Abstract
To determine functional differences between the two splice variants of PPARγ (γ1 and γ2), we sought to selectively repress γ2 expression by targeting engineered zinc finger repressor proteins (ZFPs) to the γ2-specific promoter, P2. In 3T3-L1 cells, expression of ZFP55 resulted in >50% reduction in γ2 expression but had no effect on γ1, whereas adipogenesis was similarly reduced by 50%. However, ZFP54 virtually abolished both γ2 and γ1 expression, and completely blocked adipogenesis. Overexpression of exogenous γ2 in the ZFP54-expressing cells completely restored adipogenesis, whereas overexpression of γ1 had no effect. This finding clearly identifies a unique role for the PPARγ2 isoform.
Keywords: PPARγ1, PPARγ2, adipogenesis, zinc finger protein
The nuclear hormone receptor PPARγ is essential for cellular differentiation and lipid accumulation during adipogenesis (Barak et al. 1999; Kubota et al. 1999; Rosen et al. 1999). The adipocyte-specific γ2 isoform differs from the more widely expressed γ1 in that it contains additionally 30 amino acid residues at the amino terminus (Kliewer et al. 1994; Tontonoz et al. 1994a; Zhu et al. 1995). Evidence suggests these residues contribute to a constitutive transcription activation function that is 5–10-fold greater than in γ1 (Werman et al. 1997). PPARγ2 is selectively expressed in adipose tissue (Fajas et al. 1997) and is strongly up-regulated during adipogenesis (Tontonoz et al. 1994b; Wu et al. 1998), suggesting a specific role for this isoform in fat cell differentiation. Nevertheless, a specific role for γ2 that could not be substituted by γ1 has not been clearly determined.
The ability to selectively knock out or knock down the expression of a specific gene provides a powerful approach for understanding its biological function. The targeting of individual mRNA splice variants offers an even greater level of selective control and understanding of differential isoform function. Rationally engineered transcription factors potentially provide a powerful tool for targeted regulation of endogenous genes by combining a functional transcription regulatory domain with a customized DNA binding domain that can bind to a specific sequence within the target gene. C2H2 zinc finger proteins (ZFPs) can be engineered to bind with high specificity to wide a diversity of DNA sequences (Desjarlais and Berg 1992; Choo and Klug 1994; Jamieson et al. 1994; Rebar and Pabo 1994; Greisman and Pabo 1997). Previous studies have demonstrated the utility of both engineered activator- and repressor-ZFPs in the regulation of endogenous chromosomal loci (Bartsevich and Juliano 2000; Beerli et al. 2000; Zhang et al. 2000; Liu et al. 2001). Our goal for this study was to selectively inhibit expression of the PPARγ2 isoform in the adipogenic mouse 3T3-L1 cell line by utilizing engineered zinc finger repressor proteins.
Results and Discussion
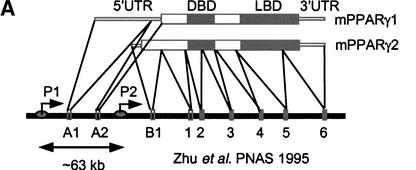
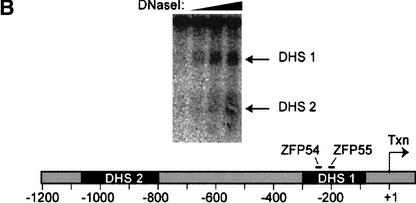
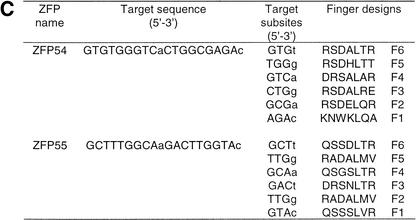
The mouse PPARγ gene spans >105 kb (Zhu et al. 1995). Coding exons 1 to 6 are conserved between the γ1 and γ2 isoforms (Fig. 1A) and transcription of these is driven by an upstream promoter (P1) that also drives expression of two untranslated γ1-specific exons, A1 and A2. The additional amino acids at the amino terminus of γ2 are encoded by an additional exon, B1, that is uniquely regulated by a separate promoter (P2) lies >63 kb downstream of P1. DNase I digestion of the endogenous PPARγ gene locus in 3T3-L1 cells revealed two DNase I hypersensitive sites in the vicinity of the proximal P2 promoter that represent regions of accessible chromatin at an endogenous locus (Fig. 1B; DHS1 and DHS2). Two six-finger ZFPs (ZFP54 and ZFP55; Fig. 1C) linked to the KRAB transcriptional repression domain (Margolin et al. 1994) were designed to bind specifically to 18-bp sequences within the DHS1 shown in Figure 1B. Each ZFP bound its cognate site on naked DNA with high affinity (Kd = 20 and 44 pM, respectively).
