Abstract
Seeds of the Arabidopsis thaliana transparent testa 1 mutant (tt1) appear yellow, due to the lack of condensed tannin pigments in the seed coat. The TT1 gene was isolated by reverse genetics using an En-1 transposon mutagenized A. thaliana population. TT1 gene expression was detected in developing ovules and young seeds only, and the gene was shown to encode a nuclear protein. Mutant seeds displayed altered morphology of the seed endothelium in which brown tannin pigments accumulate in wild-type plants, indicating that TT1 is involved in the differentiation of this cell layer. When overexpressed in transgenic A. thaliana plants, TT1 caused aberrant development and organ morphology. The protein contains a novel combination of two TFIIIA-type zinc finger motifs. Closely related motifs were detected in a number of putative proteins deduced from plant genomic and EST sequences. The new protein domain containing this type of zinc finger motifs was designated WIP, according to three strictly conserved amino acid residues. Our data indicate the existence of a small gene family in A. thaliana which is defined by the occurrence of the WIP domain. WIP genes may play important roles in regulating developmental processes, including the control of endothelium differentiation.
Keywords: C2H2/C2HC zinc finger, condensed tannins, nuclear localization, regulatory protein, tt1
Flavonoid biosynthesis supplies plants with a large variety of secondary metabolites, such as flavonols, anthocyanins, and condensed tannins. Because flavonoids are not essential for plant growth, and the loss of pigments can easily be detected, many mutants affected in flavonoid accumulation have been generated and investigated. Flavonoids serve as pigments in flowers, fruits and seeds, and as UV protectants in tissues exposed to light. In some species, they also act as signaling molecules in reproduction or symbiosis or as antimicrobial phytoalexins (Winkel-Shirley 2001). Their accumulation is highly regulated in a tissue-specific manner both during plant development and in response to various stresses. Consequently, flavonoid biosynthesis has been widely used to study the regulation of gene expression in plants (Mol et al. 1998; Weisshaar and Jenkins 1998).
Most flavonoid-related mutants identified to date in Arabidopsis thaliana show the transparent testa (tt) phenotype (Koornneef 1990). These mutants are defined by yellow to pale brown seeds which can easily be distinguished from the dark brown seeds of wild-type plants. All known tt mutations are recessive and show maternal inheritance of the seed phenotype. Some of the tt mutants are defective in structural genes of flavonoid biosynthesis (tt3, DFR; tt4, CHS; tt5, CFI; tt6, F3H; tt7, F3′H; see Fig. 1), which occur as single-copy genes in the A. thaliana genome (Shirley et al. 1995; Wisman et al. 1998; Schoenbohm et al. 2000). The encoded enzymes catalyze various steps in the formation of flavonol aglycones and anthocyanidins and are also required for synthesis of the condensed tannins responsible for seed coat pigmentation (Fig. 1; Chapple et al. 1994). In all of these tt mutants, the entire plant displays a deficiency in flavonoid accumulation.
Figure 1.
Schematic overview of flavonoid biosynthesis in A. thaliana. Essential steps leading to condensed tannins, flavonols, and anthocyanins are highlighted. Mutants corresponding to genes with known function are given in parentheses. The question mark indicates that the enzymatic activity of the BAN protein has not yet been elucidated. Dashed arrows indicate multiple steps. CHS, chalcone synthase; CFI, chalcone flavanone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; DFR, dihydroflavonol 4-reductase; LAR, leucoanthocyanidin reductase.
The mutants tt1, tt2, tt8, tt9, tt10, tt12, and tt15 appear to be altered in seed color only (Debeaujon et al. 2001). Two members of this group have been investigated in more detail. The TT12 locus encodes a protein with similarity to multidrug secondary transporters and might therefore be required for flavonoid sequestration in vacuoles of the seed coat endothelium (Debeaujon et al. 2001). The TT8 gene was shown to encode a bHLH transcription factor that is required for the expression of DFR and BANYULS (BAN) in A. thaliana siliques (Nesi et al. 2000). Mutation of the BAN locus causes anthocyanin accumulation in seeds, and it has been suggested that BAN encodes leucoanthocyanidin reductase (LAR) (Devic et al. 1999).
For the ethyl methanesulfonate-induced mutant tt1-1, no visible difference from the corresponding wild-type has been reported with respect to flavonoid accumulation in vegetative parts of the plant (Koornneef 1990; Shirley et al. 1995). Accordingly, the enzymes CHS, CFI, F3H, as well as FLS (flavonol synthase) and LDOX (leucoanthocyanidin dioxygenase), required for flavonol and anthocyanidin formation, respectively, were detected at wild-type levels in 4-day-old light-grown tt1-1 seedlings (Pelletier et al. 1999). TT1 was therefore expected to encode either a seed-specific enzyme late in the pathway leading to the production of condensed tannins, or alternatively, a seed-specific regulatory factor (Shirley et al. 1995).
