Abstract
Sonic hedgehog (Shh) is a secreted signaling molecule for tissue patterning and stem cell specification in vertebrate embryos. Shh mediates both long-range and short-range signaling responses in embryonic tissues through the activation and repression of target genes by its Gli transcription factor effectors. Despite the well-established functions of Shh signaling in development and human disease, developmental target genes of Gli regulation are virtually unknown. In this study, we investigate the role of Shh signaling in the control of Myf5, a skeletal muscle regulatory gene for specification of muscle stem cells in vertebrate embryos. In previous genetic studies, we showed that Shh is required for Myf5 expression in the specification of dorsal somite, epaxial muscle progenitors. However, these studies did not distinguish whether Myf5 is a direct target of Gli regulation through long-range Shh signaling, or alternatively, whether Myf5 regulation is a secondary response to Shh signaling. To address this question, we have used transgenic analysis with lacZ reporter genes to characterize an Myf5 transcription enhancer that controls the activation of Myf5 expression in the somite epaxial muscle progenitors in mouse embryos. This Myf5 epaxial somite (ES) enhancer is Shh-dependent, as shown by its complete inactivity in somites of homozygous Shh mutant embryos, and by its reduced activity in heterozygous Shh mutant embryos. Furthermore, Shh and downstream Shh signal transducers specifically induce ES enhancer/luciferase reporters in Shh-responsive 3T3 cells. A Gli-binding site located within the ES enhancer is required for enhancer activation by Shh signaling in transfected 3T3 cells and in epaxial somite progenitors in transgenic embryos. These findings establish that Myf5 is a direct target of long-range Shh signaling through positive regulation by Gli transcription factors, providing evidence that Shh signaling has a direct inductive function in cell lineage specification.
Keywords: Sonic hedgehog, Gli, Myf5, somites, muscle specification
Sonic hedgehog (Shh) is a member of the Hedgehog (HH) family of signaling molecules. In vertebrates, Shh functions as a morphogen to mediate long-range and short-range signaling for dorsal–ventral patterning in the specification of neural, mesodermal, and endodermal stem cell lineages (Chiang et al. 1996; Ericson et al. 1996; Goodrich et al. 1997; Hebrok et al. 2000; Ramalho-Santos et al. 2000), for limb patterning (Riddle et al. 1993), for granular cell proliferation (Wechsler-Reya and Scott 1999), and for suppression of tissue apoptosis (Teillet et al. 1998). Mutations that disrupt Shh signaling cause human birth defects (Muenke and Cohen 2000) and cancer susceptibility in mice and humans (Goodrich and Scott 1998; Hahn et al. 1999). Hedgehog signaling in Drosophila controls cuticular patterning in embryos and tissue patterning during later development (Dahmann and Basler 2000). HH signals are transduced to responding embryonic cells through HH binding to the Patched receptor (Ingham et al. 1991; Marigo et al. 1996a; Stone et al. 1996; Fuse et al. 1999), which is a negative regulator of HH signaling in the absence of ligand. HH binding to Patched blocks its repression of another transmembrane protein, Smoothened (Smo), which induces signal transduction by activating the nuclear translocation of Ci/Gli, zinc-finger transcription factors (van den Heuvel and Ingham 1996). In vertebrates, HH signal transduction is mediated through three Gli transcription factor effectors (Hui et al. 1994). Drosophila Ci has dual functions. As an unprocessed form, Ci mediates positive transcriptional responses to Hedgehog signaling activity, whereas its processed forms function as a transcription repressor (Porter et al. 1995; Aza-Blanc et al. 1997; Chen et al. 1999). In the vertebrate limb, Gli3 is differentially processed to form a transcription repressor under conditions of low Shh signaling, providing a mechanism controlling the differential activity of Gli3 in response to long-distance and short-distance Shh signaling (Wang et al. 2000). Long-distance HH signaling in embryos requires its autoprocessing and N-terminal cholesterol modification (Lewis et al. 2001; Zeng et al. 2001) as well as heparan-sulfate proteoglycans to promote its extracellular transmission (Bellaiche et al. 1998). Graded Shh signaling acts to mediate positive and negative regulation of neural tube patterning of neural cell fates (Roelink et al. 1995; Ericson et al. 1997; Briscoe and Ericson 1999), although the role of the Gli proteins in these processes remains unclear.
Although the importance of Shh signaling in development and human disease is well established, the transcriptional targets of Shh signaling through Gli transcription factors are virtually unknown. In response to signal transduction processes initiated by HH signaling, Gli/Ci transcription factors are thought to undergo nuclear translocation (Robbins et al. 1997; Sisson et al. 1997; Kogerman et al. 1999), leading to their binding to target sequence motifs in the regulatory elements of target genes (Kinzler and Vogelstein 1990; Von Ohlen et al. 1997). Gli1 and Gli2 function as positive transcription factor regulators, whereas processed Gli3 functions as a negative regulator (Sasaki et al. 1999; Wang et al. 2000). In Drosophila and vertebrates, components of the HH signal transduction pathway itself are subject to direct positive and negative feedback regulation. Patched expression is positively regulated by HH signaling through Gli/Ci regulation, providing a negative feedback mechanism for Shh signal transduction (Alexandre et al. 1996; Goodrich et al. 1996; Marigo et al. 1996b). In vertebrates, Gli1 also is positively and directly regulated by HH signaling through essential Gli-binding sites (Lee et al. 1997; Dai et al. 1999), providing a mechanism for positive regulation of HH signaling. Only a few developmental regulatory genes have been identified as direct targets of Gli/Ci regulation. In Drosophila, HH signaling regulates Wingless in cuticle patterning (Heemskerk and DiNardo 1994) through Ci-binding sites (Von Ohlen et al. 1997). In vertebrate embryos, HNF3-β is an early developmental regulator of notochord and floor plate formation (Ang and Rossant 1994; Weinstein et al. 1994; Dufort et al. 1998), and its expression in the floor plate is controlled by Shh and Gli2 regulation (Ding et al. 1998; Matise et al. 1998) through a floor plate enhancer that has an essential Gli-binding site (Sasaki et al. 1997).
Shh signaling influences a diversity of developmental processes, including proliferation (Wechsler-Reya and Scott 1999) and apoptosis (Teillet et al. 1998; Borycki et al. 1999), which has led to controversy as to whether Shh has inductive or trophic signaling functions. Specifically, it has been unknown whether Shh signaling, through its Gli transcription factor effectors, regulates target genes that control cell lineage specification or, alternatively, regulates target genes that mediate the survival and proliferation of specific embryonic cell lineages. To understand the functions of Shh in muscle cell lineage specification, we have investigated the role of Shh signaling in the control Myf5, a bHLH transcription factor gene that controls the specification of myogenic stem cell lineages in the vertebrate embryo (Rudnicki et al. 1993; Tajbakhsh et al. 1996b). Myf5 is activated at different anatomical sites in the embryo under the control of distinct cis-acting regulatory elements (Hadchouel et al. 2000; Summerbell et al. 2000; Carvajal et al. 2001), responding to different developmental signals (Munsterberg et al. 1995; Borycki et al. 1998; Tajbakhsh et al. 1998). Genetic and explant induction experiments have established that Shh produced by the notochord is an essential signal for the activation of Myf5 in epaxial muscle progenitors of the dorsal somite, but not in muscle progenitors at other sites of Myf5 activation and myogenic specification in the embryo (Borycki et al. 1999). In Shh mutant embryos, Myf5 fails to be activated specifically in the epaxial region of the somite, in the absence of localized defects in cell proliferation and apoptosis. However, these studies did not resolve whether Myf5 is a direct target of Shh signaling through Gli regulation, or alternatively, whether Myf5 regulation is a secondary response to Shh signaling.
