use 133 promoters from EPD (Périer et al. 1998) as a training set for CONPRO. Because the promoters from EPD are experimentally defined, the corresponding mRNAs in the Unigene clusters tend to be well studied, and full-length transcripts are often available. Unfortunately, most mRNA sequences in GenBank cannot be assumed to represent full-length transcripts (Kan et al. 2000). The full-length transcripts are, on average, 45 bp longer than mRNAs in the current databases (Suzuki et al. 2000). To simulate CONPRO's performance based on a typical GenBank mRNA entry, we remove the first 50 bp from the 5′ end of the corresponding 133 mRNAs (full-length transcripts). These truncated mRNAs then are used for promoter prediction. By aligning the truncated mRNAs to genomic sequences, we find the genomic position of the genes. If we simply take the 5′ end of the alignment as the promoter prediction, 68% of the promoters are correctly predicted, but 32% of all predictions are false. False predictions occur when the 50-bp segment that was removed covers an exon–intron junction resulting in an alignment that misses an upstream exon. To reduce false predictions, we use the six promoter prediction tools to look for promoters in the upstream 1.5-kb regions of the truncated mRNA alignments.
On the training set of 133 promoters, the two programs with the best performance are TSSG and PromFD (Table 1). They yield the largest number of true predictions, whereas their rate of false predictions is relatively low. Although the two methods correctly predict 56% (TSSG) or 66% (PromFD) of the 133 promoters, the ratio of true predictions to false predictions, 8 : 1 or 2 : 1, respectively, is still too high. As a result, the predictive power for either method is not sufficient. To reduce the number of false predictions, we seek a consensus among five existing promoter prediction methods: PROSCAN1.7, NNPP2.0, PromFD1.0, TSSG, and TSSW. Our method, called CONPRO, correctly predicts 73% of all the 133 promoters on this training set. The consensus prediction has very high confidence. The ratio of true predictions to false predictions, 12 : 1, is much higher than the previous methods. The improved predictive power of the consensus method will reduce the number of false-positives that need to be experimentally evaluated and will greatly assist the study of gene functions.
Table 1.
Searching Promoters for Genes with Known mRNAs
| Programs | Training set of 133 promoters | Test set of 120 promoters | ||||
|---|---|---|---|---|---|---|
| true prediction (sensitivity) | false prediction (FP/AllP) | undetected | true prediction (sensitivity) | false prediction (FP/AllP) | undetected | |
| PROSCAN1.7 | 32 (24%) | 18 (36%) | 83 | 30 (25%) | 22 (42%) | 68 |
| NNPP2.0 | 56 (42%) | 41 (42%) | 37 | 26 (22%) | 50 (66%) | 44 |
| PromFD1.0 | 88 (66%) | 43 (33%) | 36 | 69 (58%) | 57 (45%) | 23 |
| promoter2.0 | 8 (6%) | 100 (93%) | 25 | 14 (12%) | 92 (88%) | 14 |
| TSSG | 75 (56%) | 10 (12%) | 48 | 62 (52%) | 18 (23%) | 40 |
| TSSW | 57 (43%) | 29 (34%) | 47 | 58 (48%) | 20 (26%) | 42 |
| CONPRO | 97 (73%) | 8 (8%) | 28 | 85 (71%) | 8 (9%) | 27 |
Sensitivity is defined as the percentage of the promoters that are detected by the methods. FP is false predictions and AllP is all predictions. The numbers in the parentheses of FP/AllP are the percentage of false predictions in all the predictions. The number of promoters that are not detected is also listed. The CONPRO can detect promoters for about 71% of the human genes with mRNA known. For the promoters predicted, about 91% of them are true promoters.
For an independent test of CONPRO, we use 120 promoters derived from full-length human cDNAs (Suzuki et al. 2000). The corresponding full-length cDNAs are truncated by 50 bp at the 5′ end as described above. The truncated full-length cDNAs are aligned to genomic sequence, and upstream 1.5 kb regions are searched for promoters. The results on the test set are very similar to the results on the training set (Table 1). CONPRO correctly predicts 71% of 120 promoters with 91% of all predictions being true predictions, an accuracy that is better than any previous methods. We concluded that, if the functionally cloned mRNA of the gene is known, CONPRO can identify promoters for >70% of such genes, and 91% of all predictions are true promoters.
