Abstract
The budding yeast kinetochore is composed of an inner and outer protein complex, which binds to centromere (CEN) DNA and attaches to microtubules. We performed a genetic synthetic dosage lethality screen to identify novel kinetochore proteins in a collection of chromosome transmission fidelity mutants. Our screen identified several new kinetochore-related proteins including YLR381Wp/Ctf3p, which is a member of a conserved family of centromere-binding proteins. Ctf3p interacts with Mcm22p, Mcm16p, and the outer kinetochore protein Ctf19p. We used chromatin immunoprecipitation to demonstrate that Ctf3p, Mcm22p, and Mcm16p bind to CEN DNA in a Ctf19p-dependent manner. In addition, Ctf3p, Mcm22p, and Mcm16p have a localization pattern similar to other kinetochore proteins. The fission yeast Ctf3p homolog, Mis6, is required for loading of a CENP-A centromere specific histone, Cnp1, onto centromere DNA. We find however that Ctf3p is not required for loading of the budding yeast CENP-A homolog, Cse4p, onto CEN DNA. In contrast, Ctf3p and Ctf19p fail to bind properly to the centromere in a cse4-1 mutant strain. We conclude that the requirements for CENP-A loading onto centromere DNA differ in fission versus budding yeast.
Keywords: S. cerevisiae, kinetochore, centromere, chromosome segregation
High fidelity chromosome transmission, which is necessary for eukaryotic cell survival, requires coordination of many events including DNA replication, sister chromatid cohesion, and kinetochore function. The kinetochore, which is composed of centromere (CEN) DNA and associated proteins, mediates attachment of chromosomes to the spindle. The kinetochore also provides a site for centromeric cohesion and generates signals to arrest cell cycle progression if metaphase has not been achieved properly (for review, see Kitagawa and Hieter 2001). Shortly after spindle pole body (SPB) duplication, sister centromeres separate transiently and oscillate along the spindle axis until anaphase when permanent sister separation occurs (Goshima and Yanagida 2000; He et al. 2000; Tanaka et al. 2000; Pearson et al. 2001). The mechanism by which kinetochores assemble, attach to microtubules, and migrate toward SPBs in anaphase is not well understood.
In the budding yeast Saccharomyces cerevisiae, the minimal required CEN DNA element (CDE) consists of two palindromic sequences, CDEI and CDEIII, flanking an A-T rich CDEII sequence. CDEIII, which is essential, is bound by the inner kinetochore complex CBF3 (for reviews, see Clarke 1998; Ortiz and Lechner 2000; Pidoux and Allshire 2000). CBF3 is composed of four essential proteins: Ndc10p/Ctf14p/Cep2p, Cep3p, Ctf13p, and Skp1p. Skp1p and its interacting partner Sgt1p have roles at the kinetochore in G2 and in SCF-mediated degradation in G1 (Bai et al. 1996; Connelly and Hieter 1996; Kitagawa et al. 1999). CEN DNA is thought to be wrapped around a specialized nucleosome containing a conserved histone H3-like protein, Cse4p (Cnp1 in fission yeast and CENP-A in higher eukaryotes), in place of a core histone H3 (for review, see Sullivan 2001). In addition to its conserved C-terminal histone fold domain, Cse4p contains a unique and essential N-terminal domain that interacts with members of the yeast outer kinetochore (Keith et al. 1999; Ortiz et al. 1999; Chen et al. 2000). The outer kinetochore protein complex, composed of Mcm21p, Okp1p, and Ctf19p, interacts with CEN DNA in a CBF3 dependent manner (Hyland et al. 1999; Ortiz et al. 1999). The process by which the outer kinetochore proteins assemble onto the inner kinetochore is largely unknown. Ndc10p is required for both Cse4p and the outer kinetochore proteins to bind CEN DNA efficiently (Ortiz et al. 1999). In the fission yeast Schizosaccharomyces pombe, the Cse4p homolog Cnp1 requires an essential centromere protein, Mis6, to localize and bind to centromere DNA (Takahashi et al. 2000). The specific requirements for Cse4p loading onto the centromere in budding yeast are not known.
Many other proteins have been placed at the kinetochore via chromatin immunoprecipitation (ChIP) assays and localization studies (He et al. 2001; Kitagawa and Hieter 2001). A new protein complex consisting of Spc24p, Spc25p, Nuf2p, and Ndc80p was found to localize to CEN DNA (Janke et al. 2001; Wigge and Kilmartin 2001). This Ndc80p protein complex may be part of the missing link from the kinetochore to the SPB. Other potential kinetochore proteins exist based on mutant phenotypes suggestive of kinetochore function. At least three mutant collections, the chromosome transmission fidelity (ctf), minichromosome maintenance (mcm) and chromosome loss (chl) mutants, which share a common phenotype of chromosome loss, contain potential kinetochore mutants (Maine et al. 1984; Larionov et al. 1987; Spencer et al. 1990). Secondary screens applied to the ctf mutant collection successfully identified Ctf13p, Ndc10p/Ctf14p, and Ctf19p as kinetochore proteins (Doheny et al. 1993; Kroll et al. 1996; Hyland et al. 1999). In addition to a high rate of chromosome loss, a subset of mcm, chl, and ctf mutants display phenotypes reminiscent of kinetochore mutants (Doheny et al. 1993; Kouprina et al. 1993; Roy et al. 1997; Sanyal et al. 1998; Poddar et al. 1999; Ghosh et al. 2001). Thus new kinetochore proteins remain to be identified from these chromosome loss mutant collections.
We have taken a genetic approach, termed synthetic dosage lethality (SDL), to identify novel kinetochore proteins. A protein isolated in our screen, YLR381Wp/Ctf3p, shares sequence similarity with the S. pombe Mis6 centromere-binding protein (Saitoh et al. 1997). We present evidence that Ctf3p and two interacting proteins, Mcm22p and Mcm16p, are new outer kinetochore proteins that interact with and localize to CEN DNA. Unlike the role of Mis6 in fission yeast, Ctf3p is not required for Cse4p/CENP-A to bind centromere DNA, suggesting a different mechanism of kinetochore assembly in budding yeast.
Results
Synthetic dosage lethality screen
An SDL assay is based on the premise that increasing the level of a protein (such as a kinetochore protein) has no detrimental effect on the growth of a wild-type strain but may cause lethality in a target mutant strain (such as a kinetochore mutant) that has reduced activity of an interacting protein (Kroll et al. 1996; Measday and Hieter 2002). To isolate additional kinetochore mutants represented in the ctf collection, we performed an SDL screen by overexpressing CTF13, NDC10/CTF14, and CTF19 in all ctf mutant strains in which the mutated gene locus was unknown. ctf mutants that did not tolerate overexpression of kinetochore proteins, but were able to grow with vector alone, were identified as SDL mutants. Overexpression of CTF13 and/or NDC10/CTF14 was lethal in 11 mutants out of 69 transformed, whereas overexpression of CTF19 was not detrimental to any ctf mutants in the subset tested (Table 1). From the 11 mutant strains isolated in our screen, five carried alleles of ctf3, four carried alleles of ctf5 and two were the single member mutants s20 and s155. In addition, five mutants grew very poorly with increased dosage of kinetochore proteins: three strains carrying alleles of ctf3 and two single member mutants, s165 and s166 (Table 1).