Most of the factors known to regulate flavonoid biosynthesis in various species act as transcriptional activators (Quattrocchio et al. 1993). The majority of them belong to the bHLH and R2R3–MYB families of transcription factors. However, factors from the homeodomain and WRKY families are also involved in regulating certain aspects of flavonoid accumulation in A. thaliana (Kubo et al. 1999; Eulgem et al. 2000). Furthermore, some regulatory proteins have been identified that are thought to act upstream of transcription factors controlling flavonoid biosynthesis (Winkel-Shirley 2001).
Flavonoid content and composition have been shown to influence seed dormancy, storability and quality. These are agronomically important traits with regard to human and animal nutrition (Shirley 1998; Debeaujon et al. 2000). To understand and potentially modify these traits, more information is required on regulatory genes controlling the complex mechanisms of cell-specific flavonoid biosynthesis during plant development and differentiation. Here we describe the isolation of TT1 by transposon tagging employing an En-1 mutagenized A. thaliana population. We demonstrate that TT1 is involved in endothelium development in the seed coat and that it defines a novel group of plant zinc finger proteins.
Results
Isolation of the TT1 gene and complementation of tt1-1
Mutant plants producing yellow seeds but showing normal anthocyanin accumulation in leaves and stems were isolated from an A. thaliana population mutagenized with the maize (Zea mays) transposon Enhancer-1 (En-1). These mutants were used for test crosses with established tt mutants. One homozygous mutant plant (6K60.3) showed no genetic complementation of the tt phenotype in a cross with tt1-1. Cosegregation analysis in the progeny of the heterozygous parent plant of 6K60.3 revealed that the TT1 gene was tagged by an En-1 insertion (data not shown). The new tt1 allele was designated tt1-2∷En.
DNA flanking the insertion was amplified by PCR and used to screen the IGF-BAC library. BAC F3N5 and four additional positive BAC clones were identified, all of which localized close to the centromere of chromosome 1. On the basis of a MaeIII polymorphism between Landsberg erecta (Ler) and Columbia (Col) sequences of the flanking DNA, marker ms100 was generated and mapped. The position of ms100 correlated well with the position of the tt1 locus on the genetic map (Fig. 2A).
Figure 2.
Cloning of the TT1 gene. (A) Structure of the TT1 region on chromosome 1. The tt1 locus (black arrowhead) was localized to Chr. 1 (thick line) around 55 cM on the classical genetic map (Koornneef 1990; Franzmann et al. 1995) close to the centromere (black circle). Five BAC clones (black bars; BAC designation on the right) which hybridized to tt1-2∷En DNA sequence and which contained marker ms100 mapped between markers agp6e and mi63. Map positions (cM, small number below the line) were taken from the Lister and Dean (1993) recombinant inbred line map maintained at NASC. In the lower part, a schematic map of the construct used for complementation is shown (L, R, left and right T-DNA border, respectively; nptII, kanamycin resistance marker). (B) Complementation of the tt1-1 mutation. Reaction tubes containing seeds from the four lines as indicated. tt1-1∷F3N5-S/S12, line transformed with the construct shown in (A); tt1-1∷TATA, line transformed with the vector control.
A 12-kb SpeI fragment of BAC F3N5 containing the initially identified flanking DNA sequence was used for in planta transformation of tt1-1 plants. Presence of this fragment restored the plant's ability to produce seeds with the brown color of the wild-type (Fig. 2B), indicating that the 12-kb fragment contained the TT1 gene.
Structure of the TT1 gene and characterization of two tt1 alleles
The entire nucleotide sequence of the 12-kb SpeI fragment used for complementation was determined (GenBank accession no. AF190297). Sequence analysis indicated the presence of a single gene containing two exons as well as several kb of repetitive DNA (Fig. 3A). The complete cDNA corresponding to the putative TT1 gene was obtained by reverse transcription PCR (RT–PCR) and sequenced (AF190298). The transcriptional start site, which was located 41 bp upstream of the ATG codon, is preceded by a putative TATA box around position −40. The deduced TT1 protein comprises 303 amino acid residues including two zinc finger motifs (Fig. 3B).
Figure 3.
Molecular characterization of TT1. (A) Structure of the DNA fragment used for complementation of tt1-1. Schematic drawing of the 12-kb F3N5 SpeI fragment (lower panel) compared to BAC F11O6 (upper panel). The F11O6 sequence (AC018460; The Arabidopsis Genome Initiative 2000) from position 69,769 to 58,180 confirmed sequencing of the 12-kb fragment (AF190297). Exons are indicated by boxes, and arrows represent a repeated sequence element consisting of the 3′ part and 3.3 kb downstream of the TT1 gene. This element is duplicated in the 12-kb fragment, and is present in a number of additional copies with more than 90% identity between positions 18,000 and 65,000 of BAC F11O6. (B) Deduced amino acid sequence of TT1. Various amino acid motifs are highlighted: shaded, zinc finger; bold characters, potential nuclear localization signals; underlined, stretch of acidic residues; underlined with a dashed line, serine-rich region; boxed, hDLNh motif. (C) Structure of the tt1-2∷En allele. Nucleotides corresponding to the 3-bp target site duplication are underlined. En-1 indicates the orientation of the transposable element. Bold letters highlight transposon sequence. Numbers above the arrowheads indicate nucleotide positions in the cDNA (AF190298). Amino acid changes caused by the insertion are given in lowercase letters. (D) Consequences of the tt1-1 mutation. The upper part shows the DNA sequence and the splicing result of tt1-1, the lower part the TT1 sequence and the wild-type splicing product. Exons and the encoded amino acids are shown in bold, residues belonging to the zinc finger motif are shaded. Donor (GT) and acceptor (AG) intron splice sites are underlined. The three bases changed in tt1-1 are highlighted.