Building Gene Models for Genes When No Functionally Cloned mRNA Is Available
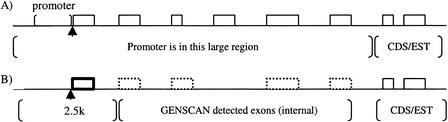
An EST sequence or a complete CDS entry in GenBank almost certainly does not represent the full-length transcript of a gene. The alignment of an EST or a complete CDS to the genomic sequence provides only an anchor for the location of the gene in the genome, but much of the gene is likely to be missing from the alignment. The promoter of the gene could be 20 kb, or even further, upstream of the alignment anchor. As has been shown previously, existing algorithms perform badly in searching for promoters in extended genomic regions (20 kb) because of their high rate of false-positive prediction (approximately one false prediction per kilobase; Fickett and Hatzigeorgiou 1997). Our strategy for improving specificity is to restrict the extent of sequence that needs to be examined. To accomplish this, we use gene modeling to identify as much as possible of the missing upstream components of the gene (Fig. 1).
Figure 1.
GENSCAN detecting the missing exons. (A) After aligning expressed sequence tags (ESTs) to the genomic sequence, the promoter of the gene may be far upstream. A search for the promoter in such large regions is an error-prone process. (solid-line boxes) Exons identified by aligning ESTs to genomic sequence. (arrow) The true transcription start site (TSS). (B) GENSCAN is used to find the missing exons of the gene (dotted-line boxes). Even the external exon (thick-line box) of the gene might not be found by GENSCAN, but we successfully located the promoter to a 2.5-kb region. Finding the promoter in this 2.5-kb region is a much easier job.
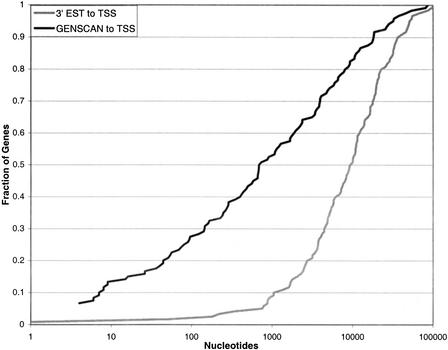
GENSCAN uses a hidden semi-Markov model to identify sets of exons that are likely to form a complete gene (Burge and Karlin 1997). We use GENSCAN to extend the EST/complete CDS alignment-defined gene anchor by scanning a 70-kb genomic region containing the alignment. The 70-kb cutoff is heuristically determined and it limits problems of gene fusion that sometimes occur in GENSCAN predictions. The output of GENSCAN then is filtered to select a gene model that overlaps the EST/complete CDS alignment anchor. In the predicted gene, only high-quality exons that are continuous from the EST/complete CDS are considered as the extension of the anchored gene (Fig. 1). Using the gene model generated by GENSCAN, we can limit the search for a promoter to a much narrower region. In the case of 3′ EST anchored genes, the TSS for nearly two-thirds (63%) of the 120 genes in our test set fall within 2.5 kb of the 5′ most predicted exon (Fig. 2). Without gene modeling, only 20 (17.5%) genes have TSS within 2.5 kb from the loci that are 3′ EST aligned; most TSSs are more than 10 kb away from the alignment.
Figure 2.
Shown in the figure is the cumulative fraction of genes in the Suzuki120 test set as a function of the number of nucleotides in the genome from the true transcription start site (TSS) to the (dark line) 5′ most high-quality GENSCAN predicted exon or (gray line) 3′ expressed sequence tag (EST) match.
The use of GENSCAN models allows us to search a considerably smaller region (2.5 kb) for promoters with only a modest decrease in search sensitivity. Furthermore, the predicted promoters almost certainly initiate the transcription of the gene modeled by GENSCAN. To identify promoters in this 2.5-kb region, we use TSSG, PromFD, TSSW, PROSCAN, and NNPP to predict candidate promoters, and these candidate promoters are compared to generate a consensus prediction.