To investigate the function of Gli transcription factors in Myf5 regulation, we identified an Myf5 epaxial somite (ES) enhancer and characterized its response to Shh signaling using a combination of transgenic and cell transfection approaches. This ES enhancer also was identified among a collection of regulatory elements in large-scale transgenic expression surveys of the 650-kb domain of the mouse MRF4/Myf5 locus (Zweigerdt et al. 1997; Hadchouel et al. 2000; Summerbell et al. 2000; Carvajal et al. 2001). These enhancers control the activation and maintenance of Myf5 and MRF4 expression in limb, branchial arch, and epaxial and hypaxial muscle progenitors. The ES enhancer is unique in the MRF4/Myf5 locus for its transcriptional activity in the epaxial muscle progenitors of the dorsal somite. In this study, we have investigated in detail the developmental activities of the ES enhancer in somites of developing wild-type embryos and homozygous and heterozygous Shh mutant embryos (Chiang et al. 1996), as well as in Shh-responsive 3T3 cells (Taipale et al. 2000), a culture model of somitic mesoderm progenitors (Taylor and Jones 1979). These studies show that Shh signaling controls the activation of the Myf5 ES enhancer, both in epaxial muscle progenitor cells during somite formation and in 3T3 cells responding to Shh induction. Furthermore, Shh regulation of the ES enhancer in somites and in 3T3 cells is mediated through an essential Gli transcription factor binding site. These findings establish that Myf5 is a direct target of long-range Shh signaling to control the specification of somite progenitor cells of the epaxial myogenic lineage. Furthermore, in contrast to the limb and neural tube, where long-range Shh signaling mediates negative regulation (Briscoe and Ericson 1999; Wang et al. 2000), we show that Myf5 in the dorsal somite responds to Shh signaling through positive regulation by Gli transcription factors, establishing that long-range Shh signaling has a direct transcriptional function in Myf5 regulation for specification of epaxial muscle progenitors.
Results
An Myf5 transcription enhancer specific for somite epaxial muscle progenitors
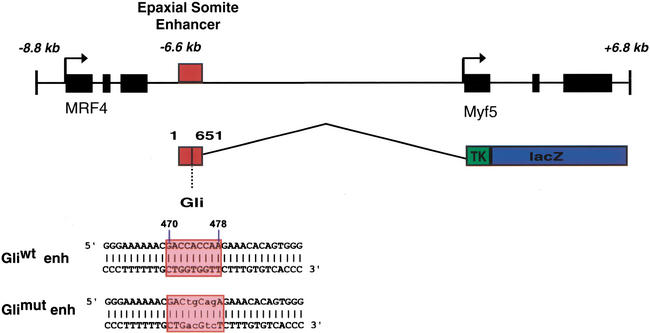
A BAC clone of the MRF4/Myf5 locus was recovered from a mouse genomic DNA library, and subclones were characterized by sequence analysis and functional enhancer assays using transient transgenesis with a lacZ gene reporter expressed under the control of a heterologous viral tk promoter (Goldhamer et al. 1992). These studies identified a 651-bp enhancer located 6.6 kb 5′ of the Myf5 transcription start site (Tajbakhsh et al. 1996a) in the MRF4/Myf5 intragenic region (Fig. 1). This 651-bp Myf5 enhancer activates reporter expression in epaxial somite muscle progenitors as well as brain cells in 9.5-days-postcoitum (dpc) embryos (Fig. 2) and has extensive (86%) identity with the equivalent region 3′ of rat MRF4 (Pin et al. 1997), including a conserved sequence related to Gli binding-site sequences (Kinzler and Vogelstein 1990).
Figure 1.
Organization of the Myf5 ES enhancer in the MRF4/Myf5 locus and the lacZ transgene. A 651-bp Myf5 ES enhancer element is located 6.6 kb upstream of the Myf5 transcription site, adjacent to the 3′ end of the MRF4 gene. The transcriptional activity of the ES enhancer was assayed using a lacZ transgene with a viral tk promoter, as shown. A candidate Gli-binding site (Gliwt enh) is located at 470 bp in the enhancer. The core region of the Gli-binding site in the Myf5 ES enhancer (red box) is variant at position 477 from the consensus Gli-binding site defined by site selection (Kinzler and Vogelstein 1990) and at postions 472 and 477 from the Gli site in the HNF-3β floor plate enhancer (Sasaki et al. 1997). Nucleotide substitution mutations introduced into this Gli sequence (Glimut enh) disrupt its Gli-binding and functional activity.
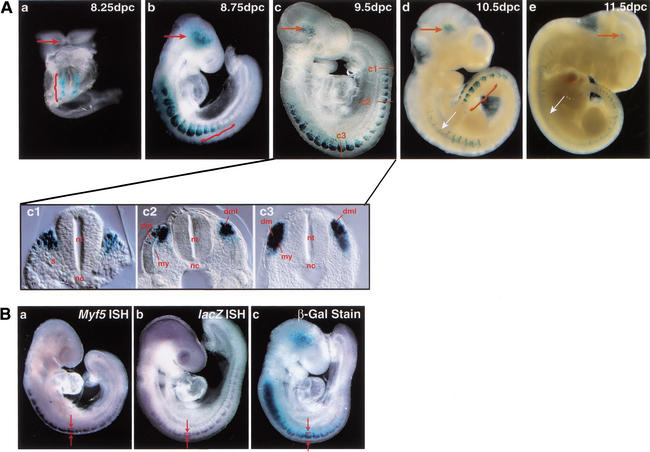
Figure 2.
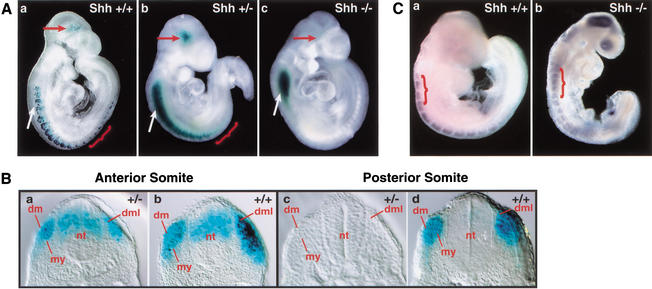
The Myf5 ES enhancer/lacZ transgene is activated in the brain and in epaxial somites in coordination with somite formation. (A) ES enhancer/lacZ transgene expression was assayed by histoenzymatic β-gal staining of whole-mount preparations of 8.25–11.5-dpc embryos recovered from E1 germ-line females, heterozygous for the ES enhancer/lacZ transgene. Specific staining was detected in the brain (red arrow) at all stages and in all somites along the anterior to posterior embryo axis at early stages (panels a–c), and predominantly in eight newest-born posterior somites in later stages (panels d,e). Ectopic dorsal neural tube staining is abundant in the E1 germ-line transgenic embryos (white arrow), but not in transient transgenic embryos (see Fig. 6A). At 8.25 dpc (a), staining is observed in the occipital somites, which are the first somites formed in the embryo, and at 8.75 dpc (b) and 9.5 dpc (c), staining is observed in somites along the entire axis, including the newest-formed posterior somites (red brackets). By 10.5 dpc, staining is predominant in the eight newest-formed posterior somites (red bracket), and only scattered staining is observed in more mature, anterior somites. A 9.5-dpc embryo (c) was sectioned transversely along the anterior to posterior embryo axis, in the most posterior, newly formed epithelial somite (panel c1), an immediately anterior somite that has formed a dermotome and dorsal medial lip (c2), and an anterior somite that has initiated myotome formation (c3). (s) Sclerotome; (dm) dermomyotome; (my) myotome; (dml) dorsal medial lip; (nt) neural tube; (nc) notochord. (B) ES enhancer/lacZ transgene expression was assayed in embryos at 9.0 dpc by whole-mount in situ hybridization (ISH) using Myf5 antisense DIG probes (panel a) and lacZ antisense DIG probes (panel b), and by whole-mount β-gal histoenzymatic staining (panel c). Arrows and brackets demarcate the boundaries of lacZ transgene and Myf5 transcript expression in the epaxial somite, as assayed by these different methods. lacZ transcripts expressed by the ES enhancer/lacZ transgene are localized only to the dorsal-most aspect of the epaxial somite (b), whereas β-gal enzyme activity in more anterior somites has a broader dorsal to ventral distribution, extending from the dorsal epaxial domain to a more ventral position where the myotome is forming.
Histoenzymatic analysis of β-galactosidase (β-gal) expression established that the Myf5 ES enhancer/lacZ transgene is activated in somites of embryos as early as 8.25 dpc, when somites are first formed and chromosomal Myf5 is first activated in epaxial muscle progenitors. Myf5 activation is coordinated with somite formation, and expression persists in newly formed somites until 11.5 dpc, when somite formation is complete (Fig. 2A). Expression can also be detected in the brain at these stages of development (Daubas et al. 2000), as well as in the anterior neural tube in the two germ-line transgenic lines, E1 and E2, investigated. Neural tube expression was not observed in transient transgenic embryos (see Fig. 6A below). Expression in the brain is localized to the region of the medial longitudinal fasciculus and mammillotegmental tracts, where Myf5 is expressed (Daubas et al. 2000), although β-gal stained cells occupy a broader domain than reported by in situ hybridization assays of Myf5 transcripts. In 8.25- to 9.5-dpc embryos, β-gal staining is abundant in somites along the entire anterior to posterior axis, including the newest formed somites in the posterior embryo (Fig. 2A, panels a–c). This reporter gene activity recapitulates chromosomal Myf5 expression and precedes the expression of all of the other muscle regulatory factors, including MRF4 (Ott et al. 1991; Tajbakhsh et al. 1997). At later developmental stages, β-gal activity is preferentially localized to the eight newest-formed somites, where Myf5 is first activated (Tajbakhsh et al. 1996a), and activity is greatly diminished in the more mature anterior somites, in which only a few stained cells are detected in the myotome and dermotome (Fig. 2A, panels d,e). These somite expression patterns establish that the ES enhancer controls the transcriptional activation of Myf5 in newly formed somites, but not the maintenance of its expression during somite maturation.