Figure 1.
Structure of PPARγ gene and location of accessible chromatin in P2 promoter. (A) Genomic structure of mPPARγ showing exon splicing (Zhu et al. 1995). (B) Location of DNase I hypersensitive sites (DHS1 and DHS2) in the proximal P2 promoter, along with positions of ZFPs binding sites. (C) ZFP target sequences and finger designs. (Target sequence) The promoter DNA sequence to which each ZFP was designed. (Finger designs) The residues in each position from −1 to +6 of the recognition helix of each finger targeted against the cognate basepair triplet subsite.
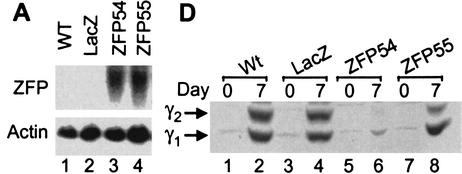
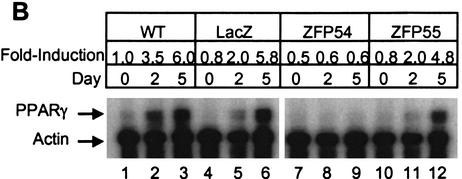
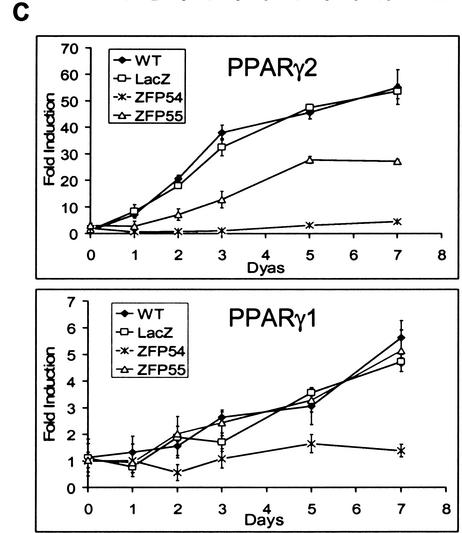
The ZFP54 and ZFP55 repressor proteins were expressed retrovirally in 3T3-L1 cells to similar levels (Fig. 2A). Nontransduced wild-type cells and stable pools of infected cells expressing each ZFP, as well as control cells retrovirally transduced to express LacZ, were induced to differentiate and initiate the adipogenic pathway. Total PPARγ mRNA level was determined at 0, 2, and 5 d post induction. Over the 5-d time course PPARγ expression increased ∼5.8- to 6.0-fold in both the wild-type and LacZ control cells (Fig. 2B). Up-regulation of total PPARγ expression was reduced slightly in the presence of ZFP55 (4.8-fold compared to 6.0-fold in wild type) but completely inhibited by ZFP54. However, a time course analysis of expression of the individual PPARγ isoforms revealed that although ZFP54 effectively inhibited expression of both isoforms, ZFP55 selectively repressed PPARγ2 by ∼50% and had little effect on PPARγ1 compared with wild-type cells (Fig. 2C). Another ZFP–KRAB fusion protein that binds to a sequence that is absent in the PPARγ gene has no effect on PPARγ expression or adipogenesis (data not shown). The effects of these ZFPs are confirmed at the protein level whereby PPARγ2 expression is virtually knocked out in the presence of ZFP54, whereas only a low but detectable level of PPARγ1 remains (Fig. 2D). In the presence of ZFP55 only PPARγ2 expression is reduced substantially by ZFP55. Because P1 and P2 are >63 kb apart, it was predicted that ZFPs targeted to P2 would selectively inhibit PPARγ2 expression. This was the case with ZFP55, which gave 50% knockdown of PPARγ2 only, however ZFP54 suppressed both isoforms almost completely. ZFP54 and ZFP55 bind to opposite strands of the DNA and have slightly different DNA binding affinities, which may contribute to their differential effects on P2.