Table 1.
Summary of synthetic dosage lethality screen
|
ctf
mutant
|
Synthetic dosage
lethality
|
||
|---|---|---|---|
| Complementation group or single
member number
|
Total no.allelesa
|
CTF13
|
NDC10/CTF14
|
| ctf3 | 11 | ctf3-45 | ctf3-2, -14, -34, -45, -52 |
| ctf5 | 5 | ctf5-70, -110, -131 | ctf5-31, -70, -110, -131 |
| s20 | 1 | s20 | s20 |
| s155 | 1 | s155 | s155 |
| Slow growth | |||
| CTF13NDC10/CTF14 | |||
| ctf3 | 11 | ctf3-2, -52 | ctf3-135 |
| s165 | 1 | s165 | |
| s166 | 1 | s166 | |
Total number of alleles in ctf collection.
Identification of ctf mutants from SDL screening
To clone the genes corresponding to each of the ctf mutants isolated in the SDL screen, we rescued the chromosome loss phenotype originally used to isolate the ctf mutant collection (Koshland and Hieter 1987; Spencer et al. 1990). Three known, or putative, kinetochore proteins were identified as the gene loci mutated in ctf5 (mcm21), s20 (chl4/ctf17-20/mcm17) and s155 (mcm16-155) mutants (Table 2). Mcm21p/Ctf5p is a member of the outer kinetochore complex that interacts with Ctf19p and Okp1p (Ortiz et al. 1999). Chl4p/Ctf17p/Mcm17p is predicted to function at the kinetochore (Kouprina et al. 1993; Roy et al. 1997). A different allele of ctf17 (ctf17-61) was identified previously in a CTF13 overexpression SDL screen (Kroll et al. 1996). Finally, Mcm16p has been implicated as a kinetochore protein and mcm16Δ cells lose chromosome III at a rate 50-fold higher than wild-type strains (Sanyal et al. 1998).
Table 2.
Identification of ctf mutants isolated in SDL screen
| Complementation group or single
member number
|
Rescue
gene
|
Function
|
|---|---|---|
| ctf3 | YLR381W | Unknown |
| ctf5 | MCM21 | Kinetochore protein |
| s20 | CHL4/CTF17/ MCM17 | Putative kinetochore protein |
| s155 | MCM16 | Putative kinetochore protein |
| s165 | IRR1/SCC3 | Cohesin |
| s166 | SMC1 | Cohesin |
Two mutants isolated in the SDL screen, s165 and s166, were rescued by genes encoding the cohesin proteins, Irr1p/Scc3p and Smc1p, respectively (Table 2). A cohesin complex composed of at least four essential cohesin proteins, Mcd1p/Scc1p, Irr1p/Scc3p, Smc1p, and Smc3p, maintains sister chromatid arm cohesion from S phase until the onset of anaphase (for review, see Koshland and Guacci 2000; Nasmyth et al. 2000; Amon 2001). Cohesin proteins are more densely distributed at the centromere and coimmunoprecipitate with CEN DNA (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999; Hartman et al. 2000; Panizza et al. 2000). Lethality of a cohesin mutant due to overexpression of a kinetochore protein, however, has not been shown previously.
The gene locus mutated in ctf3 mutants was identified as YLR381W, an open reading frame (ORF) of unknown function (Table 2). BLAST analysis revealed that YLR381Wp/Ctf3p shares 20% identity and 33% similarity along its entire length with the S. pombe Mis6 centromere-binding protein (Fig. 1) (Saitoh et al. 1997). Mis6 was shown previously to share similarity with human and rat LRPR1, a hormone-regulated leucine-rich protein (Slegtenhorst-Eegdeman et al. 1995; Roberts et al. 1996; Saitoh et al. 1997). Interestingly, human LRPR1 has recently been found to localize constitutively to centromeres (Song-Tao Liu and Tim Yen, pers. comm.). In addition, a BLAST search with Mis6 revealed similarity to a Neurospora crassa protein that we have called N. crassa Mis6 (Fig. 1). Thus, Ctf3p is a member of a protein family that contains at least two centromere-localized proteins.
Figure 1.
Saccharomyces cerevisiae Ctf3p, Schizosaccharomyces pombe Mis6, human LRPR1, and Neurospora crassa Mis6 homolog (GenBank accession no. CAC28775) protein alignments. Black boxes are identical amino acids, gray boxes are similar amino acids.
Genome-wide two-hybrid screens
To further study the function of Ctf3p, we cloned CTF3 into the Gal4 DNA binding domain vector pOBD2 and screened by yeast two-hybrid assay for protein interactions against a genomic array of ∼6000 ORFs (Uetz et al. 2000). Mcm22p was identified in two independent screens as a Ctf3p interacting protein. mcm22 mutant strains lose chromosome III 100-fold more frequently than wild-type strains and have other phenotypes implicating a role for Mcm22p at the kinetochore (Maine et al. 1984; Poddar et al. 1999). Interestingly, genome-wide two-hybrid screens identified Mcm16p and Mcm22p as interacting partners (Uetz et al. 2000). Moreover, an allele of mcm16 (s155) was isolated in our SDL screen (Table 2). We cloned MCM22 into pOBD2 and performed a genome-wide two-hybrid screen. Mcm22p interacted with Spc34p in two independent screens. Spc34p is a kinetochore protein that was first isolated in a preparation of spindle poles and associated microtubules (Wigge et al. 1998; He et al. 2001).
Ctf3p, Mcm22p, Mcm16p, and Ctf19p interact in yeast lysates
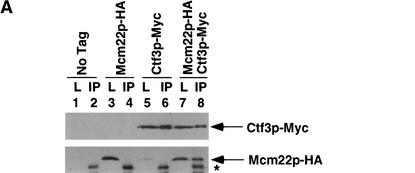
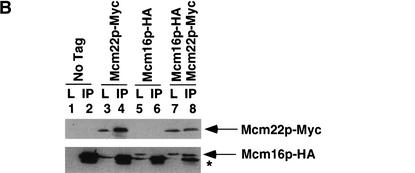
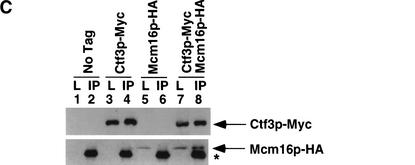
We performed coimmunoprecipitation assays from yeast lysates with protein partners that interacted in the yeast two-hybrid screens. Ctf3p, Mcm22p, and Mcm16p were epitope-tagged with either 13 Myc or triple HA and strains were constructed that expressed one HA-tagged protein and one Myc-tagged protein. Log phase cells were lysed and anti-Myc immunoprecipitations were performed. Ctf3p–Myc interacted with Mcm22p–HA (Fig. 2A, lane 8) and Mcm16p–HA (Fig. 2C, lane 8) and Mcm22p–Myc interacted with Mcm16p–HA (Fig. 2B, lane 8) only when both tagged proteins were expressed. All three proteins also interacted with each other when immunoprecipitations were done with anti-HA conjugated Sepharose beads (data not shown). Thus, Ctf3p, Mcm22p, and Mcm16p interact both by two-hybrid and coimmunoprecipitation assays.
Figure 2.