Figure 6.
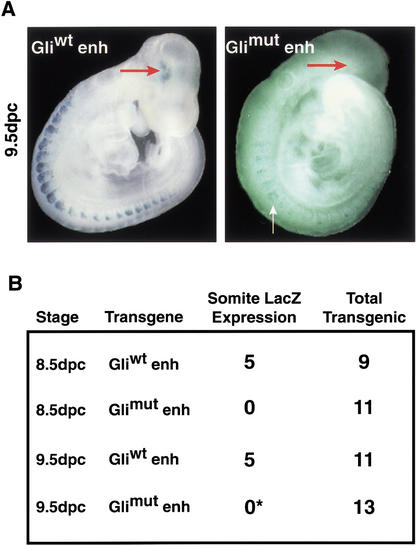
The Myf5 ES enhancer activation in somites and the brain is mediated through its Gli-binding sequence. The Myf5 ES enhancer/lacZ constructs containing either a wild-type (Gliwt enh) or mutant (Glimut enh) Gli-binding site were introduced into embryos for transient transgenesis by paranuclear injection. Embryos were recovered at 8.5 and 9.5 dpc, genotyped to identify transgenic embryos, and assayed for reporter expression by β-gal staining. (A, left panel) A 9.5-dpc embryo expressing a wild-type Gliwt enh transgene with reporter activity in somites and brain (red arrow). (Right panel) A 9.5-dpc Glimut enh transgenic embryo with ectopic transgene expression in the ventral somite (white arrow), but not in the dorsal epaxial somite. This embryo was the only one of 24 Glimut enh transgenic embryos to show ectopic staining in somites. (B) Tabular summary of the Gliwt enh and Glimut enh transgene expression in somites of 8.5- and 9.5-dpc transgenic embryos. Epaxial somite expression of the Gli mutant enhancer is completely blocked. Only one 9.5-dpc Glimut enh embryo (*), shown in A, had ectopic expression in the somite.
To investigate the dorsal–ventral pattern of ES enhancer activity in somites, β-gal-stained embryos were sectioned along their anterior to posterior axes at 9.5 dpc, when somites along the entire axis are stained (Fig. 2A, panels c1–c3). Enhancer activity is detected in the first, newest-formed somite in the posterior embryo, and this expression is localized to the cluster of dorsal medial somite cells that are known to activate chromosomal Myf5 for epaxial muscle progenitor cell specification (Tajbakhsh et al. 1997). Enhancer activity was not detected in the presomitic mesoderm at any developmental stage in transient or germ-line transgenic embryos. In more posterior somites, transgene expression is localized only to the dorsal medial lip (DML) of the dermomyotome, which is the proliferative source of progenitor cells that form the first myotomal epaxial muscles of the embryo. β-Galactosidase staining persists in these DML progenitors in more mature anterior somites that have initiated myotome formation, but abundant numbers of stained cells also populate the dorsal aspect of the myotome and the dorsal aspect of the overlying dermotome, which are formed by the dorsal and ventral migration of DML cells to these sites (Ordahl et al. 2001). Previously, β-gal staining of the dermotome ES enhancer/lacZ transgenic lines was interpreted as ectopic activity of the enhancer, which was thought to lack a negative regulatory element (Summerbell et al. 2000). Alternatively, dermotome and myotome β-gal staining could reflect persistence of β-gal enzyme in the absence of sustained transcriptional activity of the ES/lacZ transgene. To distinguish these possibilities, we compared expression of the ES enhancer/lacZ transgene using β-gal enzyme staining with whole-mount in situ hybridization assays to detect β-gal transcripts (Fig. 2B, panels a–c). These experiments show that lacZ RNA transcripts are restricted to the DML in all somites, including anterior somites. Transcripts were not detectable in the dermotome or myotome, where β-gal enzyme activity accumulates and persists during somite maturation. Similar localization of β-gal transcripts was observed in studies of the expression of lacZ transgenes controlled by the larger 23-kb sequence upstream of the Myf5 promoter, which included the ES enhancer (Hadchouel et al. 2000). These findings establish that the Myf5 ES enhancer is transcriptionally active only in the epaxial muscle progenitor cells of the DML, and its transcriptional activity is not maintained as DML cells migrate dorsally and ventrally to form both the myotome and dermotome, thus refuting the possibility that a negative regulatory element is necessary for localized ES enhancer expression in epaxial somite muscle progenitors.
The Myf5 ES enhancer has a functional Gli transcription factor binding site
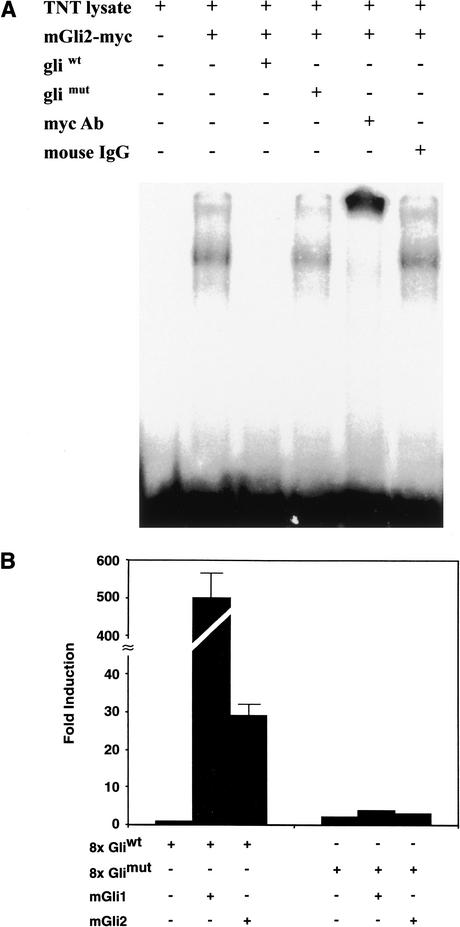
The Myf5 ES enhancer has a sequence element related to Gli-binding sequences (Fig. 1; Kinzler and Vogelstein 1990; Sasaki et al. 1997), suggesting that this enhancer is a direct target of Shh induction through Gli transcription factor regulation. As Gli binding sites in transcription enhancers have not previously been well documented, we performed electrophoretic mobility shift binding assays to test whether this Myf5 sequence element binds Gli proteins. Myc-tagged Gli2 protein expressed in a reticulocyte expression system was reacted with radiolabeled oligonucleotides of the Myf5 ES enhancer Gli sequence, and protein–DNA complexes were analyzed by electrophoretic mobility shift assays. We found that Gli2 protein produced in reticulocyte extracts forms stable DNA–protein complexes with the target Myf5 enhancer sequence, and these complexes can be supershifted with anti-myc antibodies, showing that these complexes are formed by binding of the myc-tagged Gli2 protein to the Myf5 enhancer sequence (Fig. 3A). Gli2 protein binding to the ES enhancer site is sequence-specific, based on competition with excess wild-type Gli oligonucleotide (Gliwt), but not with excess Glimut oligonucleotides with base-substitution mutations that destroy the core Gli DNA-binding sequence (see Fig. 1). Nuclear Myc-tagged Gli1 and Gli2 proteins expressed in 293 cells and C2C12 myoblasts also bind specifically with Gliwt oligonucleotides in these gel shift assays (data not shown), providing supporting evidence that Gli transcription factors can bind specifically to the Gli site in the Myf5 enhancer.
Figure 3.