Figure 2.
Ectopic expression of PPARγ2-specific ZFPs in 3T3-L1 cells. 3T3-L1 cells were infected with retroviral vectors expressing PPARγ2 promoter-specific zinc finger repressors, ZFP54 and ZFP55. The uninfected 3T3-L1 cells (wild type) or cells infected with retroviral vector expressing LacZ gene were used as negative controls. (A) Total RNA was isolated from selected cell lines and determined the levels of ZFP mRNA by Northern blot analysis using a 300 bp C-terminal fragment of ZFP as probe. The equivalence of RNA loading was verified by β-actin. (B) Total RNA was isolated on indicated days postdifferentiation and subjected to RNase protection assay using a PPARγ-specific riboprobe. Arrows indicate the protected PPARγ and β-actin mRNAs. The fold-induction was calculated by normalization of PPARγ-specific signal with β-actin signal and presented as fold-change compared to day-0 of wild-type cells. (C) The real time quantitative RT–PCR (TaqMan) analysis of PPARγ1 and PPARγ2 mRNA expression. 40 ng and 60 ng of reversed transcribed total RNA was used for PPARγ2 and PPARγ1 analysis, respectively. (D) Western blot analysis with a polyclonal antibody against PPARγ. The arrows indicate PPARγ1 and PPARγ2 proteins.
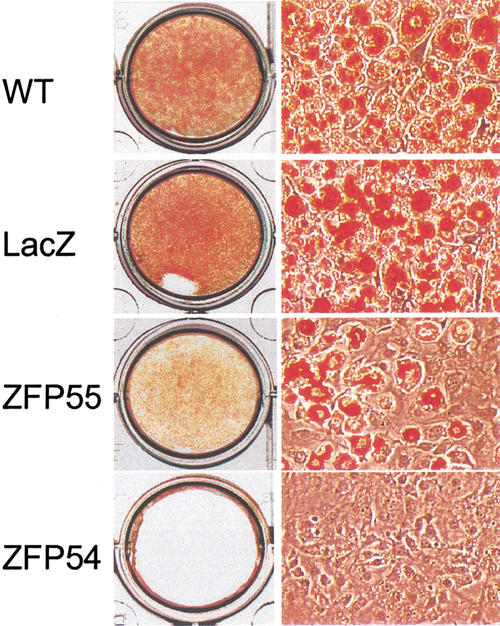
The promoter specificity exhibited by the two ZFPs facilitates examination of the functional requirements for each PPARγ isoform in adipogenesis. In complete concordance with inhibited mRNA and protein expression, we find that ZFP54 totally blocks the cellular lipid accumulation that is a marker of adipogenesis (Fig. 3). Furthermore, the cells in which only PPARγ2 is selectively repressed by 50% (ZFP55) also show a corresponding 50% loss in adipogenic capacity, supporting the idea of a specific requirement for PPARγ2 in this pathway.
Figure 3.
Effect of PPARγ2-ZFPs on adipogenesis. Cultured wild-type 3T3-L1 and retrovirally infected cells were induced to differentiate with the standard adipogenic hormones for 10 days. Cell were fixed in 3% formaldehyde and stained with Oil Red O. Stained cells were photographed with Nikon-Diaphot300 microscope (10 × 20) and 3CCD Vida Camera Systems (Optronic Engineering).
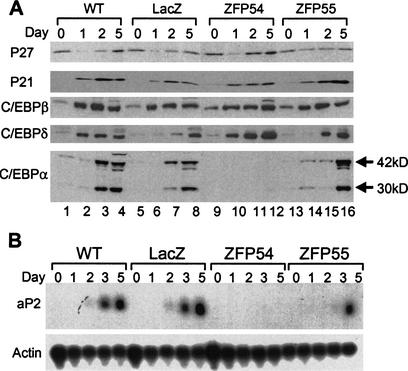
A key aspect of engineered ZFPs is the high theoretical specificity of DNA binding and gene targeting. Each ZFP is designed to recognize an 18-bp nucleotide sequence that probability dictates to be unique within the mammalian genome. To further characterize the specificity of PPARγ targeting, we analyzed the effect of ZFP expression on other genes both upstream and downstream of PPARγ in the adipogenic gene cascade. The cell cyclin-dependent kinase inhibitors p21 and p27 play key roles in cell cycle progression (Harper et al. 1993; Toyoshima and Hunter 1994). P27 is expressed abundantly in growth-arrested preadipocytes where its expression is decreased transiently on hormonal induction of differentiation but returns to initial levels as the cells exit clonal expansion. p21 is absent in growth-arrested cells but increases during S-phase of mitotic expansion (Morrison and Farmer 1999). Here we show that the expression profile of both p21 and p27 is preserved irrespective of the presence of ZFPs (Fig. 4A).