Analysis of Genes When Only Complete Coding Regions Are Available
Although it is not one of our three major categories, for some genes in GenBank only the coding sequence is annotated. In addition, when annotating genomic sequence of species other than human, functionally cloned mRNA or ESTs may not be available; therefore, coding regions identified using TBLASTN may be a major source of gene identification (Gish and States 1993). Our approach to promoter prediction in this case is similar to that described above. We use the conceptually translated coding sequence alignment to anchor a gene model and then search the region upstream of the 5′ most predicted exon for candidate promoters. We analyze the performance of this approach using the same training set of 133 promoters. The longest open reading frame (ORF) in each mRNA is considered the complete coding region of the mRNA. Although this ignores the possibility of alternative translation start site or alternative splicing, this reflects widespread practice in the molecular biology community. CONPRO aligns the coding region sequence to the genomic sequence, uses GENSCAN to generate a gene model that overlaps the alignment anchor, and searches for candidate promoters in the region 1.5 kb upstream of the 5′ most predicted exon. CONPRO finds 86 of the 133 promoters in the training set (65%), which is higher than all other methods. Of the 95 predictions made by CONPRO, 91% are correct (Table 2). The 5′ UTRs of human genes have average length of 125 bp (Suzuki et al. 2000). By comparison, if the 5′ ends of the CDS alignment are taken as predictions of promoters, using our scoring criterion of positive predictions, 53% of promoters are correctly picked up, but 47% of the predictions by this simple procedure are false. GENSCAN extension alone does help a little in this case. After the extension, 56% of the ends of 5′ most exons are in −200 to 100 relative to the true TSS. However, the false-positive rate is still very high, 44%.
Table 2.
Searching Promoters for Genes with Known Complete Coding Region
| Programs | Training set of 133 promoters | Test set of 120 promoters | ||||
|---|---|---|---|---|---|---|
| true prediction (sensitivity) | false prediction (FP/AllP) | undetected | true prediction (sensitivity) | false prediction (FP/AllP) | undetected | |
| PROSCAN1.7 | 31 (23%) | 21 (40%) | 81 | 26 (22%) | 28 (52%) | 66 |
| NNPP2.0 | 63 (47%) | 50 (44%) | 21 | 24 (20%) | 63 (72%) | 33 |
| PromFD1.0 | 53 (40%) | 44 (45%) | 48 | 60 (50%) | 63 (51%) | 10 |
| promoter2.0 | 9 (7%) | 101 (92%) | 23 | 10 (8%) | 92 (90%) | 18 |
| TSSG | 78 (57%) | 13 (14%) | 42 | 56 (47%) | 27 (33%) | 37 |
| TSSW | 58 (44%) | 36 (38%) | 39 | 56 (47%) | 38 (40%) | 26 |
| CONPRO | 86 (65%) | 9 (9%) | 38 | 75 (63%) | 10 (12%) | 35 |
The CONPRO can detect promoters for about half of the human genes with mRNA known. For the promoters predicted, about 88% of them are true promoters. Additional information is given in the legend of Table 1.
We then tested the performance of CONPRO using 120 independent promoters derived from full-length human cDNAs (Suzuki et al. 2000). Each full-length cDNA is searched for the longest ORF, and this is used as the complete coding region, as described. In this case CONPRO correctly identifies >60% of all promoters. Of the 85 promoters predicted by CONPRO, 88% are correct.