Identification of a functional Gli-binding site in the Myf5 ES enhancer. (A) Electrophoresis mobility shift assays show that Myc-tagged Gli2 protein expressed in reticulocyte lysates (TNT lysate) binds and forms a gel shift complex with a 32P-radiolabeled, 32-bp oligonucleotide of the ES enhancer wild-type Gli site (mGli2-myc). A 100-fold excess of wild-type Gli site (Gliwt) oligonucleotide competes for Gli2-myc binding to radiolabeled wild-type oligonucleotide, whereas mutant Gli site (Glimut) oligonucleotide does not compete for binding. Anti-Myc antibodies (myc Ab) supershift Gli2–myc gel shift complexes whereas control antibodies (mouse IgG) have no effect, showing that Gli2 is a major component of these gel shift complexes. (B) Luciferase reporters of 8× multimerized wild-type Gli binding site (8× Gliwt) and 8× mutant Gli binding site (8× Glimut) from the Myf5 ES enhancer were cotransfected into 3T3 cells with mouse mGli1 or mGli2 expression vectors. Results are expressed as fold induction, which is the ratio of luciferase activity induced in Gli-transfected cells relative to basal luciferase activity in control transfected 3T3 cells. Both Gli1 and Gli2 are strong transcription activators of the 8× Gliwt reporter, but not the 8× Glimut reporter, showing that the ES enhancer Gli-binding site can form active transcription complexes with Gli transcription factors synthesized in vivo.
To test whether Gli binding to the ES enhancer Gli-binding site forms an active transcription complex, Shh-responsive 3T3 cells (Taipale et al. 2000) were cotransfected with luciferase reporter genes of 8× multimerized wild-type (8× Gliwt) and mutant (8× Glimut) Gli-binding sites, with Gli1 or Gli2 expression vectors, and with a Renilla reference plasmid (Fig. 3B). The 8× Gliwt reporter is transactivated 500-fold by Gli1 and 30-fold by Gli2 expression vectors, whereas the 8× Glimut is not transactivated by either Gli1 or Gli2. These biochemical and functional studies, therefore, establish that the Myf5 ES enhancer has a functional Gli-binding site for Shh regulation.
Shh signaling controls Myf5 ES enhancer activation and endogenous Myf5 epaxial expression
To investigate whether Shh signaling regulates Myf5 ES enhancer activity, we crossed the Myf5 ES enhancer/lacZ transgene into Shh mutant mice (Chiang et al. 1996), which were then mated to recover litters of 9.0–9.5-dpc heterozygous (+/−) and homozygous (−/−) Shh mutant embryos carrying the reporter construct for whole-mount analysis of transgene expression (Fig. 4A). All Shh−/− embryos analyzed (n = 3) show a complete absence of β-gal staining in all somites, including newly formed posterior somites, where the enhancer is first activated in wild-type embryos (Fig. 4A, panel c). Brain transgene expression also was eliminated in all Shh−/− embryos, but ectopic neural tube transgene expression persisted at high levels, providing a convenient internal control for transgene activity in mutant embryos. Significantly, we found that Myf5 ES enhancer activity is greatly diminished in somites of all Shh+/− mutant embryos analyzed (n = 4). Expression was most severely lost in the posterior, newest-formed somites, which had little or no transgene expression (Fig. 4B, cf. panels c and d). Neural tube and brain expression remains high in Shh+/− embryos relative to reduced somite expression (Fig. 4B, cf. panels a and b). In heterozygous mutant embryos, β-gal staining, although reduced, continues to be restricted to the DML, consistent with the normal dorsal pattern of ES enhancer transgene activity. Dorsal dermotome and myotome morphology are normal in heterozygous Shh mutant embryos (Borycki et al. 1999), providing additional evidence that the ES enhancer activation is controlled by Shh signaling and that defective Myf5 activation in Shh mutant embryos is not a result of primary defects in somite maturation.
Figure 4.
Activation of the Myf5 ES enhancer/luciferase transgene in somites and the brain is disrupted in homozygous and heterozygous Shh mutant embryos. (A) (panel a) Wild-type embryos (Shh+/+) and embryos that were (panel b) heterozygous Shh mutant (Shh+/−) and (panel c) homozygous Shh mutant (Shh−/−) were stained for β-gal to assay lacZ transgene activity in the brain (red arrow), somites (red bracket), and neural tube (white arrow). All embryos are heterozygous for the ES enhancer/lacZ transgene. ES enhancer activity is completely absent in somites and the brain of homozygous mutant embryos and greatly reduced in heterozygous mutant embryos, even under extended staining conditions, whereas ectopic neural tube expression persists. (B) (panels a,c) Heterozygous Shh mutant (+/−) and (panels b,d) wild-type (+/+) embryos were assayed for β-gal staining and then sectioned to localize staining in somites and neural tube in anterior (a,b) and posterior (c,d) regions of the embryo. In Shh+/− embryos, lacZ transgene activity is reduced in the anterior somites (a) and is almost entirely absent in the most posterior somites (c). (dml) Dermomyotome; (s) somite; (nt) neural tube. (C) (panel a) Wild-type Shh+/+ and (panel b) homozygous mutant Shh−/− embryos at 8.75 dpc were assayed for Myf5 expression by in situ hybridization. In wild-type embryos (a), Myf5 is activated exclusively within the epaxial domain of newly formed somites, whereas in Shh−/− embryos (b), Myf5 expression is disrupted in the epaxial domain of newly formed somites. Myf5 expression distributed diffusely, at low levels, particularly in the four most anterior occipital somites (red bracket). This low-level expression is not localized to the epaxial domain in the dorsal somite, but is distributed throughout the medial and lateral aspects of the somite. Staining in the head of Shh−/− embryos is not reproducible.
To investigate whether Shh signaling is required for epaxial expression of the endogenous Myf5 gene at the time of first somite formation, whole-mount in situ hybridization assays were performed in 8.75-dpc Shh+/+ and Shh−/− embryos. In wild-type embryos at this developmental stage, Myf5 expression is restricted to the epaxial domain of newly formed somites (Fig. 4C, panel a). In Shh−/− embryos, Myf5 expression distributed diffusely, at low levels, most prominently in the four most anterior occipital somites (Fig. 4C, panel b). This low-level expression is not localized to the epaxial domain in the dorsal somite, but is distributed throughout the medial and lateral aspects of the somite, consistent with our previously reported observations of Myf5 expression in newly formed somites of Shh−/− embryos at 9.75 dpc (Borycki et al. 1999). Reduced levels of Myf5 expression were also reported in the most anterior occipital somites of Smoothened mutant embryos, which also are disrupted in Shh signaling (Zhang et al. 2001).
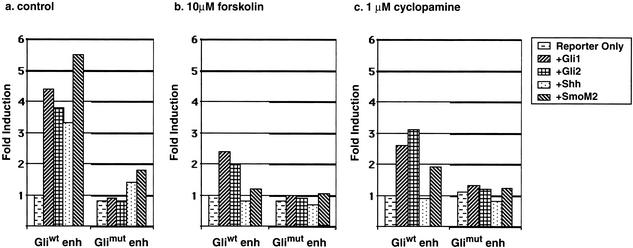
To further test whether the ES enhancer is a target of Shh signaling, we investigated the Shh signaling response of the ES enhancer in Shh-responsive 3T3 cells (Taipale et al. 2000). For these experiments, the Myf5 ES enhancer, cloned into a luciferase reporter gene with a δ-crystalline promoter, was cotransfected into 3T3 cells along with a Renilla reference reporter and expression vectors encoding Shh signaling components. Shh inducers included Shh, an oncogenic mutant form of Smoothened (SmoM2) that constitutively activates Shh signal transduction (Taipale et al. 2000), and Gli1 and Gli2. Each of these Shh signaling components was found to activate the Myf5 ES enhancer (Fig. 5a), establishing that Shh has an inductive function in Myf5 ES enhancer regulation. The role of Shh signaling in Myf5 ES enhancer activation is further supported by results of Shh inhibitor studies. The Shh induction of the Myf5 ES enhancer was completely inhibited by forskolin, an antagonist of Shh signaling (Fig. 5b; Fan et al. 1995), and by cyclopamine, a compound that disrupts the capacity of Smo to mediate Shh signal transduction (Fig. 5c; Taipale et al. 2000), further supporting the hypothesis that Shh signaling specifically induces ES enhancer activation.
Figure 5.