Figure 4.
Effects of ZFP54 and ZFP55 on the expression of cyclin-dependent kinase inhibitors, C/EBPs and aP2. Whole cells extracts and total RNA from the experiment shown in Figure 2 were subjected to the following analysis. (A) Western blot analysis of p21 and p27, and C/EBP gene family members (β, δ, and α). Arrows indicate the 42-kD and 30-kD isoforms of C/EBPα. (B) The aP2 gene expression was analyzed by Northern blot.
Transient expression of the transcription factors C/EBPβ and C/EBPδ occurs very early during adipocyte differentiation in response to the standard adipogenic hormones (IBMX/DEX/Insulin) (Cao et al. 1991; Wu et al. 1998). The proximal promoter of PPARγ2 contains a tandem array of C/EBP binding elements (Zhu et al. 1995). PPARγ is up-regulated immediately following C/EBPβ and C/EBPδ (Wu et al. 1998) and one would predict that expression of these upstream genes would not be affected by targeted repression of PPARγ. In concordance with this prediction, we found that the expression level of C/EBPβ protein was not significantly affected by the presence of either ZFPs (Fig. 4A). C/EBPδ expression also was not inhibited, but rather an increase in its expression was observed in the presence of ZFP54. This suggests a potential negative feedback mechanism whereby PPARγ may actively down-regulate C/EBPδ, in keeping with the reduction in C/EBPδ observed during progression of adipogenesis (Wu et al. 1998). Taken together these results support the notion that the inhibitory effects of ZFPs are specific.
Genes that are expressed during adipogenesis after PPARγ induction include C/EBPα and the adipocyte-specific aP2 (Wu et al. 1998; Hamm et al. 2001). As predicted, the loss of PPARγ expression in the presence of ZFP54 abrogates both C/EBPα and aP2 expression (Fig. 4A,B), supporting a key role for PPARγ in the regulatory cascade leading to the expression of these genes. However, in the presence of ZFP55 the expression of C/EBPα was delayed (Fig. 4A, lanes 3,7,15), although wild-type expression levels were achieved by day five. This suggests that although the remaining PPARγ expression in the presence of ZFP55 is sufficient to fully stimulate C/EBPα expression, it is not, in itself, sufficient to achieve wild-type levels of cellular differentiation and lipid accumulation.
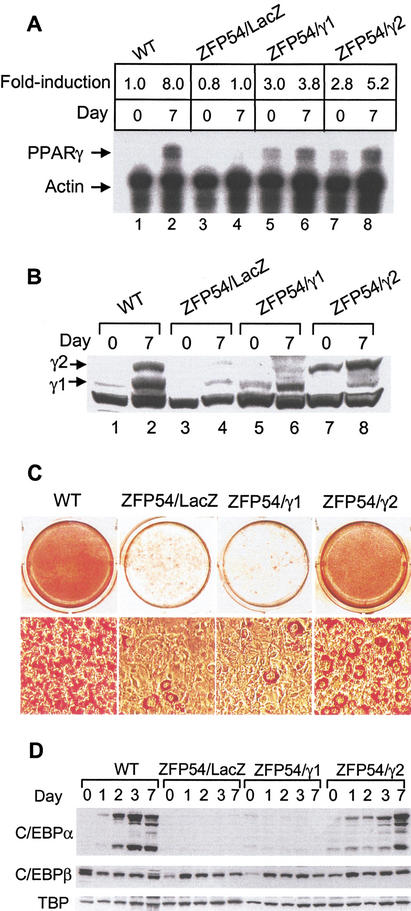
The studies by others described earlier have demonstrated an absolute requirement for PPARγ expression in adipose tissue development, but they failed to differentiate clearly between the specific regulatory capacity of PPAR γ1 and γ2 isoforms (Barak et al. 1999; Kubota et al. 1999; Rosen et al. 1999). In the present study, the abrogation of endogenous expression of both PPARγ isoforms by ZFP54 provides a unique cell-based model system in which to study the differential functions of each isoform. Selective repression of the endogenous PPARγ gene would be expected to generate what otherwise should be an adipogenically competent cell line that is devoid of only PPARγ. Rescue of these cells by exogenous expression of either PPAR γ1 or γ2 might be expected to identify isoform-specific functional differences in the capacity to potentiate adipogenesis. Here we used retrovirus to overexpress each PPARγ isoform