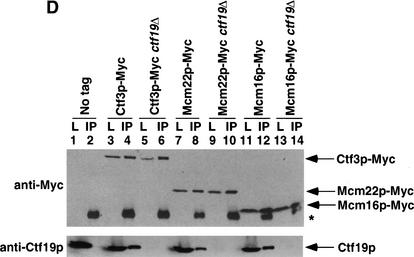
Ctf3p, Mcm22p, Mcm16p, and Ctf19p coimmunoprecipitate from yeast lysates. The indicated strains were grown to log phase, lysed and anti-Myc immunoprecipitations were performed. Forty micrograms of whole-cell lysate (L) and 25% of total immunoprecipitate (IP) were loaded on the gel. Western blots were performed to detect Myc- or HA-tagged proteins. The yeast strains used were: (A) untagged (No Tag, YVM100), Mcm22p–HA (YVM210), Ctf3p–Myc (YVM218), Mcm22p–HA Ctf3p–Myc (YVM255); (B) untagged (No Tag, YVM100), Mcm22p–Myc (YVM 290), Mcm16p–HA (YVM344), Mcm16p–HA Mcm22p–Myc (YVM346); (C) untagged (No Tag, YVM100), Ctf3p–Myc (YVM218), Mcm16p–HA (YVM344), Ctf3p–Myc Mcm16p–HA (YVM336); and (D) untagged (No Tag, YVM100), Ctf3p–Myc (YVM218), Ctf3p–Myc ctf19Δ (YVM400), Mcm22p–Myc (YVM 290), Mcm22p–Myc ctf19Δ (YVM398), Mcm16p–Myc (YVM325), Mcm16p–Myc ctf19Δ (YVM402). Asterisks are IgG antibody light chain.
We next tested for an interaction between Ctf3p, Mcm22p, Mcm16p, and members of the CBF3 kinetochore complex. We could not detect Ndc10p/Ctf14p, Cep3p, Skp1p, or Sgt1p in Ctf3p, Mcm22p, or Mcm16p immune complexes (data not shown). Genetic experiments, however, suggested an interaction between the Ctf3p complex and CBF3. ctf3, mcm22, mcm16, and ctf19 deletion strains (which are viable on their own or in combination) were synthetically lethal (SL) in combination with a ctf14-42 (ndc10) mutation (Table 3; Hyland et al. 1999). In addition, deleting CTF3, MCM22, MCM16, or CTF19 in a ctf13-30 strain lowers the nonpermissive temperature of the ctf13-30 mutant from 35°C to 32°C (Table 3).
Table 3.
Genetic interactions
| Genotype
|
Phenotype
|
|---|---|
| ctf3ΔHIS3 mcm22ΔURA3 mcm16ΔkanMX6 | viable |
| ctf3ΔHIS3 mcm22ΔURA3 mcm16ΔkanMX6 ctf19ΔTRP1 | viable |
| ctf3ΔHIS3 ctf13-30 | CSLa |
| mcm22ΔURA3 ctf13-30 | CSLa |
| mcm16ΔkanMX6 ctf13-30 | CSLa |
| ctf3ΔHIS3 mcm22ΔURA3 mcm16ΔkanMX6 ctf13-30 | CSLa |
| ctf3ΔHIS3 mcm22ΔURA3 mcm16ΔkanMX6 ctf19ΔTRP1 ctf13-30 | CSLa |
| ctf3ΔHIS3 ctf14-42 | SL |
| mcm22ΔURA3 ctf14-42 | SL |
| mcm16ΔkanMX6 ctf14-42 | SL |
| ctf3ΔTRP1 cse4-1 | CSLb |
| ctf19ΔTRP1 cse4-1 | CSLb |
SL, synthetic lethality.
CSL, conditional synthetic lethality. ctf3-30 is lethal at 35°C, nonpermissive temperature is reduced to 32°C when combined with ctf3Δ, mcm22Δ, mcm16Δ, ctf3Δmcm22Δmcm16Δ, or ctf3Δmcm22Δmcm16Δctf19Δ mutants.
cse4-1 is lethal at 38°C (Stoler et al. 1995), nonpermissive temperature is reduced to 35°C when combined with ctf3Δ or ctf19Δ mutants.
We next explored potential interactions between Ctf3p, Mcm22p, Mcm16p, and the outer kinetochore. Using a Ctf19p polyclonal antibody, we detected Ctf19p in anti-Myc Ctf3p, Mcm22p, and Mcm16p immunoprecipitates from a wild-type but not a ctf19Δ strain (Fig. 2D, cf. lanes 4 and 6, 8 and 10, 12 and 14). Ctf19p interacts with Ndc10p in vitro and the Ctf19p-interacting proteins Mcm21p and Okp1p interact with members of the CBF3 complex from yeast lysates (Ortiz et al. 1999). We conclude that Ctf3p, Mcm22p, and Mcm16p may be linked to the inner kinetochore via Ctf19p.
Ctf3p, Mcm22p, and Mcm16p associate with CEN DNA in a Ctf19p-dependent manner
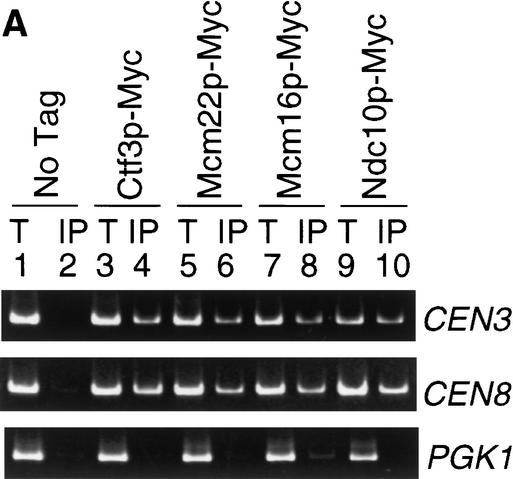
Having shown that Ctf3p, Mcm22p, and Mcm16p interact with each other and an established kinetochore protein, Ctf19p, we next tested their ability to bind CEN DNA by performing ChIP assays. Myc epitope-tagged Ctf3p, Mcm16p, Mcm22p, and Ndc10p were isolated from chromatin preparations of log phase cells, followed by PCR analysis to determine if CEN DNA was present in the immunoprecipitates (Fig. 3A). The templates for both total chromatin and immunoprecipitate were titrated to determine the linear range for PCR (data not shown). As expected, a region of CEN3 and CEN8 were amplified specifically from the Ndc10p immunoprecipitate (Fig. 3A, lane 10), but not from the untagged strain immunoprecipitate (Fig. 3A, lane 2). Similarly, Ctf3p, Mcm16p, and Mcm22p all specifically precipitated CEN DNA but not a non-CEN locus, PGK1 (Fig. 3A, lanes 4,6,8).
Figure 3.