Shh signaling activates the Myf5 epaxial enhancer through its Gli-binding site in 3T3 cells. Myf5 ES enhancer/luciferase reporter plasmids with wild-type (Gliwt) and mutant (Glimut) Gli sites were cotransfected into cultures of 3T3 cells with Gli1, Gli2, Shh, and activated Smoothened (SmoM2) expression vector plasmids. The luciferase activities of the ES enhancer/luciferase transgene in response to induction by different Shh signaling components are expressed as fold induction, which represents the luciferase activity of the wild-type or mutant ES enhancer/luciferase reporter in response to inducers relative to its activity in transfected control cells without inducers. (a) In untreated, control cultures, the wild-type ES enhancer was induced by Shh, activated SmoM2, Gli1, and Gli2; and this Shh induction was blocked by mutation of the Gli-binding site in the ES enhancer. (b) In cultures treated with 10 μM forskolin, an antagonist of Shh signaling (Fan et al. 1995), the wild-type ES enhancer was completely blocked in its response to Shh and activated SmoM2, significantly reduced in its response to Gli1 and Gli2, and the Gli mutant ES enhancer was completely unresponsive to induction by the signaling components. (c) In cultures treated with 1 μM cyclopamine, wild-type ES enhancer is completely blocked in its response to Shh, and reduced but not blocked in its activation by SmoM2, Gli1, and Gli2, consistent with activity of cyclopamine as an inhibitor of Smo function (Taipale et al. 2000).
Shh induction of the Myf5 ES enhancer is mediated through its Gli-binding site
To determine whether Shh regulates Myf5 ES enhancer activation through Gli transcription factors, we first used 3T3 transfection assays to compare the Shh induction of the Myf5 ES enhancer luciferase reporters with a wild-type (Gliwt enh) or a mutant (Glimut enh) Gli-binding site. These studies show that mutations of the Gli-binding site that block DNA binding block transactivation of the Myf5 ES enhancer by Shh, SmoM2, Gli1, and Gli2 (Fig. 5a–c). Furthermore, forskolin and cyclopamine do not further reduce the activity of the Gli mutant enhancer below basal activity in the absence of Shh signaling, providing evidence that these inhibitors directly block Shh induction and do not inhibit other activities required for reporter gene response (Fig. 5b,c).
We next used transient transgenic assays to determine whether the Gli site in the Myf5 ES enhancer was required for reporter expression in the epaxial somite. Myf5 ES enhancer/lacZ constructs containing either wild-type (Gliwt enh) or mutant (Glimut enh) Gli-binding sites were introduced into embryos by pronuclear injection, and β-gal expression in somites and the brain was assayed in 8.5- and 9.5-dpc embryos (Fig. 6) as well as at 10.5 dpc (data not shown). These studies showed that mutation of the Gli-binding site in the ES enhancer completely blocks reporter activation in the epaxial somite in all transgenic embryos examined. Therefore, the Gli-binding site in the ES enhancer transgene has an essential positive regulatory function for enhancer activation in newly formed somites at multiple developmental stages. Significantly, mutation of the Gli site in the ES enhancer does not cause misexpression of the enhancer at other tissue sites of Shh signaling in the embryo such as the neural tube, establishing that this Gli site does not function to repress ES enhancer activity in other cell lineages. Together, these transgenic and 3T3 transfection studies establish that Myf5 ES enhancer activation in the epaxial somite is mediated through long-range Shh signaling, under the positive control of the Gli transcription factors.
Discussion
This study reports the identification of a Shh-regulated enhancer that controls activation of Myf5, a master regulatory gene for specification of epaxial skeletal muscle progenitor cells (Rudnicki et al. 1993; Tajbakhsh et al. 1996b). The Myf5 ES enhancer activates reporter expression in the epaxial muscle progenitors of newly forming somites from 8.25 dpc, when somites are first formed in the developing embryo, until 11.5 dpc, when somite formation is complete. The activity of ES enhancer for epaxial somite regulation is unique in a 650-kb region around the MRF4/Myf5 locus, which has been shown to include other enhancers that direct Myf5 transgene expression to all of the other domains of muscle progenitor specification in the embryo, including branchial arches, limb bud, and the hypaxial somite (Patapoutian et al. 1993; Zweigerdt et al. 1997; Hadchouel et al. 2000; Summerbell et al. 2000). Because Myf5 is an essential regulator of epaxial muscle specification, our finding that the Myf5 ES enhancer is a direct target of Shh signaling through an essential Gli-binding site provides the first evidence that Shh has a direct inductive function in the process of cell lineage specification. In addition to epaxial somite expression, the ES enhancer also controls Myf5 expression in the longitudinal tracks of the developing brain (Daubas et al. 2000). The brain activity of the ES enhancer also is mediated through its Gli-binding site, and although the domain of ES enhancer/lacZ reporter activity is broader than the expression of Myf5 transcripts, these brain cells are near a source of Shh, indicating that the transcriptional activity of the ES enhancer in the brain also is controlled by Shh signaling.
Our findings show that Myf5 ES enhancer is under positive regulation by Shh signaling and Gli regulation through a Gli site that we identified in DNA/protein binding and functional transcription assays. This Gli-binding site is required for enhancer activity in somites and the brain at both 8.5 and 9.5 dpc, and for Shh induction in 3T3 cells. Significantly, mutation of the Gli site did not lead to ectopic enhancer activity in the somite or other embryonic tissues, indicating that the enhancer is subject to positive regulation by Gli transcription factors. Because this Gli site is essential to mediate reporter expression in the somite, other regulatory elements must also be present within the enhancer to restrict its activity to the epaxial somite, either by repressing enhancer activity in other Shh-responsive tissues such as the neural tube, or by functioning cooperatively with Glis to activate the enhancer specifically in the DML. It remains to be determined which Gli transcription factor regulates ES enhancer activation. Gli1, Gli2, and Gli3 are all expressed in somites (Hui et al. 1994). In avian embryos, the expression of the three Gli transcription factor genes initiates in newly forming somites, immediately preceding Myf5 activation, and Gli2 and Gli3 transcripts become localized to the epaxial muscle progenitor cells of the dorsal somite, at the site of Myf5 activation (Borycki et al. 2000). Genetic studies show that Gli1, Gli2, and Gli3 have redundant functions in both neural and somite development in the mouse (Mo et al. 1997; Motoyama et al. 1998; Park et al. 2000), indicating that these Gli genes could also be redundant for Myf5 regulation. Identification of the specific Glis that function together with somite transcription factors to activate the ES enhancer in epaxial somite progenitors will provide a basis for an understanding of the molecular mechanisms that control the tissue specificity of Shh signaling responses.
The Myf5 ES enhancer is dependent on Shh signaling for its activation in epaxial myogenic progenitor cells of somites throughout the embryo axis, as shown by the loss of the ES enhancer/lacZ transgene expression in somites of Shh−/− embryos. The somites of Shh mutant embryos form normally and undergo dermotome epithelialization and DML formation, but fail to express Myf5 specifically in the epaxial somite domain. Significantly, Shh−/− embryos exhibit normal cell survival and proliferation in the dorsal somite (Borycki et al. 1999). Therefore, the loss of transgene expression in the Shh mutant embryos cannot be attributed to defects in the survival or proliferation of Myf5-expressing cells in the newly formed somite. During later stages of somite maturation in Shh−/− mutant embryos, the dorsal aspects of the dermotome and myotome are absent. Similarly, in Myf5−/− mutant embryos, dorsal somite cell migration and dermotome and myotome formation are also blocked (Tajbakhsh et al. 1996b). Our findings and those of recent lineage marking studies (Ordahl et al. 2001) now show that the DML cells contribute dorsal growth of both the myotome and the dermotome. Thus, we can now explain the absence of the medial dermotome and myotome in Shh−/− embryos as a specific defect in the Shh-controlled specification of the DML cells to form these lineages, rather than a degenerative defect in somite maturation. It also is noteworthy that the activity of the ES enhancer is greatly reduced in Shh+/− embryos, in which somite formation and maturation are entirely normal, further supporting the specificity of Shh signaling for Myf5 epaxial somite regulation. Previously, we noted that Myf5 transcript expression, as assayed by in situ hybridization, was unaffected in somites of Shh+/− embryos (Borycki et al. 1999), which contrasts with our current finding that ES enhancer activity, assayed by the β-gal staining assay, is significantly reduced in Shh+/− embryos. These differing results may reflect improved quantitative sensitivity of the β-gal-staining assay for transgene activity. Alternatively, transcriptional or posttranscriptional compensatory mechanisms may modulate Myf5 expression in response to reduced levels of Shh, and these mechanisms may not be operative for the ES enhancer transgene and its β-gal transcript.