on the background of ZFP54 expressing 3T3-L1 cells. Data in Figure 5A,B demonstrate the comparable levels of expression of exogenous PPARγ1 and γ2 mRNA and proteins at day 0 in the presence of ZFP54. The exogenously expressed PPARγ1 isoform was completely incapable of inducing adipogenesis in the presence of ZFP54 (Fig. 5C). In marked contrast, expression of PPARγ2 effectively restored cellular differentiation and lipid accumulation. In addition, PPARγ2 fully restored C/EBPα expression at 7 d postinduction, whereas PPARγ1 was totally ineffective (Fig. 5D). Neither isoform had any effect on the expression of C/EBPβ or TBP (Fig. 5D). This result clearly demonstrates for the first time a regulatory function for PPARγ2 in adipogenesis that cannot be achieved by PPARγ1 in the absence of exogenous ligand.
Figure 5.
Ectopic expression of PPARγ2, but not γ1 restores adipogenesis in ZFP54 cells. The L1/ZFP54 cells were infected with the retroviral constructs expressing either PPARγ1 (pBMN/neo-γ1) or PPARγ2 (pBMN/neo-γ2). The retroviral vector expressing LacZ was used as empty vector control. The doubly infected cell lines were designated as ZFP54/γ1, ZFP54/γ2, and ZFP54/LacZ, respectively. After the selection with puromycin and geneticin, cells were subjected to the differentiation protocol as described previously. (A) Total RNA was isolated from the indicated cell lines and PPARγ mRNA level was quantitated by RNase protection assay. (B) PPARγ1 and γ2 protein levels by Western blot analysis. (C) Cells were differentiated with the adipogenic hormones for 10 d and stained with Oil Red O. (D) C/EBPα and β protein expression by Western blot analysis. The membrane was stripped and reblotted with a rabbit polyclonal antibody against TBP.
It is unclear how the 30 amino acid residues unique to the PPARγ2 isoform confer additional regulatory function. However, in addition to rendering the constitutive activation function of the PPARγ2 amino terminus up to 10-fold greater than that of PPARγ1(Werman et al. 1997), clinical studies have identified a human allelic variant of at least one of these residues (Pro12Ala) that has been variously associated with decreased receptor activity, lower body mass index, obesity, improved insulin sensitivity, and decreased risk of type 2 diabetes (Beamer et al. 1998; Deeb et al. 1998; Altshuler et al. 2000).
The effective restoration of the adipogenic cascade by adding back only the product of the targeted gene, PPARγ, indicates that in this study the use of engineered repressor ZFPs does not result in the general disruption of cellular metabolic processes. This observation, which demonstrates a precise specificity for ZFP targeting, represents an important advance in the rapid establishment of cell-based gene knockdown models and such an approach may, in future, be readily transferable to whole animal transgenic studies.
Materials and methods
Design, synthesis, and DNA binding affinity of zinc finger proteins
Pairs of 3-finger ZFPs were generated as described previously (Zhang et al. 2000) then linked to form each 6-finger ZFP. The DNA binding affinity of each ZFP was determined using a gel shift technique essentially as described earlier (Zhang et al. 2000) except that 10 μM zinc was used.