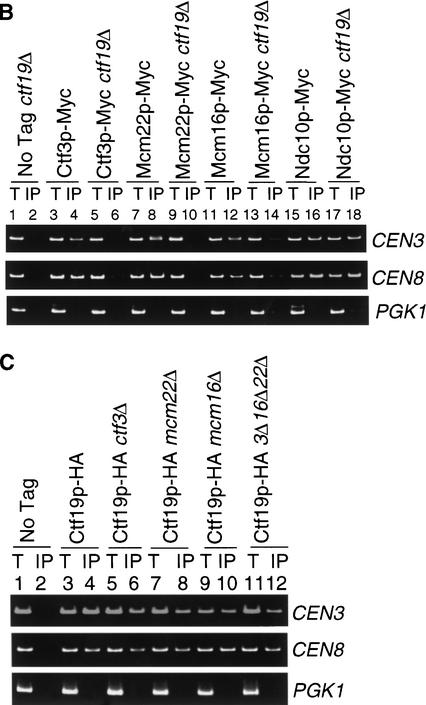
Ctf3p, Mcm22p, and Mcm16p coimmunoprecipitate CEN DNA in a Ctf19p dependent manner. Anti-Myc or anti-HA ChIP assays were performed either from log phase cells (A) or Nz arrested cells (B,C) (treated with 15 μg/mL Nz for 100 min). The templates used for PCR (see Materials and Methods) were total chromatin (T) or immunoprecipitate (IP). The yeast strains used were: (A) untagged (No Tag, YVM100), Ctf3p–Myc (YVM218), Mcm22p–Myc (YVM290), Mcm16p–Myc (YVM325), Ndc10p–Myc (YVM499); (B) untagged (No Tag, YVM100), Ctf3p–Myc (YVM218), Ctf3p–Myc ctf19Δ (YVM400), Mcm22p–Myc (YVM 290), Mcm22p–Myc ctf19Δ (YVM398), Mcm16p–Myc (YVM325), Mcm16p–Myc ctf19Δ (YVM402), Ndc10p–Myc (YVM499), Ndc10p–Myc ctf19Δ (YVM729); and (C) untagged (No tag, YVM100), Ctf19p–HA (YPH132; Hyland et al. 1999), Ctf19p–HA ctf3Δ (YVM479), Ctf19p–HA mcm22Δ (YVM481), Ctf19p–HA mcm16Δ (YVM491), Ctf19p–HA ctf3Δmcm16Δmcm22Δ (YVM 496; noted as 3Δ16Δ22Δ in the Figure).
Because Ctf19p is present in immunoprecipitations of Ctf3p, Mcm16p, and Mcm22p (Fig. 2D), we tested if the interaction of Ctf3p, Mcm22p, and Mcm16p with CEN DNA depends on Ctf19p. Ctf3p, Mcm22p, and Mcm16p were immunoprecipitated from chromatin preparations from either wild-type or ctf19Δ strains. Cells were arrested with nocodazole (Nz), a drug that depolymerizes microtubules, to synchronize cells in G2 phase. ctf19Δ strains respond in a wild-type manner to Nz. It was important to ensure an equivalent population of cells in the wild-type versus ctf19Δ strain because a log phase ctf19Δ cell population has an increased number of G2 cells (Hyland et al. 1999). Ctf3p, Mcm22p, Mcm16p, and Ndc10p were still able to bind CEN DNA after Nz treatment, suggesting that intact microtubules are not required for their CEN interaction (Fig. 3B, lanes 4,8,12,16). Interestingly, Ctf3p, Mcm22p, and Mcm16p all showed significant reductions in association with CEN DNA in a ctf19Δ strain (Fig. 3B, cf. lanes 4 and 6, 8 and 10, 12 and 14). The reduced CEN binding was not attributable to a problem with immunoprecipitation as Western blot analysis showed that Ctf3p, Mcm16p, and Mcm22p were immunoprecipitated efficiently from ctf19Δ strains (data not shown). The inner kinetochore protein Ndc10p was still able to bind CEN DNA in a strain lacking Ctf19p (Fig. 3B, cf. lanes 16 and 18).
To address the reciprocal protein–CEN dependencies, we performed ChIP to test whether Ctf19p is able to bind CEN DNA in the absence of Ctf3p, Mcm22p, and Mcm16p. Cells were synchronized by Nz treatment to arrest cells in G2. ctf3Δ, mcm22Δ, and mcm16Δ mutants, which have wild-type FACs profiles in log phase cells, all respond in a wild-type manner to spindle damage induced by Nz treatment suggesting a normal spindle checkpoint response (data not shown). Ctf19p has been shown previously to interact with CEN DNA in Nz-treated cells (Hyland et al. 1999). HA epitope-tagged Ctf19p (Hyland et al. 1999) was immunoprecipitated from chromatin preparations and found to interact with CEN DNA in ctf3Δ, mcm16Δ, mcm22Δ strains, and in the viable triple ctf3Δmcm16Δmcm22Δ mutant (Fig. 3C, cf. lane 4 with lanes 6,8,10,12). Thus Ctf19p, which binds CEN DNA in an Ndc10p-dependent manner (Ortiz et al. 1999), does not require Ctf3p, Mcm16p, or Mcm22p to interact with CEN DNA. Taken together, these data strongly suggest that Ctf3p, Mcm22p, and Mcm16p form a separate complex that is located distal to the Ctf19p and CBF3 protein complexes.
Localization of Ctf19p, Ctf3p, Mcm22p, Mcm16p, and Spc34p
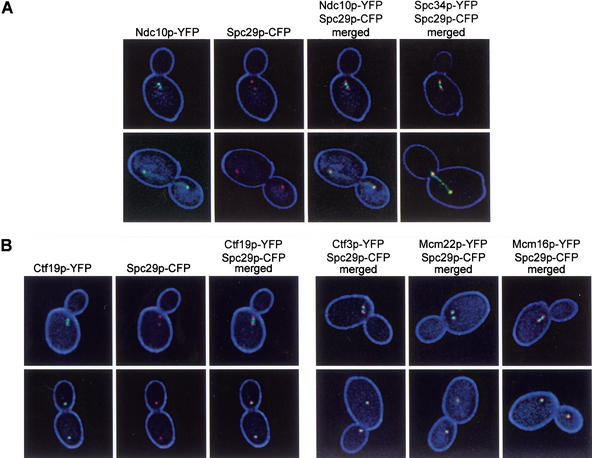
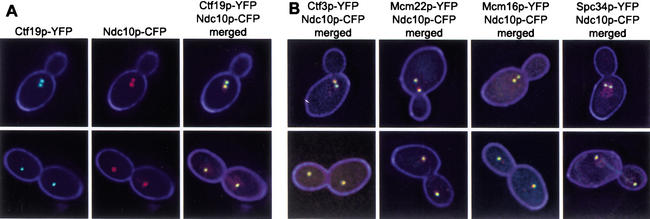
The interaction of Ctf3p, Mcm22p, and Mcm16p with CEN DNA suggests their presence at the kinetochore. Recent cell imaging data has shown that in budded cells, kinetochore proteins and CEN DNA lie adjacent to the interior side of the SPB (Goshima and Yanagida 2000; He et al. 2000, 2001; Pearson et al. 2001). Yellow fluorescent protein (YFP)-tagged kinetochore proteins were expressed in a strain containing a cyan fluorescent protein (CFP)-tagged SPB protein (Spc29p–CFP). Ndc10p was imaged as a control for the localization of an authentic kinetochore protein. In cells with short spindles (preanaphase), Ndc10p–YFP lay adjacent to the interior side of Spc29p–CFP (Fig. 4A). In cells with long spindles (postanaphase), Ndc10p–YFP partially colocalized with Spc29p–CFP and stained along the spindle (Fig. 4A), as seen by others (Goh and Kilmartin 1993; Goshima and Yanagida 2000). Indirect immunofluorescence studies showed previously that Ctf19p localized near the SPB (Hyland et al. 1999). Ctf19p–YFP localized adjacent to the nuclear side of the SPB in preanaphase cells. In postanaphase cells, Ctf19p–YFP and Spc29p–CFP signals overlapped considerably (Fig. 4B). Ctf3p–YFP, Mcm22p–YFP, and Mcm16p–YFP showed a similar localization pattern to Ctf19p–YFP (Fig. 4B). In addition, we tested localization of the Mcm22p two-hybrid interacting protein, Spc34p. Spc34p–YFP also localized next to the SPB in preanaphase cells and, similar to Ndc10p–YFP, stained along the spindle in postanaphase cells (Fig. 4A) (Wigge et al. 1998; He et al. 2001).