In addition to the Shh-dependent activation of Myf5 in the epaxial somite progenitors, through the ES enhancer, Shh-independent signals activate Myf5 in other muscle progenitor lineages in the embryo. This Shh-independent regulation is mediated by the other Myf5 transcription enhancers that have been identified in a 650-kb region around the MRF4/Myf5 locus (Zweigerdt et al. 1997; Hadchouel et al. 2000; Summerbell et al. 2000; Carvajal et al. 2001). These enhancers direct Myf5 activation in the myogenic progenitor cell domains that form and express Myf5 in wild-type and Shh mutant embryos, including the progenitor cells of the hypaxial somite, the limb, and the branchial arches (Borycki et al. 1999). These transcription enhancers are controlled by developmental signals such as Wnts (Tajbakhsh et al. 1998). Our earlier findings established that Myf5 is activated in all of these muscle progenitor cell domains, except the epaxial somite domain, in 9.75-dpc and 10.50-dpc Shh mutant embryos (Borycki et al. 1999). In this study, we examined Myf5 expression in earlier 8.75-dpc Shh mutant embryos, which are forming the first somites. These findings confirm our earlier work showing that Myf5 expression is specifically disrupted in the dorsal epaxial domain somites of 8.75-dpc Shh mutant embryos, which, as previously reported, express low levels of Myf5 in the medial and lateral somite domains. Based on analysis of medial and lateral somite marker genes, medial–lateral somite patterning is disrupted in Shh mutant embryos such that lateral somite genes, including hypaxial Myf5, are expressed medially, suggesting that low-level Myf5 expression in somites of Shh mutant embryos reflects activity of the hypaxial Myf5 enhancer on these lateral signals. In 8.75-dpc embryos, reduced levels of Myf5 expression also are observed in the most anterior four occipital somites of Shh mutant embryos as well as Smoothened (Smo) mutant embryos, which are defective in all HH signaling (Zhang et al. 2001). A distal Myf5 enhancer located at −58 to −48 kb is active in occipital somites and could serve this function (Hadchouel et al. 2000). This same enhancer regulates the Myf5 expression in the myotome of the medial somite and may contribute to the low level of Myf5 expression observed in the medial somites of Shh−/− embryos (Kruger et al. 2001).
Epaxial muscle progenitors in the dorsal somite are located at a significant distance from the notochord source of Shh, indicating that epaxial Myf5 expression is controlled by long-range Shh signaling through the ES enhancer. The ES enhancer apparently is regulated positively and directly by Shh signaling, as this signaling activity is mediated through a Gli-binding site that is essential for ES enhancer activity. Disruption of this Gli-binding site does not lead to ectopic expression of the ES enhancer, indicating that this Gli-binding site is not used to repress Myf5 expression in other Shh-responsive tissues such as the neural tube. Long-range transport of Shh in somites has not been investigated, but in the limb, long-range Shh signaling requires N-terminal processing and cholesterol modification (Lewis et al. 2001; Zeng et al. 2001). The levels of Shh in the responding dorsal somite would need to be sufficiently high to elicit a positive regulatory response, which would presumably include limiting the processing of Gli3 into its repressor form, which is thought to occur under conditions of low-level Shh signaling (Wang et al. 2000).
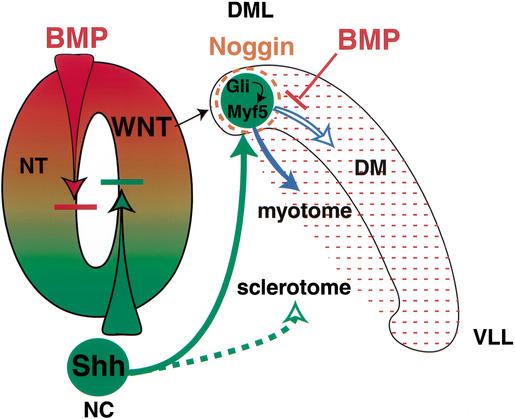
Our ES enhancer transgene expression results show that lacZ transcripts are localized to the DML in both newly formed and mature somites of wild-type embryos, indicating that the Shh signaling is restricted to the DML and that enhancer activity is not maintained as cells, marked by persistent β-gal enzyme, migrate from the DML to form the dorsal aspects of the dermotome and the myotome. DML cells that form the myotome maintain Myf5 transcription, likely through the activity of a myotome-specific maintenance enhancer located −58 to −48 kb 5′ of Myf5 (Hadchouel 2000). Presumably the sustained expression of Myf5, under the control of this maintenance enhancer, would promote the specification of myotome progenitors to the myogenic lineage. In DML cells that form the dermotome, the ES enhancer is inactive and Myf5 transcription ceases as these cells become committed to the dermotome lineage. Specific mechanisms, therefore, must exist to enhance and localize long-distance Shh signaling for Gli activation of the Myf5 ES enhancer specifically in DML progenitors and to suppress its response to Shh signaling as DML cells migrate to the dorsal myotome and dermotome. One possibility is that the Shh signaling response of the ES enhancer in the DML is controlled by opposing Shh and BMP signals, which are known to function together in the specification of the epaxial and hypaxial progenitor cells in the medial and lateral somite (Fig. 7). When BMP signaling in the somite is blocked, Myf5 expression is ectopically expressed throughout the dermomyotome; and when BMP signaling in the somite is enhanced, Myf5 expression in the DML is repressed and hypaxial genes are activated ectopically in the epaxial domain (Pourquie et al. 1996). Furthermore, the DML is protected from the repressive activity of BMP on Shh signaling by the localized expression of Noggin, a potent BMP inhibitor that is activated in the DML in response to Wnt signaling from the dorsal neural tube (Hirsinger et al. 1997; Marcelle et al. 1997; Reshef et al. 1998). In the mouse, Noggin expression is essential for epaxial somite specification (McMahon et al. 1998). According to this mechanism, the localized expression of Noggin or another BMP inhibitor in the DML would enhance the response of these cells to low levels of Shh signals from the ventral notochord. Shh signaling and ES enhancer activity would be inhibited as DML cells migrate outside of the BMP inhibitor domain to form the myotome and dermotome. The discovery that the Myf5 ES enhancer is a direct target of Shh signaling and Gli regulation provides a molecular approach to test this and other models of Shh function in embryo patterning and cell lineage specification.
Figure 7.
A signal enhancement model for localized Shh signaling for epaxial muscle specification and dermotome and myotome differentiation. This model proposes that medial lateral patterning of the dermomyotome in the dorsal somite is controlled by opposing Shh and BMP signals in the dorsal somite. Opposing Shh and BMP signals also control dorsal ventral patterning of the neural tube (NT) specification (Roelink et al. 1995), and the specification of sclerotome progenitors in the ventral somite (McMahon et al. 1998). In the dorsal somite, epaxial myogenic specification is controlled by long-distance Shh induction of Myf5 through positive Gli regulation of the Myf5 ES enhancer. Positive Gli regulation of Myf5 in the DML is controlled by the localized expression of Noggin, which inhibits the repressive activity of BMP signals prevalent in the dermomyotome, thus allowing high signal transduction activity in response to low levels of Shh from the distant notochord (NC). Subsequently, Myf5-expressing DML cells migrate dorsally into a repressive high BMP environment that represses ES enhancer activity and Myf5 expression, allowing these cells to assume a dermotome fate. DML cells that migrate ventral laterally also lose ES enhancer activity, but these myotome progenitor cells activate a myotome-specific, Myf5-maintenance enhancer (Hadchouel et al. 2000) to maintain high-level Myf5 transcription to promote their specification to the myotome lineage and their differentiation to form the epaxial myotomal muscles.
Materials and methods
Identification of an Myf5 epaxial somite transcription enhancer
A mouse BAC 129/SvJ genomic DNA library (Genome Systems) was screened with Myf5 PCR primer probes to identify clones including the MRF4/Myf5 locus. Subclones that included the linked MRF4 and Myf5 genes were analyzed by DNA sequencing and transgenic analysis using lacZ reporter genes and transient transgenic injection. Transgene expression was assayed by whole-mount histoenzymatic assays of β-gal as well as by whole-mount in situ hybridization with DIG lacZ probe, as described previously (Goldhamer et al. 1995; Borycki et al. 1999). The lacZ reporter gene has a viral tk promoter driving transcription of a lacZ gene encoding β-gal with a nuclear localization domain (Goldhamer et al. 1992). DNA sequence and transgenic analyses identified a 651-bp BamHI–EcoR1 fragment in the MRF4/Myf5 intragenic region, located −6.6 kb 5′ of the Myf5 promoter, immediately 3′ of the MRF4 gene and 5′ of a previously identified branchial arch enhancer (Patapoutian et al. 1993). This region was highly conserved with rat intragenic sequence (Pin et al. 1997) and contained a candidate Gli-binding site, identified with the PatSearch 1.1 transcription site search program. Two germ-line transgenic lines carrying the −6.6-kb epaxial somite (ES) enhancer, E1 and E2, were produced in CD1 mice for developmental and genetic studies. Transgene expression was identical in both germ-line transgenic lines, including epaxial somite and brain expression, and ectopic expression in the anterior neural tube, which was not observed in transient transgenic embryos. Embryos were recovered from pregnant females between 8.0 and 11.5 dpc for β-gal staining in whole-mount histoenzymatic assays (Goldhamer et al. 1995). Embryos were sectioned transversely along the anterior to posterior embryo axis using a vibratome. For genetic studies, heterozygous Shh mutant mice (Chiang et al. 1996) in a CD1 background were crossed with Myf5 ES enhancer transgenic lines heterozygous for the transgene, to generate homozygous Shh mutant mice that carried the Myf5 ES enhancer transgene. These lines were bred to produce homozygous and heterozygous Shh mutant embryos for β-gal staining to assay Myf5 ES enhancer transgene activities.