Analysis of Genes When Only 3′ ESTs Are Known
Identifying promoters for human genes is a more challenging problem when the only experimental evidence available is a 3′ EST sequence. Almost all 3′ ESTs are partial sequences that do not represent the full-length transcripts. As expected, most promoters are far upstream of the aligned genomic location at which a 3′ EST aligns (Fig. 1). Using the scoring criterion for positives, with the 5′ end of a 3′ EST alignment to the genome as a prediction of promoters, we can predict only 2% of all promoters (e.g., 98% false-positive rate). Our approach to locate a promoter given a 3′ EST is to use genomic alignment of the 3′ EST sequence to anchor a gene. As described above, GENSCAN is used to predict a gene model in a 70-kb region around the 3′ EST alignment. GENSCAN extension alone can get much better estimates of the 5′ end of the genes. If the 5′ end of the 5′ most predicted exon is taken as promoter prediction, 23% promoters can be picked up by GENSCAN extension, with 77% false predictions. The TSS typically falls within a region 2.5 kb upstream of the 5′ most exon in the gene model (Fig. 2). This region is searched for candidate promoters by using each promoter prediction program, and CONPRO compares these predictions to generate a consensus prediction.
To examine the performance of CONPRO when a 3′ EST sequence is available, we use a 500-bp sequence from the 3′ end of functionally cloned mRNA or full-length cDNA as simulated 3′ EST. As a training set, we use the 118 promoters from EPD for which the mRNA alignment is completely contained in a single genomic contig. The results of promoter searching are presented in Table 3. Again, among the six existing programs we used, none of them provides sufficient predictive power to be of practical use in genome annotation. PromFD1.0 and TSSG are still the best two methods. TSSG correctly predict 33% of the 118 promoters, whereas PromFD can predict 32% correctly. The ratio of true predictions to false predictions is 3.5 : 1 for TSSG and 1 : 1 for PromFD. On this training set, CONPRO can successfully identify 38% of the 118 promoters with a ratio of true predictions to false predictions of 7.5 : 1.
Table 3.
Searching Promoters for Genes with Known 3′ ESTs
| Programs | Training set of 118 promoters | Test set of 120 promoters | ||||
|---|---|---|---|---|---|---|
| true prediction (sensitivity) | false prediction (FP/All P) | undetected | true prediction (sensitivity) | false prediction (FP/All P) | undetected | |
| PROSCAN1.7 | 15 (13%) | 17 (53%) | 99 | 12 (10%) | 21 (64%) | 97 |
| NNPP2.0 | 33 (28%) | 43 (57%) | 56 | 20 (17%) | 67 (77%) | 45 |
| PromFD1.0 | 38 (32%) | 33 (46%) | 78 | 36 (30%) | 61 (63%) | 62 |
| promoter2.0 | 6 (5%) | 99 (94%) | 13 | 7 (6%) | 104 (94%) | 9 |
| TSSG | 39 (33%) | 11 (22%) | 68 | 32 (27%) | 30 (48%) | 58 |
| TSSW | 32 (27%) | 19 (37%) | 67 | 27 (23%) | 31 (53%) | 62 |
| CONPRO | 45 (38%) | 6 (12%) | 67 | 44 (37%) | 8 (15%) | 68 |
CONPRO predicts promoters for about 37% human genes. Of all the predictions made by CONPRO, about 85% are correct. Additional information is given in the legend of Table 1.
For an independent test of CONPRO, we use 120 promoters derived from full-length cDNAs (Suzuki et al. 2000). The 3′ ESTs are simulated as described above from full-length cDNAs. The searching results on the test set are similar to the training set. More than half of the predictions of the individual methods are false predictions. CONPRO can reduce the rate of false predictions and is able to predict 37% of the 120 promoters with six true-positives for every false-positive. We conclude that CONPRO predictions provide more reliable promoter locations for 37% of novel genes when only 3′ EST sequence is available.
Analysis of Genes When 5′ EST Sequence Is Available
The last category of genes is the set of Unigene clusters that include 5′ EST sequences. When an experimentally defined 5′ EST is available, there is an obvious approach to promoter prediction: take the 5′ end of the EST as TSS. Using the criterion for scoring positives, 38% of the 5′ ends of the 5′ EST sequence alignment are true-positive predictions and 62% are false-positives.