Figure 4.
Localization of kinetochore–YFP fusions compared to Spc29p–CFP. All strains are homozygous diploids containing genomic carboxy-terminal CFP and YFP fusions. YFP signal appears green and CFP signal appears red. Overlapping of YFP and CFP signal appears yellow. (A) Ndc10p–YFP Spc29p–CFP (YVM1176) — YFP, CFP, and merged images are shown as an example; Spc34p–YFP Spc29p–CFP (YVM1093)—only merged YFP/CFP image shown. (B) Ctf19p–YFP Spc29p–CFP (DHY201)—YFP, CFP, and merged images are shown as an example, Ctf3p–YFP Spc29p–CFP (DHY202), Mcm22p–YFP Spc29p–CFP (DHY203), Mcm16p–YFP Spc29p–CFP (DHY204)—only merged YFP/CFP images shown.
Colocalization of Ctf19p, Ctf3p, Mcm16p, Mcm22p, and Spc34p with Ndc10p
Because Ctf3p, Mcm22p, Mcm16p, and Spc34p have a similar cellular localization to kinetochore proteins, we tested for colocalization with CFP-tagged Ndc10p. As expected, Ctf19p–YFP colocalized with Ndc10p–CFP in short spindle and long spindle stage cells (Fig. 5A). Ctf3p, Mcm22p, Mcm16p, and Spc34p YFP fusions were also found to colocalize with Ndc10p–CFP (Fig. 5B). Colocalization of Ndc10p–GFP with another kinetochore protein, Mtw1p–CFP, has also been shown (Goshima and Yanagida 2000). Thus, localization to the interior side of the SPBs plus colocalization with SPB proximal Ndc10p (Ndc10p shows additional staining along the spindle) may serve as a signature localization pattern for the identification of novel kinetochore proteins.
Figure 5.
Colocalization of kinetochore–YFP fusions with Ndc10p–CFP. All strains are homozygous diploids containing genomic C-terminal CFP and YFP fusions. YFP signal appears green and CFP signal appears red. Overlapping of YFP and CFP signal appears yellow. (A) Ctf19p–YFP Ndc10p–CFP (DHY192)—YFP, CFP, and merged images are shown as an example. (B) Ctf3p–YFP Ndc10p–CFP (DHY196), Mcm22p–YFP Ndc10p–CFP (DHY198), Mcm16p–YFP Ndc10p–CFP (DHY197), Spc34p–YFP Ndc10p–CFP (DHY193)—only merged YFP/CFP images shown.
Interactions between Cse4p and the outer kinetochore
The S. pombe Ctf3p homolog, Mis6, is required for the CENP-A histone H3 variant, Cnp1, to localize to and bind centromere DNA (Takahashi et al. 2000). Evidence suggests that the budding yeast CENP-A homolog, Cse4p, is an essential subunit of a specialized centromere specific histone (for review, see Sullivan 2001). We tested for potential interactions between Cse4p and the Ctf3p complex, as well as Ctf19p, which interacts with Cse4p by two-hybrid (Ortiz et al. 1999; Chen et al. 2000). Coimmunoprecipitation experiments from log phase yeast lysates failed to detect any interaction between Cse4p and Ctf3p, Mcm22p, Mcm16p or Ctf19p (data not shown). However, genetic studies identified an interaction between CSE4 and the outer kinetochore. The nonpermissive temperature of a cse4-1 mutant (Stoler et al. 1995) is lowered when combined with a deletion of CTF3 or CTF19. ctf3Δ cse4-1 and ctf19Δ cse4-1 strains are lethal at 35°C compared to cse4-1 mutants alone which are viable up to 38°C (Table 3). This suggests that a strain defective for Cse4p is further compromised for kinetochore function in the absence of Ctf3p or Ctf19p.
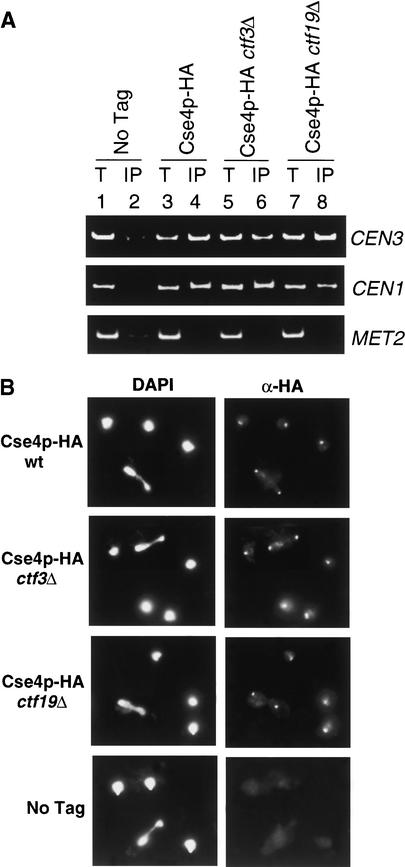
Assembly of Cse4p, Ctf3p, and Ctf19p onto CEN DNA
To further study budding yeast Cse4p/CENP-A assembly requirements, we tested whether Cse4p can interact with CEN DNA in the absence of Ctf3p or Ctf19p. We expressed endogenous Cse4p–HA in a wild-type, ctf3Δ or ctf19Δ mutant strain and performed ChIP assays in Nz-arrested cells to synchronize cells in G2 (Fig. 6A). Cse4p was still able to bind CEN DNA in ctf3Δ (Fig. 6A, lane 6) and ctf19Δ strains (Fig. 6A, lane 8). Cse4p did not bind the noncentromeric locus MET2 (Fig. 6A, lanes 4,6,8). We also tested whether the localization of Cse4p is altered in a ctf3Δ or ctf19Δ mutant strain. Previous studies have shown that Cse4p localizes to discrete foci adjacent to the nuclear face of the SPB (Meluh et al. 1998; Chen et al. 2000; Pearson et al. 2001). We found that the wild-type localization pattern of Cse4p–HA was not altered in ctf3Δ or ctf19Δ log phase cells (Fig. 6B). Thus, unlike Mis6, Ctf3p is not required to recruit Cse4p/CENP-A onto CEN DNA.
Figure 6.
Cse4p interacts with and localizes to CEN DNA in the absence of Ctf3p or Ctf19p. (A) Anti-HA ChIP assays were performed from Nz-arrested cells (treated with 15 μg/mL Nz for 100 min). The templates used (see Materials and Methods) were total chromatin (T) or immunoprecipitate (IP). (B) Anti-HA Cse4p immunofluorescence was performed as described (Materials and Methods). Total DNA was stained with DAPI. Each panel shows Cse4p–HA localization in an undivided cell (single DAPI mass) and a dividing cell (bilobed DAPI mass). No anti-HA signal was seen in the untagged control cells. All strains are homozygous diploids: untagged, (No Tag, YVM1142); Cse4p–HA wild-type (wt, YVM1141); Cse4p–HA ctf3Δ (YVM1143); Cse4p–HA ctf19Δ (YVM1145).