Plasmids and stable cell lines
The retroviral expression vector pBMN (Garry Nolan Laboratory, Stanford University) was constructed by inserting an internal ribosome entry site (IRES) along with either puroror neor. The retroviral plasmids pBMNpuro–ZFP54 and pBMNpuro–ZFP55 were constructed by inserting ZFP54 and ZFP55 cDNA fragments into pBMN–puro vector at EcoRI and XhoI sites. pBMN/neo-γ1 was generated by inserting the full-length mPPARγ1 cDNA (nucleotide positions −6 to +1415) into pBMN–neo vector at BamHI and NotI sites. pBMN/neo-γ2 construct was derived from mPPARγ2/pSport (a gift from Dr. B. Spiegelman, Harvard Medical School, Boston, MA) by specifically ligating the blunt-ended PPARγ2 cDNA into pBMN/neo vector at the blunt-ended EcoRI site. To generate infectious recombinant viruses, Phoenix-ECO cells (Garry Nolan Laboratory, Stanford University, CA) were cultured in DMEM medium containing 10% FBS. Cells were transfected with 15 μg of retroviral plasmids by Lipofectamine 2000 kit (GIBCO BRL) following the protocol as recommended by the manufacturer. Viral supernatant was harvested at 72 h posttransfection and added onto ∼75% confluent 3T3-L1 cells in the presence of 8 μg/mL polybrene (Sigma) for 2 h. Infected cells were selected by adding 2 μg/mL puromycin (Sigma) for 5 d. For double infection, L1/ZFP54 cells were infected with either pBMN/neo-γ1, or pBMN/neo-γ2, and cells were selected with both 600 μg/mL geneticin (GIBCO BRL) and 2 μg/mL puromycin for 14 d. Pools of stably infected cells were used in these experiments.
Cell culture and differentiation
Uninfected 3T3-L1 and virally infected 3T3-L1 cells were cultured and maintained for 2 d postconfluence in DMEM containing 10% CS. Differentiation protocol was conducted as described previously (Camp et al. 2001)
Mapping of DNase I accessible chromatin regions
DNase I mapping was performed essentially as described previously (Liu et al. 2001). Undifferentiated 3T3-L1 cell nuclei were partially digested with DNase I and the genomic DNA was extracted followed by XbaI digestion at −1588 bp in the P2 promoter. The probe for Southern blotting of the proximal P2 promoter was an XbaI/EcoRI fragment spanning from −1588 to −1007.
The real-time quantitative PCR (TaqMan)
Gene-specific primers and probes were designed using the Primer Express software (Perkin Elmer Life Sciences). The real-time quantitative RT–PCR reaction was performed essentially following the manufacturer's protocol. Briefly, reaction mixture contained 5.5 mM MgCl2, 500 μM dNTP, 2.5 μM random hexamers, 200 nM FAM-probe, and 600 nM of both forward and reverse primers in a final volume of 25 μL, and was analyzed in ABI PRISM 7700 sequence detection system (Perkin Elmer Life Sciences). Relative quantitation of PPARγ mRNA levels was plotted as fold-change compared to day 0 of wild-type 3T3-L1 cells. 18S ribosomal RNA was used for normalization. TaqMan reverse transcriptase reactions were performed in triplicates and the experiments were repeated independently at least three times.
RNA isolation, Northern blot, and RNase protection assay
Total RNA was isolated from cultured cells using Ultraspec RNA system (Biotecx Laboratories, Inc.). ZFP, aP2, and rat β-actin cDNA were labeled with [α-32P] dCTP (Amersham) using random primed labeling kit (GIBCO BRL). Northern blot analysis and RNase protection assay (RPA) were performed as described previously (Camp et al. 1999).
Immunoblot analysis
Cells were lysed in HNTG cell lysate buffer (50 mM Hepes, 150 mM NaCl, 10% glycerol, 1% Triton, 1.5 mM MgCl2, and 1 mM EDTA) at 4°C for 15 min, followed by centrifugation at 15,000 rpm at 4°C for 10 min. The supernatant was collected and the protein concentration was determined by BCA protein assay (Pierce). Western blot analysis was carried out as described previously (Camp et al. 1999). Rabbit polyclonal antibodies against C/EBPα, C/EBPβ, C/EBPδ, TBP, and monoclonal antibodies against p21 and p27 were purchased from Santa Cruz.
Acknowledgments
We thank Neelam Srivastava, Katherine Pasquetti, and Hong Qi for technical assistance, and Casey Case, Todd Leff, and Yen Choo for valuable discussion and criticism. We dedicate this work to the memory of our friend and colleague, Alan P. Wolffe.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL heidi.camp@pfizer.com; FAX (734) 622-5668.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.953802.