To evaluate the performance of CONPRO in this case, we again generate training and test data sets. It is controversial as to how best the 5′ EST should be simulated using a functionally cloned mRNA as a reference. To circumvent this issue, we select a 5′ EST at random from the Unigene cluster containing the mRNA corresponding to the promoter in our training and test sets. In the training set of EPD promoters, 102 EPD promoters are associated with an mRNA and the Unigene cluster containing 5′ EST sequences. The selected 5′ EST is aligned to genomic sequences and GENSCAN is again used to build a gene model overlaps with the EST alignment. After GENSCAN extension, if the 5′ end of the 5′ most exon is taken as promoter prediction, 43% of all promoters can be picked up, with a 57% false-positive rate. The upstream 2.5-kb region of the 5′ most predicted exon is searched for promoters. The best two programs are TSSG and PromFD, with 51% and 46% of the 102 promoters detected, respectively (Table 4). However, the ratio of true predictions to false predictions is 1 : 1 for PromFD and 4 : 1 for TSSG. Using CONPRO, we have identified promoters for 58% of the 102 genes with the ratio of true predictions to false predictions of 8 : 1.
Table 4.
Searching Promoters for Genes with Known 5′ ESTs
| Programs | Training set of 102 promoters | Test set of 110 promoters | ||||
|---|---|---|---|---|---|---|
| true prediction (sensitivity) | false prediction (FP/All P) | undetected | true prediction (sensitivity) | false prediction (FP/All P) | undetected | |
| PROSCAN1.7 | 25 (25%) | 17 (40%) | 75 | 18 (16%) | 25 (58%) | 78 |
| NNPP2.0 | 41 (40%) | 46 (53%) | 31 | 20 (18%) | 71 (78%) | 30 |
| PromFD1.0 | 47 (46%) | 45 (49%) | 48 | 42 (38%) | 52 (55%) | 47 |
| promoter2.0 | 5 (5%) | 92 (95%) | 5 | 8 (7%) | 86 (91%) | 16 |
| TSSG | 52 (51%) | 13 (20%) | 37 | 33 (30%) | 32 (49%) | 45 |
| TSSW | 35 (34%) | 28 (44%) | 39 | 31 (28%) | 41 (57%) | 38 |
| CONPRO | 59 (58%) | 7 (11%) | 39 | 41 (37%) | 7 (15%) | 62 |
CONPRO can find promoters for about 37% human genes. Of all the predictions made by CONPRO, about 85% are correct. Additional information is given in the legend of Table 1.
For an independent test set, we use 110 promoters derived from full-length human cDNAs that are associated with Unigene clusters containing a 5′ EST. Using this test set, CONPRO identifies promoters for 37% of these genes, and 85% predictions are true-positives. We conclude that gene modeling and consensus promoter search yields more true-positives and far fewer false predictions compared with simply taking the 5′ end of a 5′ EST as the TSS.
Prediction of Promoters in the Human Genome
Having a reliable method for promoter prediction, we can search for promoters in the extended regions of the human genome. We use the Golden path data set for human genomic sequence (downloaded from http://genome.ucsc.edu, release of April 2001) and expressed sequence data from human Unigene clusters (build 125). Only the genes aligning with large contigs are considered for further promoter prediction. For the 17,624 human Unigene clusters containing mRNA, we align them individually to the genomic sequence. The upstream 1.5-kb regions of the aligned genomic loci are searched for promoters. CONPRO detects promoters for 6440 human genes in this category. Greater than 90% of these promoters are expected to be true promoters. For the 68,589 human Unigene clusters containing only EST sequences, promoters are found for 6627 human genes with only 3′ ESTs known, and 677 human genes with 5′ ESTs known.
CONPRO predicts promoters with very high confidence. Of 7304 promoters of novel genes for which only ESTs are known, ∼85% are likely to be correct, and >90% of 6440 promoters for functionally cloned mRNA anchored predictions are likely to be correct. We have set up a Web server: http://stl.bioinformatics.med.umich.edu/conpro/. The server takes mRNA or EST sequences as input and looks for the corresponding promoter of the gene in the human genome. The set of 13,744 promoters identified in this study is also available from the Web server.