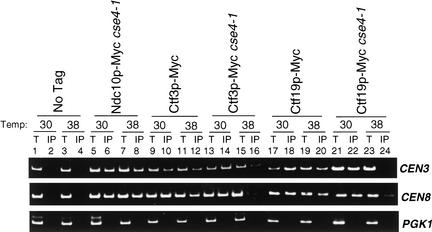
Having shown that Cse4p interacts with CENs in the absence of Ctf3p or Ctf19p, we asked whether Ctf3p or Ctf19p require Cse4p for CEN association. We constructed Ndc10p, Ctf3p, and Ctf19p Myc epitope-tagged strains in the cse4-1 temperature sensitive mutant (Stoler et al. 1995). At the permissive temperature, Ctf3p, Ctf19p, and Ndc10p all bound CEN3 and CEN8 DNA but not a non-CEN locus, PGK1 in the cse4-1 strain (Fig. 7, lanes 6,14,22). On shifting to the nonpermissive temperature, however, Ctf3p and Ctf19p showed reduced binding to CEN DNA in the cse4-1 mutant, but not in a wild-type background (Fig. 7, cf. lanes 12 and 16, 20 and 24). Western blot analysis showed that Ctf3p and Ctf19p were still immunoprecipitated in the cse4-1 mutant at nonpermissive temperature (data not shown). Therefore, Ctf3p and Ctf19p both require functional Cse4p to interact efficiently with CEN DNA. Interestingly, Ndc10p is still able to associate with CEN DNA in the cse4-1 mutant at nonpermissive temperature (Fig. 7, cf. lanes 6 and 8).
Figure 7.
Ctf3p and Ctf19p interact poorly with CEN DNA in a cse4-1 mutant. Wild type and cse4-1 mutant strains were grown at permissive temperature (Temp, 30°C) to early log phase. The culture was then split in two with half remaining at 30°C and the other half grown at the nonpermissive temperature (38°C) for 3 h. Anti-Myc ChIP assays were performed. PCR templates (see Materials and Methods) were total chromatin (T) and immunoprecipitate (IP). The strains used were: cse4-1 (No Tag, Stoler et al. 1995); Ndc10p–Myc cse4-1 (YVM1047); Ctf3p–Myc (YVM218); Ctf3p–Myc cse4-1 (YVM1053); Ctf19p–Myc (IPY313); Ctf19p–Myc cse4-1 (YVM1049).
Discussion
SDL screening as a method to identify kinetochore proteins
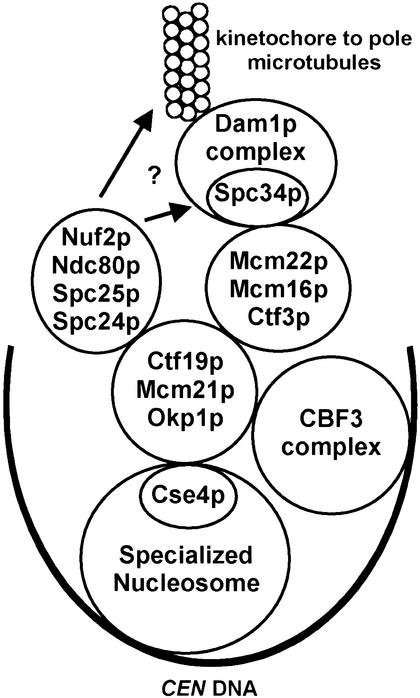
We used SDL as a method to screen for novel kinetochore proteins from a collection of mutants with defects in chromosome transmission. All ctf mutants that were identified as SDL on overexpression of a CBF3 component contain mutations in proteins involved in kinetochore function (Tables 1, 2). We isolated mutants in outer kinetochore proteins (ctf3, mcm21/ctf5, mcm16-155), a putative kinetochore protein (chl4/ctf17-20/mcm17) and two cohesin proteins (irr1/scc3-165 and smc1-166). Mcm16p interacts with Ctf3p in yeast lysates and both proteins interact with the outer kinetochore protein Ctf19p, which lies in a complex with Mcm21p (Figs. 2, 8; Hyland et al. 1999; Ortiz et al. 1999). Although cohesin and kinetochore proteins have not been shown to interact physically, cohesin proteins bind to CEN DNA (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999). Thus SDL screens can serve as a powerful method to identify members of a protein complex starting with a mutant collection and a wild-type gene (Kroll et al. 1996; Measday and Hieter 2002).
Figure 8.
New interactions at the budding yeast kinetochore. The proposed Cse4p specialized nucleosome and the CBF3 inner kinetochore complex bind to CEN DNA (curved black line). The outer kinetochore subcomplexes are shown within circles and interactions between kinetochore subcomplexes are depicted by touching circles. Some of the protein–protein interactions shown may not be direct. We propose that the Ctf3p subcomplex interacts with CEN DNA via the Ctf19p subcomplex. Links between the kinetochore and the spindle microtubules may occur via the Dam1p and Ndc80p complexes.
Ctf3p, Mcm22p, Mcm16p, and Ctf19p may link the kinetochore to the spindle microtubules
Our studies add Ctf3p, Mcm22p, and Mcm16p to the outer kinetochore complex. Genetic evidence suggests that the Ctf3p subcomplex forms a larger complex with Ctf19p. Firstly, a quadruple deletion of ctf3 mcm22 mcm16 and ctf19 is viable (Table 3). Secondly, the nonpermissive temperature of a ctf13-30 strain is reduced to the same degree in a ctf13-30 ctf3Δ (or any single mutant combination) strain as in a ctf13-30 ctf3Δ mcm22Δ mcm16Δ ctf19Δ strain (Table 3). In an effort to understand if Ctf3p, Mcm22p, Mcm16p, and Ctf19p are functionally redundant, we assayed the rate of loss of a nonessential chromosome fragment in strains carrying kinetochore mutant combinations. We found that the rate of chromosome fragment loss was equivalent in a single ctf3, mcm22, mcm16, or ctf19 deletion strain to the rate of loss in the quadruple ctf3 mcm22 mcm16 ctf19 deletion strain (data not shown). This result suggests that Ctf3p, Mcm22p, Mcm16p, and Ctf19p function in the same complex and that removal of one protein is equivalent to removal of all four.
The Mcm22p–Spc34p two-hybrid interaction suggests a possible role for the Ctf3p complex (Fig. 8). Based on CEN interaction and localization studies, Spc34p may provide a link from the kinetochore to the spindle microtubules (Fig. 4A; Wigge et al. 1998; He et al. 2001). Indeed, Spc34p has been shown to be part of a larger complex containing the microtubule-associated protein Dam1p (Cheeseman et al. 2001a). Using various lysis conditions we were unable to detect coimmunoprecipitation between Mcm22p and Spc34p from log phase or Nz-arrested cells (data not shown). Perhaps these interactions will only be detected in the presence of intact microtubules. Genetic interactions between the dam1-1 mutant and the outer kinetochore ctf19Δ, mcm16Δ, mcm21Δ, and mcm22Δ mutants also suggest a connection from the outer kinetochore to Dam1p (Cheeseman et al. 2001a,b; Jones et al. 2001). Interestingly, genetic and two-hybrid interactions have been detected between Mcm21p and Spc24p, a member of the Ndc80p kinetochore complex (Janke et al. 2001). Thus, the outer kinetochore proteins may be linked to the microtubules via both the Ndc80p complex and the Dam1p complex (Fig. 8).