References
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPARγ is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- Bartsevich VV, Juliano RL. Regulation of the MDR1 gene by transcriptional repressors selected using peptide combinatorial libraries. Mol Pharmacol. 2000;58:1–10. doi: 10.1124/mol.58.1.1. [DOI] [PubMed] [Google Scholar]
- Beamer BA, Yen CJ, Andersen RE, Muller D, Elahi D, Cheskin LJ, Andres R, Roth J, Shuldiner AR. Association of the Pro12Ala variant in the peroxisome proliferator-activated receptor-γ2 gene with obesity in two Caucasian populations. Diabetes. 1998;47:1806–1808. doi: 10.2337/diabetes.47.11.1806. [DOI] [PubMed] [Google Scholar]
- Beerli RR, Dreier B, Barbas CF., III Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp HS, Whitton AL, Tafuri SR. PPARγ activators down-regulate the expression of PPARγ in 3T3-L1 adipocytes. FEBS Lett. 1999;447:186–190. doi: 10.1016/s0014-5793(99)00268-9. [DOI] [PubMed] [Google Scholar]
- Camp HS, Chaudhry A, Leff T. A novel potent antagonist of peroxisome proliferator-activated receptor gamma blocks adipocyte differentiation but does not revert the phenotype of terminally differentiated adipocytes. Endocrinology. 2001;142:3207–3213. doi: 10.1210/endo.142.7.8254. [DOI] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes & Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Choo Y, Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc Natl Acad Sci. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb SS, Fajas L, Nemoto M, Pihlajamaki J, Mykkanen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARγ2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- Desjarlais JR, Berg JM. Redesigning the DNA-binding specificity of a zinc finger protein: A data base-guided approach. Proteins. 1992;12:101–104. doi: 10.1002/prot.340120202. [DOI] [PubMed] [Google Scholar]
- Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, Najib J, Laville M, Fruchart JC, Deeb S, et al. The organization, promoter analysis, and expression of the human PPARγ gene. J Biol Chem. 1997;272:18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- Hamm JK, Park BH, Farmer SR. A role for C/EBPβ in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3-L1 preadipocytes. J Biol Chem. 2001;276:18464–18471. doi: 10.1074/jbc.M100797200. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Jamieson AC, Kim SH, Wells JA. In vitro selection of zinc fingers with altered DNA-binding specificity. Biochemistry. 1994;33:5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, Umesono K, Evans RM. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, et al. PPARγ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol. Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- Liu PQ, Rebar EJ, Zhang L, Liu Q, Jamieson AC, Liang Y, Qi H, Li PX, Chen B, Mendel MC, et al. Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions. Activation of vascular endothelial growth factor A. J Biol Chem. 2001;276:11323–11334. doi: 10.1074/jbc.M011172200. [DOI] [PubMed] [Google Scholar]
- Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJ., III Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison RF, Farmer SR. Role of PPARγ in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J Biol Chem. 1999;274:17088–17097. doi: 10.1074/jbc.274.24.17088. [DOI] [PubMed] [Google Scholar]
- Rebar EJ, Pabo CO. Zinc finger phage: Affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPARγ2: Tissue-specific regulator of an adipocyte enhancer. Genes & Dev. 1994a;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994b;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- Werman A, Hollenberg A, Solanes G, Bjorbaek C, Vidal-Puig AJ, Flier JS. Ligand-independent activation domain in the N terminus of peroxisome proliferator-activated receptor gamma (PPARγ). Differential activity of PPARγ-1 and -2 isoforms and influence of insulin. J Biol Chem. 1997;272:20230–20235. doi: 10.1074/jbc.272.32.20230. [DOI] [PubMed] [Google Scholar]
- Wu Z, Xie Y, Morrison RF, Bucher NL, Farmer SR. PPARγ induces the insulin-dependent glucose transporter GLUT4 in the absence of C/EBPα during the conversion of 3T3 fibroblasts into adipocytes. J Clin Invest. 1998;101:22–32. doi: 10.1172/JCI1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Spratt SK, Liu Q, Johnstone B, Qi H, Raschke EE, Jamieson AC, Rebar EJ, Wolffe AP, Case CC. Synthetic zinc finger transcription factor action at an endogenous chromosomal site. Activation of the human erythropoietin gene. J Biol Chem. 2000;275:33850–33860. doi: 10.1074/jbc.M005341200. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qi C, Korenberg JR, Chen XN, Noya D, Rao MS, Reddy JK. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPARγ) gene: Alternative promoter use and different splicing yield two mPPARγ isoforms. Proc Natl Acad Sci. 1995;92:7921–7925. doi: 10.1073/pnas.92.17.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]