DISCUSSION
In this study, we propose a new angle to look at an old problem: de novo promoter prediction. The traditional approach to promoter prediction is to search the genome for sequences with the characteristics of known promoter regions without making reference to expressed sequence data. Because the human genome is very large (International Human Genome Sequencing Consortium 2001; Venter et al. 2001), and previous promoter prediction tools make many false predictions (Fickett and Hatzigeorgiou 1997; Scherf et al. 2000), this approach has not been useful in annotating promoters for the whole human genome. We introduce a new source of data, transcripts of genes, which has not been used previously in promoter prediction. Using expressed sequence data to anchor genes, we can limit the genomic sequence regions that needed to be searched to locate a promoter and thereby increase the specificity of our predictions.
Our approach has two big advantages. One is that we limit the search to regions of the genome that have very high probability of containing promoters; therefore, the number of false predictions is reduced while the search sensitivity is still satisfying. The second advantage is that the promoters we identified are associated with the genes of the transcript. This is very important for biological studies of promoters, both for individual gene or global expression studies using microarrays.
Another reason CONPRO outperforms previous promoter prediction tools is that we use multiple programs instead of a single program to predict the promoters. The five methods we used explore promoters from different aspects and really complement each other. If several of the five complementary methods have the same prediction, it is less likely that the prediction is false. Therefore, we substantially reduce the number of false-positives while CONPRO still has sensitivity comparable to or better than previous tools.
CONPRO is very successful in identifying the promoters for genes when location information derived from mRNA or ESTs is available. For genes in which a functionally cloned but possibly truncated mRNAs is available, it can correctly predict the promoters for >70% the genes, and ∼91% of the predictions are true promoters. If no functionally cloned mRNA is available and all we know is EST sequences, we still can correctly identify promoters for ∼37% of such genes. Of the promoters we find for these EST genes, 85% of them are correct. Applying our method to the human genome and human genes in Unigene clusters, considering only the genes aligning with large genomic contigs, we find promoters for 13,744 human genes, of which 7304 are novel genes with only ESTs known, and 6440 of the 13,744 genes have mRNA known.
In our view, gene identification and promoter identification are tightly interwoven. However, previous de novo gene prediction tools are not suitable for promoter identification. The reason is gene prediction tools tend to split and fuse genes very often. In addition, it is difficult for them to find the 5′ UTRs of the genes. The best of these tools, GENSCAN, only gets the 5′ gene boundary in the −200 to 100 region relative to true TSS for 32% of the 120 genes in the test set. In other words, 68% of the 5′ gene boundaries, if used as promoter predictions, are incorrect.
Current estimates suggest that the human genome contains on the order of 35,000 genes. Some genes may not be represented in the EST collections. Furthermore, we did not consider mRNAs or ESTs aligned to short genomic contigs in the published human genome. Finally, given a genomically aligned gene transcript, the sensitivity of our method was approximately half of all human genes. The 13,744 candidate promoters we identified here therefore is in reasonable agreement with the number of predictions expected assuming that the human genome contains on the order of 35,000 genes.
We have not considered the phenomenon of multiple or alternative promoters for a transcription unit. Our estimated sensitivity for detection of a promoter associated with a transcription unit (85%–90%) may be higher if alternative promoters were taken into consideration. Because alternative promoters may mediate differential or tissue specific gene expression, the identification of alternative promoters remains a high priority in the subject on which we are actively working.
METHODS
Human Genome Sequence and Genes
The human genome sequence is downloaded from Golden path assembled human genome release April 2001 (http://genome.ucsc.edu). The transcript sequences (mRNA or ESTs) of human genes are downloaded from Unigene data set build 125. For the first pass of aligning the Unigene data set to genomic sequences, our group has developed a fast method called multi (D.J. States, unpubl.) designed to rapidly identify near identity sequence matches between large collections of query and target sequences. Only EST matches to XNU masked genomic sequence containing a run of at least 20 identities are considered further. Final alignments of EST (or mRNA) sequences to the genome are generated using sim4 (Florea et al. 1998) to define the exon boundaries more precisely.