Ctf3p and Mis6 comparison
Ctf3p is a member of a family of proteins conserved from yeast to humans (Fig. 1). Three of the four family members, Ctf3p, Mis6, and human LRPR1, have known or implicated roles at the centromere (Saitoh et al. 1997; Goshima et al. 1999; Partridge et al. 2000; Takahashi et al. 2000; T. Liu and T. Yen, pers. comm.). The discrepancy between the requirement for Mis6 in Cnp1 centromere binding and the dispensability of Ctf3p in Cse4p CEN binding may reflect specific differences between Ctf3p/Mis6 or Cse4p/Cnp1, or a general difference in the organization and size of S. pombe and S. cerevisiae centromere DNA. Moreover, Mis6 is essential, whereas Ctf3p is not. Ctf3p may be a component of a larger complex required for Cse4p localization and deletion of CTF3 alone cannot detect this requirement. In addition, Cse4p contains a unique N terminus not present in its CENP-A homologs (see below). Thus the roles of Ctf3p/Mis6 and Cse4p/Cnp1 may overlap but may also differ in functions specific to budding or fission yeast.
Cse4p and outer kinetochore assembly at the centromere
Interactions between Cse4p and the outer kinetochore have been detected. The unique N terminus of Cse4p interacts by two-hybrid with Ctf19p (Ortiz et al. 1999; Chen et al. 2000). MCM21 is a dosage suppressor of cse4-23 and cse4-39 mutants and mcm21Δ and cse4-39 are synthetically lethal (Chen et al. 2000). cse4 mutants also display synthetic lethal interactions with okp1-5, ctf19Δ and mcm22Δ strains (Chen et al. 2000). We found that the nonpermissive temperature of a cse4-1 mutant was lowered when combined with a ctf3Δ or a ctf19Δ strain (Table 3). We have also found a requirement for Cse4p in loading outer kinetochore proteins onto CEN DNA. Ctf3p and Ctf19p do not bind CEN DNA efficiently in a cse4-1 mutant strain at nonpermissive temperature (Fig. 7). Cse4p may not be required for inner kinetochore assembly because Ndc10p still binds CEN DNA on disruption of Cse4p. However, we cannot exclude the possibility that Cse4-1p is still bound to the centromere at nonpermissive temperatures, enabling Ndc10p binding (Fig. 7). It has been shown that Cse4p requires functional Ndc10p to bind CEN DNA (Ortiz et al. 1999). From our ChIP data, combined with the data from Ortiz et al. (1999), we can tentatively place kinetochore assembly in the following order: Ndc10p, Cse4p, Ctf19p, and the Ctf3 complex (Ctf3p, Mcm22p, and Mcm16p) (Fig. 8). Further studies will demonstrate how this large network of yeast kinetochore proteins is connected to the spindle microtubules and if a similar pattern of organization occurs with human homologs of yeast kinetochore proteins.
Materials and methods
SDL assay
The inducible plasmids used for SDL screening were: pGAL1-CTF13 (pKF88) (Kroll et al. 1996), pGAL1-NDC10/CTF14 (pKH2) (Measday and Hieter 2002), and pGAL1-CTF19 (pKH21) (Hyland et al. 1999). Plasmids were transformed into ctf mutants on glucose plates (noninducing conditions) and colony purified. Sixty-nine ctf mutants were transformed: 38 representing alleles from six ctf complementation groups and 31 representing single member mutants (mutants not yet assigned to ctf complementation groups and designated “s” with a number). Four isolates of each transformant were struck onto galactose plates (inducing conditions) and glucose plates (as a control for growth) at 25°C, 30°C, and 35°C. Growth was monitored from two to four days for glucose plates and three to seven days for galactose plates at the above temperatures.
Epitope tagging
Thirteen Myc and three HA genomic C-terminal tags were designed according to Longtine et al. (1998). The Cse4p–HA genomic tag was generated according to Meluh et al. (1998).
Protein sequence alignments
Sequence similarities were first identified using the NCBI (National Center for Biotechnology Information) BLAST server. Alignments were done using CLUSTAL W (Thompson et al. 1994) and MULTICLUSTAL (Yuan et al. 1999). Shading was done using the Boxshade 3.21 web site http://www.ch.embnet.org/software/BOX_form.html.
Genomic two-hybrid
CTF3 and MCM22 were cloned into pOBD2 (http://depts.washington.edu/∼yeastrc/th_2.htm) and two-hybrid screens were performed as described previously (Uetz et al. 2000).
Coimmunoprecipitation from yeast lysates
Cells were grown to log phase, and IPs were performed as described previously (Tyers et al. 1992). Two milligrams of yeast lysate and 20 μL of anti-Myc (9E10) affinity matrix (Covance) were used per IP. Antibodies used for Western blot analysis were: anti-HA (12CA5, Boehringer Mannheim) (1:1000), anti-Myc (9E10, Covance/BAbCO) (1:1000), and anti-Ctf19p (see below) (1:5000).
Ctf19p polyclonal antibody
The N-terminal 189 amino acids of Ctf19p were cloned into pRSETC (Invitrogen) to create a Ctf19p–His6 fusion protein. Ctf19p–His6 was expressed in Escherichia coli strain BL21(λDE3) and purified on a nickel NTA agarose column (QIAGEN). Purified fusion protein was injected into rabbits for polyclonal antibody production (Covance Research Products).
ChIP assays
ChIP experiments were performed as described previously (Tanaka et al. 1997; Hecht and Grunstein 1999) with the following alterations. For all ChIP assays, cells were cross-linked with 1% formaldehyde for 30 min at room temperature. Cell extracts were sonicated 6 times for 10 sec each (chromatin sheared to an average size of 500 bp). IPs were performed with 25μL of anti-Myc (9E10) or anti-HA (HA.11) affinity matrix (Covance). At least 1.5 mg of lysate was used per IP. The templates used for PCR reactions ranged from 1/200 to 1/30 of total chromatin and 1/30 to 1/15 of total IP depending on the linear range for PCR. Primers used for PCR analysis were: CEN1 (bp 151374 to 151676 of chromosome I; Meluh and Koshland 1997), CEN3 (base pairs 114,315 to 114,558 of chromosome III; Meluh and Koshland 1997), CEN8 (base pairs 105,499 to 105,748 of chromosome VIII), MET2 (base pairs 117,972 to 118,264 of chromosome XIV; Zeng et al. 1999) and PGK1 (base pairs 138,949 to 139,237 of chromosome III; Meluh and Koshland 1997).
Immunofluorescence
Cse4p–HA was localized by indirect immunofluorescence microscopy as described previously (Hyland et al. 1999) with the following modifications. Cells were grown to log phase and fixed with 3.7% formaldehyde for 15 min at 25°C. The primary antibody used was anti-HA (12CA5, Boehringer Mannheim) (1:1000) and the secondary antibody used was goat-anti-mouse IgG coupled with fluorescein (Cappel) (1:1000).