Luciferase reporter genes and DNA transfection assays
Myf5 ES enhancer/luciferase reporter plasmids were generated by subcloning the 651-bp Myf5 ES enhancer sequence, with a wild-type (Gliwt enh) or a mutant (Glimut enh) Gli-binding site, into the pδ51lucII luciferase reporter, which has a δ-crystalline promoter with low basal activity in DNA transfection assays. 8× Gli/luciferase reporter plasmids were generated by subcloning into the pδ51lucII luciferase reporter 8× multimers of the wild-type (8× Gliwt, 5′-AACGACCACCAAGAAACA-3′) and mutant (8× Glimut, 5′-AACGACtgCagAGAAACA-3′) Gli-binding site present in the 651-bp Myf5 ES enhancer. These multimers were generated as direct repeats by oligonucleotide synthesis. Mouse Gli2 expression vector was prepared by subcloning Gli2 (Sasaki et al. 1999) into pGEM myc tag vectors, producing C-terminal-tagged pGEM Gli2–myc.
DNA transfection assays were performed by plating confluent 3T3 cultures into 6- or 12-well plates at a 1:6 dilution in DMEM supplemented with 10% calf serum. After 18 h, cells were cotransfected with Renilla luciferase plasmid (pRL-TK, Promega; 5% w/w DNA), and Myf5 ES enhancer luciferase or 8× Gli luciferase reporter plasmids (45%), and Gli1, Gli2, Shh, or SmoM2 expression plasmids (50%), using Fugene 6 (Roche) transfection reagent (1 μg/well in 6-well plates or 500 ng/well in 12-well plates) in a 3:1 ratio (v/w) of reagent to DNA. Two h after addition of DNA, cells were rinsed and cultured in 0.5% serum medium either containing forskolin or cyclopamine or without (control). After 24 h cells were lysed and luciferase assays were performed as described previously (Dhoot et al. 2001). Luciferase activity in each culture sample was normalized to Renilla luciferase activity to correct for variations in transfection efficiency.
Electrophoretic mobility shift assays
Gli2 protein was synthesized by TNT coupled transcription/translation (Promega), using pGEM–Gli2–myc plasmid as a template. For electrophoretic mobility shift assays, 5 μL of Gli2-myc translation extracts were incubated on ice for 45 min with 5 × 104 cpm of 32P-labeled double-stranded oligonucleotide of the Gli wild-type (Gliwt, 5′-GGGAAAAAACGACCACCA AGAAACACAGT-3′) and Gli mutant (Glimut, 5′-GGGAAAA AACGACtgCagAGAAACACAGT-3′) binding sites in 20 μL containing 20 mM Tris (pH 7.9), 80 mM NaCl, 1 mM β-mercaptoethanol, 0.1 mg/mL of bovine serum albumin, and 10% (v/v) glycerol. In competition assays, 100-fold excess of unlabeled competitor Gliwt or Glimut oligonucleotide was added during preincubation. In supershift assays, 4 μg of Myc monoclonal antibody (9E10) was added to the binding mixture on ice for 1 h prior to the addition of radiolabeled probe. Bound complexes resolved by electrophoresis on 5% polyacrylamide gels in 0.25× TBE buffer, and radioactivity was detected by autoradiography.
Acknowledgments
The authors thank the Penn Center for Developmental Biology Mouse Transgenic Users Group and Diane Zhou for production of transient transgenic embryos, LiYa Yu for excellent technical assistance, and the Penn Institute for Human Gene Therapy Mouse Transgenic Core for production of germ-line transgenic mice. The authors also thank P. Beachy for providing Activated Smoothened (SmoM2) expression vector and samples of cyclopamine, Chin Chiang for providing Shh mutant mice, and H. Sasaki for providing Gli1 and Gli2 expression vectors. This work was funded by grants to C.P.E. from the March of Dimes and the NICHD.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL emersonc@mail.med.upenn.edu; FAX (215) 898-9871.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.940702.
References
- Alexandre C, Jacinto A, Ingham PW. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes & Dev. 1996;10:2003–2013. doi: 10.1101/gad.10.16.2003. [DOI] [PubMed] [Google Scholar]
- Ang SL, Rossant J. HNF-3β is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, The I, Perrimon N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature. 1998;394:85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- Borycki A-G, Mendham L, Emerson CP., Jr Control of somite patterning by Sonic hedgehog and its downstream signal response genes. Development. 1998;125:777–790. doi: 10.1242/dev.125.4.777. [DOI] [PubMed] [Google Scholar]
- Borycki AG, Brunk B, Tajbakhsh S, Buckingham M, Chiang C, Emerson CP., Jr Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development. 1999;126:4053–4063. doi: 10.1242/dev.126.18.4053. [DOI] [PubMed] [Google Scholar]
- Borycki A, Brown AM, Emerson CP., Jr Shh and Wnt signaling pathways converge to control Gli gene activation in avian somites. Development. 2000;127:2075–2087. doi: 10.1242/dev.127.10.2075. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Ericson J. The specification of neuronal identity by graded Sonic hedgehog signalling. Semin Cell Dev Biol. 1999;10:353–362. doi: 10.1006/scdb.1999.0295. [DOI] [PubMed] [Google Scholar]
- Carvajal JJ, Cox D, Summerbell D, Rigby PWJ. A BAC transgenic analysis of the Mrf5/Myf5 locus reveals interdigitated elements that control activation and maintenance of gene expression during muscle development. Development. 2001;128:1857–1858. doi: 10.1242/dev.128.10.1857. [DOI] [PubMed] [Google Scholar]
- Chen CH, von Kessler DP, Park W, Wang B, Ma Y, Beachy PA. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98:305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Basler K. Opposing transcriptional outputs of Hedgehog signaling and engrailed control compartmental cell sorting at the Drosophila A/P boundary. Cell. 2000;100:411–422. doi: 10.1016/s0092-8674(00)80677-7. [DOI] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- Daubas P, Tajbakhsh S, Hadchouel J, Primig M, Buckingham M. Myf5 is a novel early axonal marker in the mouse brain and is subjected to post-transcriptional regulation in neurons. Development. 2000;127:319–331. doi: 10.1242/dev.127.2.319. [DOI] [PubMed] [Google Scholar]
- Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP., Jr Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, Hui CC. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- Dufort D, Schwartz L, Harpal K, Rossant J. The transcription factor HNF3β is required in visceral endoderm for normal primitive streak morphogenesis. Development. 1998;125:3015–3025. doi: 10.1242/dev.125.16.3015. [DOI] [PubMed] [Google Scholar]
- Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87:661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- Ericson J, Briscoe J, Rashbass P, van Heyningen V, Jessell TM. Graded Sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. Cold Spring Harb Symp Quant Biol. 1997;62:451–466. [PubMed] [Google Scholar]
- Fan CM, Porter JA, Chiang C, Chang DT, Beachy PA, Tessier-Lavigne M. Long-range sclerotome induction by Sonic hedgehog: Direct role of the amino-terminal cleavage product and modulation by the cyclic AMP signaling pathway. Cell. 1995;81:457–465. doi: 10.1016/0092-8674(95)90398-4. [DOI] [PubMed] [Google Scholar]
- Fuse N, Maiti T, Wang B, Porter JA, Hall TM, Leahy DJ, Beachy PA. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc Natl Acad Sci. 1999;96:10992–10999. doi: 10.1073/pnas.96.20.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhamer DJ, Faerman A, Shani M, Emerson CP., Jr Regulatory elements that control the lineage-specific expression of myoD. Science. 1992;256:538–542. doi: 10.1126/science.1315077. [DOI] [PubMed] [Google Scholar]
- Goldhamer DJ, Brunk BP, Faerman A, King A, Shani M, Emerson CP., Jr Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development. 1995;121:637–649. doi: 10.1242/dev.121.3.637. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Scott MP. Hedgehog and patched in neural development and disease. Neuron. 1998;21:1243–1257. doi: 10.1016/s0896-6273(00)80645-5. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Johnson RL, Milenkovic L, McMahon J, Scott M. Conservation of the hedgehog/patched signaling pathway from flies to mice: Induction of a mouse patched gene by hedgehog. Genes & Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Hadchouel J, Tajbakhsh S, Primig M, Chang TH, Daubas P, Rocancourt D, Buckingham M. Modular long-range regulation of myf5 reveals unexpected heterogeneity between skeletal muscles in the mouse embryo. Development. 2000;127:4455–4467. doi: 10.1242/dev.127.20.4455. [DOI] [PubMed] [Google Scholar]