Promoter Sets
The training set for our promoter prediction analyses is a set of nonredundant human and mouse promoters in EPD63 (Périer et al. 1998) for which an unambiguous alignment in the finished genome sequence can be established with at least 5 kb of flanking region upstream. Based on the EPD release 63 and the available genome sequences, this results in a set of 133 promoters including 120 human promoters and 13 mouse promoters. The mRNAs corresponding to the 133 promoters are found in the Unigene data set. The 5′ EST training set consists of 5′ EST from the Unigene clusters containing the EPD sequences. Twenty-one of the 133 clusters have no 5′ EST. Therefore, the 5′ EST training set has 102 promoters. The 3′ EST training set was simulated using the 3′ 500 bp of the functionally cloned mRNA. For 15 sequences, the genomic contigs do not include the 3′ end of the gene and therefore were eliminated. This results in a 3′ EST training set of 118 sequences.
Because the previously published methods for promoter prediction (PromFD, PROSCAN, TSSW, TSSG, and NNPP) were trained on promoters in EPD, we develop an independent test set of promoters derived by aligning full-length cDNAs with genomic sequence. The full-length human cDNAs are selected from the set of oligo-capped cDNA cloned by Suzuki (Suzuki et al. 2000). Sixty-three percent of clones in such libraries are full length (Sugahara et al. 2001); we selected a subset of 954 clustered full-length cDNA from this set of 10,000 clones (Suzuki et al. 2000). They are much more likely to be real full length. The derived promoters with sequence similar to EPD promoters are excluded, as are full-length cDNAs significantly shorter than the existing functionally cloned mRNA. Finally, full-length cDNAs not contained in a genomic contig are eliminated. This results in a test set of 120 promoters. Unigene clusters containing 5′ ESTs are found for 110 of the 120 full-length cDNAs.
Promoter Prediction Programs
Among the recently developed, actively maintained promoter prediction programs, we select six that are available through Internet access. They are PROSCAN1.7, NNPP, PromFD1.0, promoter2.0, TSSG, and TSSW. PromFD has been downloaded and installed locally. The other five programs are used directly from their Web servers. For the application in this study, we have modified PromFD so that it produced both promoters and the score of the prediction. Furthermore, the threshold and window shifting steps of PromFD also have been adjusted. The window shifting steps are reduced from 150 to 20 bp.
Because of the restrictions on the PromoterInspector Web site (Scherf et al. 2000), only 35 genomic fragments length 2.5 kb containing EPD promoters are tested, generating 11 predictions of which five are correct according to the criterion in the previous promoter prediction literature (Fickett and Hatzigeorgiou 1997). Similarly, using 20 experimentally defined promoters on chromosome 22 (Scherf et al. 2001), PromoterInspector made 10 predictions near the promoter loci, of which five are correct. Because the sensitivity is relatively low and Web site restrictions prevented convenient use of PromoterInspector, this tool is not included in our analysis.
CONPRO Prediction
We develop CONPRO, which combines existing methods, TSSG, TSSW, NNPP, PROSCAN, and PromFD, for predicting promoters in the upstream regions of genes. For each program, the highest score prediction is taken as the prediction of the program in the region. If three predictions fall in a 100-bp region, this is considered a consensus prediction. If no three-way consensus is achieved, we then check whether TSSG and PromFD predictions fall within the window. For CONPRO, the TSS is calculated as mean of the individual predictions. Consistent with the previous promoter prediction literature, we score a prediction of TSS as positive if it is within the range from 200 bp upstream to 100 bp downstream of the true TSS (Fickett and Hatzigeorgiou 1997).
WEB SITE REFERENCES
http://genome.ucsc.edu; site from which to download the Golden path data set for human genomic sequence.
http://stl.bioinformatics.med.umich.edu/conpro; site at which CONPRO is available for searching promoters in the human genome.
Acknowledgments
We thank Tom Blackwell and the rest of the group for insightful discussions and Richard McEachin for careful reading of the manuscript. We also thank Dr. Gary Stormo at Washington University for providing the source codes for PromFD. We are grateful to the groups maintaining the Web servers of NNPP, TSSG, TSSW, promoter2.0, and PROSCAN (at CIT of NIH). This work was supported in part through grants from the National Institutes of Health (HG-R01–01391), the Department of Energy (DE-FG02–94ER61910), and the Merck Foundation for Genome Research (Grant 225).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dstates@umich.edu; FAX (734) 615-6553.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.198002.