Fluorescence microscopy
Strains were genomically tagged with YFP or CFP according to: http://depts.washington.edu/∼yeastrc/fm_home3.htm. Cells containing YFP and CFP fusions were grown overnight on solid YPD supplemented with extra adenine at 30°C and then fixed in SD complete liquid containing 3.7% formaldehyde and incubated for 10 min at 30°C. Cells were washed once with PBS and resuspended in PBS containing 1.3 mg/mL concanavalin A tagged with Alexa-633 (Molecular Probes) and incubated for 1 min. Cells were washed, mounted, and imaged with a DeltaVision microscope as described previously (Drees et al. 2001).
Acknowledgments
We would like to acknowledge Bryan Sundin for his help with microscopy experiments. We thank Brenda Andrews, Kristin Baetz, and Melanie Mayer for critical reading of this manuscript. V.M. was supported by an NCIC Postdoctoral Fellowship; I.P. was supported by an NCIC Student Fellowship and a U.B.C. Killam Predoctoral Fellowship; P.H. was supported by a CIHR Senior Scientist Award. This work was supported by a National Institutes of Health grant (CA16519) to P.H. and a National Institutes of Health grant (NCRR RR11823) to T.D and S.F.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hieter@cmmt.ubc.ca; FAX (604) 875-3840.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.949302.
References
- Amon A. Together until separin do us part. Nat Cell Biol. 2001;3:E12–E14. doi: 10.1038/35050642. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., Brew, C., Wolyniak, M., Desai, A., Anderson, S., Muster, N., Yates, J.R., Huffaker, T.C., Drubin, D.G., and Barnes, G. 2001a. Implication of a novel multi-protein Dam1p complex in outer kinetochore function. J. Cell Biol. (in press). [DOI] [PMC free article] [PubMed]
- Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001b;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Riley DJ, Chen PL, Lee WH. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L. Centromeres: Proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr Opin Genet Dev. 1998;8:212–218. doi: 10.1016/s0959-437x(98)80143-3. [DOI] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiaekinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees BL, Sundin B, Brazeau E, Caviston JP, Chen GC, Guo W, Kozminski KG, Lau MW, Moskow JJ, Tong A, et al. A protein interaction map for cell polarity development. J Cell Biol. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SK, Poddar A, Hajra S, Sanyal K, Sinha P. The IML3/MCM19 gene of Saccharomyces cerevisiaeis required for a kinetochore-related process during chromosome segregation. Mol Genet Genomics. 2001;265:249–257. doi: 10.1007/s004380000408. [DOI] [PubMed] [Google Scholar]
- Goh PY, Kilmartin JV. NDC10: A gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes & Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- Hyland KM, Kingsbury J, Koshland D, Hieter P. Ctf19p: A novel kinetochore protein in Saccharomyces cerevisiaeand a potential link between the kinetochore and mitotic spindle. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Lechner J, Shevchenko A, Magiera MM, Schramm C, Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, He X, Giddings TH, Winey M. Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc Natl Acad Sci. 2001;98:13675–13680. doi: 10.1073/pnas.241417098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith KC, Baker RE, Chen Y, Harris K, Stoler S, Fitzgerald-Hayes M. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Hieter P. Evolutionary conservation between budding yeast and human kinetochores. Nat Rev Mol Cell Biol. 2001;2:678–687. doi: 10.1038/35089568. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- Koshland D, Hieter P. Visual assay for chromosome ploidy. Methods Enzymol. 1987;155:351–372. doi: 10.1016/0076-6879(87)55024-8. [DOI] [PubMed] [Google Scholar]
- Koshland DE, Guacci V. Sister chromatid cohesion: The beginning of a long and beautiful relationship. Curr Opin Cell Biol. 2000;12:297–301. doi: 10.1016/s0955-0674(00)00092-2. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Kirillov A, Kroll E, Koryabin M, Shestopalov B, Bannikov V, Zakharyev V, Larionov V. Identification and cloning of the CHL4 gene controlling chromosome segregation in yeast. Genetics. 1993;135:327–341. doi: 10.1093/genetics/135.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov VL, Karpova TS, Zhouravleva GA, Pashina OB, Nikolaishvili NT, Kouprina NY. The stability of chromosomes in yeast. Curr Genet. 1987;11:435–443. doi: 10.1007/BF00384604. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maine G, Sinha P, Tye B-K. Mutants of S. cerevisiaedefective in the maintenance of mini-chromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday, V. and Hieter, P. 2002. Synthetic dosage lethality. Methods Enzymol. (in press). [DOI] [PubMed]
- Megee PC, Mistrot C, Guacci V, Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes & Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: Cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Ortiz J, Lechner J. The budding yeast kinetochore: Less simple than expected. Protoplasma. 2000;211:12–19. [Google Scholar]
- Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes & Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes & Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC. Centromeres: Getting a grip of chromosomes. Curr Opin Cell Biol. 2000;12:308–319. doi: 10.1016/s0955-0674(00)00094-6. [DOI] [PubMed] [Google Scholar]
- Poddar A, Roy N, Sinha P. MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiaeare required for chromosome transmission. Mol Microbiol. 1999;31:349–360. doi: 10.1046/j.1365-2958.1999.01179.x. [DOI] [PubMed] [Google Scholar]
- Roberts RG, Kendall E, Vetrie D, Bobrow M. Sequence and chromosomal location of a human homologue of LRPR1, an FSH primary response gene. Genomics. 1996;37:122–124. doi: 10.1006/geno.1996.0528. [DOI] [PubMed] [Google Scholar]
- Roy N, Poddar A, Lohia A, Sinha P. The mcm17 mutation of yeast shows a size-dependent segregational defect of a mini-chromosome. Curr Genet. 1997;32:182–189. doi: 10.1007/s002940050264. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Sanyal K, Ghosh SK, Sinha P. The MCM16 gene of the yeast Saccharomyces cerevisiaeis required for chromosome segregation. Mol Gen Genet. 1998;260:242–250. doi: 10.1007/s004380050892. [DOI] [PubMed] [Google Scholar]
- Slegtenhorst-Eegdeman KE, Post M, Baarends WM, Themmen AP, Grootegoed JA. Regulation of gene expression in Sertoli cells by follicle-stimulating hormone (FSH): Cloning and characterization of LRPR1, a primary response gene encoding a leucine-rich protein. Mol Cell Endocrinol. 1995;108:115–124. doi: 10.1016/0303-7207(94)03468-9. [DOI] [PubMed] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes & Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Sullivan KF. A solid foundation: Functional specialization of centromeric chromatin. Curr Opin Genet Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3–Cdc28 kinase complex of S. cerevisiaeis regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiaecontains conserved centromere components and has a function in chromosome segregation. J Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomycesspindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Amend A, Borkowski J, DeMarco R, Bailey W, Liu Y, Xie G, Blevins R. MULTICLUSTAL: A systematic method for surveying CLUSTAL W alignment parameters. BioInformatics. 1999;15:862–863. doi: 10.1093/bioinformatics/15.10.862. [DOI] [PubMed] [Google Scholar]
- Zeng X, Kahana JA, Silver PA, Morphew MK, McIntosh JR, Fitch IT, Carbon J, Saunders WS. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]