- Hahn H, Wojnowski L, Miller G, Zimmer A. The patched signaling pathway in tumorigenesis and development: Lessons from animal models. J Mol Med. 1999;77:459–468. doi: 10.1007/s001099900018. [DOI] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, St Jacques B, McMahon AP, Melton DA. Regulation of pancreas development by hedgehog signaling. Development. 2000;127:4905–4913. doi: 10.1242/dev.127.22.4905. [DOI] [PubMed] [Google Scholar]
- Heemskerk J, DiNardo S. Drosophila hedgehog acts as a morphogen in cellular patterning. Cell. 1994;76:449–460. doi: 10.1016/0092-8674(94)90110-4. [DOI] [PubMed] [Google Scholar]
- Hirsinger E, Duprez D, Jouve C, Malapert P, Cooke J, Pourquie O. Noggin acts downstream of Wnt and Sonic hedgehog to antagonize BMP4 in avian somite patterning. Development. 1997;124:4605–4614. doi: 10.1242/dev.124.22.4605. [DOI] [PubMed] [Google Scholar]
- Hui CC, Slusarski D, Platt KA, Holmgren R, Joyner AL. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev Biol. 1994;162:402–413. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Taylor AM, Nakano Y. Role of the Drosophila patched gene in positional signalling. Nature. 1991;353:184–187. doi: 10.1038/353184a0. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, Toftgard R, Zaphiropoulos PG. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- Kruger M, Mennerich D, Fees S, Schafer R, Mundlos S, Braun T. Sonic hedgehog is a survival factor for hypaxial muscles during mouse development. Development. 2001;128:743–752. doi: 10.1242/dev.128.5.743. [DOI] [PubMed] [Google Scholar]
- Lee J, Platt KA, Censullo P, Ruiz i Altaba A. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- Lewis PM, Dunn MP, McMahon JA, Logan M, Martin JF, St-Jacques B, McMahon AP. Cholesterol modification of Sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell. 2001;105:599–612. doi: 10.1016/s0092-8674(01)00369-5. [DOI] [PubMed] [Google Scholar]
- Marcelle C, Stark MR, Bronner-Fraser M. Coordinate actions of BMPs, Shh and Noggin mediate patterning of the dorsal somite. Development. 1997;124:3955–3963. doi: 10.1242/dev.124.20.3955. [DOI] [PubMed] [Google Scholar]
- Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996a;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- Marigo V, Scott MP, Johnson RL, Goodrich LV, Tabin CJ. Conservation in hedgehog signaling: Induction of a chicken patched homolog by Sonic hedgehog in the developing limb. Development. 1996b;122:1225–1233. doi: 10.1242/dev.122.4.1225. [DOI] [PubMed] [Google Scholar]
- Matise MP, Epstein DJ, Park HL, Platt KA, Joyner AL. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- McMahon JA, Takada S, Zimmerman LB, Fan CM, Harland RM, McMahon AP. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes & Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, Chik KW, Shi XM, Tsui LC, Cheng SH, et al. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui CC. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat Genet. 1998;20:54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- Muenke M, Cohen MM., Jr Genetic approaches to understanding brain development: Holoprosencephaly as a model. Ment Retard Dev Disabil Res Rev. 2000;6:15–21. doi: 10.1002/(SICI)1098-2779(2000)6:1<15::AID-MRDD3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Munsterberg AE, Kitajewski J, Bumcrot DA, McMahon AP, Lassar AB. Combinatorial signaling by Sonic hedgehog and wnt family members induces myogenic bHLH gene expression in the somite. Genes & Dev. 1995;9:2911–2922. doi: 10.1101/gad.9.23.2911. [DOI] [PubMed] [Google Scholar]
- Ordahl CP, Berdougo E, Venters SJ, Denetclaw WFJ. The dermomyotome dorsomedial lip drives growth and morphogenesis of both the primary myotome and dermomyotome epithelium. Development. 2001;128:1731–1744. doi: 10.1242/dev.128.10.1731. [DOI] [PubMed] [Google Scholar]
- Ott MO, Bober E, Lyons G, Arnold H, Buckingham M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development. 1991;111:1097–1107. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- Patapoutian A, Miner JH, Lyons GE, Wold B. Isolated sequences from the linked Myf-5 and MRF4 genes drive distinct patterns of muscle-specific expression in transgenic mice. Development. 1993;118:61–69. doi: 10.1242/dev.118.1.61. [DOI] [PubMed] [Google Scholar]
- Pin CL, Ludolph DC, Cooper ST, Klocke BJ, Merlie JP, Konieczny SF. Distal regulatory elements control MRF4 gene expression in early and late myogenic cell populations. Dev Dyn. 1997;208:299–312. doi: 10.1002/(SICI)1097-0177(199703)208:3<299::AID-AJA2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Porter JA, von Kessler DP, Ekker SC, Young KE, Lee JJ, Moses K, Beachy PA. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature. 1995;374:363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- Pourquie O, Fan CM, Coltey M, Hirsinger E, Watanabe Y, Breant C, Francis-West P, Brickell P, Tessier-Lavigne M, Le Douarin NM. Lateral and axial signals involved in avian somite patterning: A role for BMP4. Cell. 1996;84:461–471. doi: 10.1016/s0092-8674(00)81291-x. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Melton DA, McMahon AP. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 2000;127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- Reshef R, Maroto M, Lassar AB. Regulation of dorsal somitic cell fates: BMPs and Noggin control the timing and pattern of myogenic regulator expression. Genes & Dev. 1998;12:290–303. doi: 10.1101/gad.12.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Roelink H, Porter JA, Chiang C, Tanabe Y, Chang DT, Beachy PA, Jessell TM. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of Sonic hedgehog autoproteolysis. Cell. 1995;81:445–455. doi: 10.1016/0092-8674(95)90397-6. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3β floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Nishizaki Y, Hui C-C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: Implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- Summerbell D, Ashby PR, Coutelle O, Cox D, Yee S-P, Rigby PWJ. The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Development. 2000;127:3745–3751. doi: 10.1242/dev.127.17.3745. [DOI] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Bober E, Babinet C, Pournin S, Arnold H, Buckingham M. Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Dev Dyn. 1996a;206:291–300. doi: 10.1002/(SICI)1097-0177(199607)206:3<291::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Buckingham M. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf5 null mice. Nature. 1996b;384:266–270. doi: 10.1038/384266a0. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Borello U, Vivarelli E, Kelly R, Papkoff J, Duprez D, Buckingham M, Cossu G. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development. 1998;125:4155–4162. doi: 10.1242/dev.125.21.4155. [DOI] [PubMed] [Google Scholar]
- Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- Teillet M, Watanabe Y, Jeffs P, Duprez D, Lapointe F, Le Douarin NM. Sonic hedgehog is required for survival of both myogenic and chondrogenic somitic lineages. Development. 1998;125:2019–2030. doi: 10.1242/dev.125.11.2019. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Ingham PW. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- Von Ohlen T, Lessing D, Nusse R, Hooper JE. Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc Natl Acad Sci. 1997;94:2404–2409. doi: 10.1073/pnas.94.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JJ. The winged-helix transcription factor HNF-3 β is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- Zeng X, Goetz JA, Suber LM, Scott WJ, Jr, Schreiner CM, Robbins DJ. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature. 2001;411:716–720. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R asymmetry by the mouse node. Cell. 2001;105:781–792. [PubMed] [Google Scholar]
- Zweigerdt R, Braun T, Arnold HH. Faithful expression of the Myf-5 gene during mouse myogenesis requires distant control regions: A transgene approach using yeast artificial chromosomes. Dev Biol. 1997;192:172–180. doi: 10.1006/dbio.1997.8759. [DOI] [PubMed] [Google Scholar]