REFERENCES
- Audic S, Claverie JM. Detection of eukaryotic promoters using Markov transition matrices. Comput Chem. 1997;21:223–227. doi: 10.1016/s0097-8485(96)00040-x. [DOI] [PubMed] [Google Scholar]
- Benham CJ. Sites of predicted stress-induced DNA duplex destabilization occur preferentially at regulatory loci. Proc Natl Acad Sci. 1993;90:2999–3003. doi: 10.1073/pnas.90.7.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chen QK, Hertz GZ, Stormo GD. PromFD 1.0: A computer program that predicts eukaryotic pol II promoters using strings and IMD matrices. Comput Appl Biosci. 1997;13:29–35. doi: 10.1093/bioinformatics/13.1.29. [DOI] [PubMed] [Google Scholar]
- Fickett JW, Hatzigeorgiou AG. Eukaryotic promoter recognition. Genome Res. 1997;7:861–878. doi: 10.1101/gr.7.9.861. [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Hutchinson GB. The prediction of vertebrate promoter regions using differential hexamer frequency analysis. Comput Appl Biosci. 1996;12:391–398. doi: 10.1093/bioinformatics/12.5.391. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Kan Z, Gish W, Rouchka E, Glasscock J, States DJ. UTR reconstruction and analysis using genomically aligned EST sequences. ISMB. 2000;8:218–227. [PubMed] [Google Scholar]
- Knudsen S. promoter2.0: For the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- Liu R, Blackwell TW, States DJ. A structure based similarity measure for nucleic acid sequence comparison. In: Istrail S, et al., editors. Proceedings of the Second Annual Conference of Computational Biology (RECOMB98). New York: ACM Press; 1998. pp. 173–181. [Google Scholar]
- ————— Conformational models for binding site recognition by the E. coli MetJ transcription factor. Bioinformatics. 2001;17:622–633. doi: 10.1093/bioinformatics/17.7.622. [DOI] [PubMed] [Google Scholar]
- Périer RC, Junier T, Bonnard C, Bucher P. The eukaryotic promoter database EPD. Nucleic Acids Res. 1998;26:353–357. doi: 10.1093/nar/26.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge DS. Predicting Pol II promoter sequences using transcription factor binding sites. J Mol Biol. 1995;249:923–932. doi: 10.1006/jmbi.1995.0349. [DOI] [PubMed] [Google Scholar]
- Reese, M.G. and Eeckman, F.H. 1995. Novel neural network algorithms for improved eukaryotic promoter site recognition. Accepted talk for The Seventh International Genome Sequencing and Analysis Conference. Hyatt Regency, Hilton Head Island, SC.
- Scherf M, Klingenhoff A, Werner T. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: A novel context analysis approach. J Mol Biol. 2000;297:599–606. doi: 10.1006/jmbi.2000.3589. [DOI] [PubMed] [Google Scholar]
- Scherf M, Klingenhoff A, Frech K, Quandt K, Schneider R, Grote K, Frisch M, Gailus-Durner VV, Seidel A, Brack-Werner R, et al. First pass annotation of promoters on human chromosome 22. Genome Res. 2001;11:333–340. doi: 10.1101/gr.154601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V, Salamov A. The Gene-Finder computer tools for analysis of human and model organisms genome sequences. ISMB. 1997;5:294–302. [PubMed] [Google Scholar]
- Sugahara Y, Carninci P, Itoh M, Shibata K, Konno H, Endo T, Muramatsu M, Hayashizaki Y. Comparative evaluation of 5′-end-sequence quality of clones in CAP trapper and other full-length-cDNA libraries. Gene. 2001;263:93–102. doi: 10.1016/s0378-1119(00)00557-6. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ishihara D, Sasaki M, Nakagawa H, Hata H, Tsunoda T, Watanabe M, Komatsu T, Ota T, Isogai T, et al. Statistical analysis of the 5′ untranslated region of human mRNA using “Oligo-Capped” cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]