Genomic sequence analysis of the tt1 mutant alleles revealed that in tt1-2∷En the 8-kb En-1 element was inserted in the first exon (Fig. 3C), and that tt1-1 contained a mutation near the splice acceptor site of exon 2 (Ler, AF190299; tt1-1, AF190300). Analysis of the tt1-1 cDNA (AF251686) revealed that an aberrant acceptor splice site downstream of the original one was used in tt1-1 (Fig. 3D). This causes a frame shift replacing 142 amino acids of TT1, including most of the zinc finger motifs, with 23 residues from an alternative frame. The data from two independent tt1 alleles which both contain mutant versions of the newly isolated gene provided final proof that this gene was TT1.
Accumulation pattern of TT1 mRNA
Expression of the TT1 mRNA was studied in various organs of A. thaliana plants. TT1 mRNA was not detectable by RNA gel blot techniques using polyA+ RNA or by in situ localization with radioactively or digoxigenin-labeled riboprobes (E. Schmelzer, unpubl.). However, the TT1 mRNA was detected by RT–PCR in flowers and developing siliques, but not in roots, stems, or young or senescent leaves (Fig. 4A). At the level of mRNA accumulation, no difference was detected between wild-type and tt1-1 plants (data not shown).
Figure 4.
Monitoring TT1 expression. (A) TT1 expression detected by RT–PCR in various organs of Ler plants. Ro, roots; yL, young leaves; oL, old leaves; St, stem; Fl, flowers; Si, siliques; Co, no template. The TT1 fragment was detected by ethidium bromide staining after 35 amplification cycles. The ACTIN2 transcript (AC2, 25 cycles) was used as a control. (B,C) Histochemical staining of flowers and siliques of transgenic A. thaliana plants containing the TT1-GUS transgene. Panel B represents flower developmental stage 15, and panel C stage 16 (Smyth et al. 1990). Similar results were obtained with material from several independent transgenic lines. (D,E) Longitudinal sections of siliques of TT1-GUS plants stained for GUS activity. Siliques (stage 15 of flower development) from several independent transgenic lines were analyzed and two representative pictures were selected.
To obtain a more detailed view of the pattern of TT1 expression, 1.1 kb of 5′ upstream region of the TT1 gene were fused at the ATG start codon to the uidA open reading frame (ORF) encoding β-glucuronidase (GUS). This chimeric construct was stably introduced into A. thaliana and GUS activity was determined. Histochemical staining of floral organs showed GUS activity only in developing ovules and young seeds (Fig. 4B,C). Sections through stained siliques revealed GUS activity in the endothelium, the innermost cell layer of the seed coat, and to a lesser extent in the other cell layers of the testa (Fig. 4D,E). Staining was detected in developing seeds during stages 13 to 17 of flower development. These results are in good agreement with the RT–PCR data. The temporal and spatial patterns of TT1 expression in developing seeds are consistent with a role of TT1 in seed coat development or pigmentation.
Phenotypic effects of the tt1 mutation
Based on the expression data, we reinvestigated the phenotypic effects of the tt1 mutation on various aspects of flavonoid biosynthesis and seed development in mutant plants in comparison with the wild-type. RNA gel blot analysis of stems, flower buds, flowers, and dark-adapted or UV-treated leaves of adult tt1-1 and Ler plants revealed no differences in the transcript levels of CHS, CFI, F3H, FLS and DFR. HPTLC analysis of methanolic extracts from leaves, flower buds, and developing and mature seeds indicated that the contents and relative composition of flavonol glycosides in these organs were identical in wild-type Ler and tt1-1 mutant plants (data not shown).
Yellow seed color, attributed to the lack of condensed tannins in the seed coat, has been the only visible difference to wild-type described to date for the tt1-1 mutant. Comparative TLC analysis of extracts from acid-treated mature seeds confirmed this observation. Release of cyanidin, which was detected after hydrolysis of condensed tannins, was visible only in extracts of Ler but not of tt1-1 seeds (Fig. 5A). This lack of cyanidin indicates that the mutant does not accumulate significant amounts of condensed tannins in the seeds. These results also show that alterations in flavonoid accumulation in tt1-1 plants are restricted to the seed coat.
Figure 5.
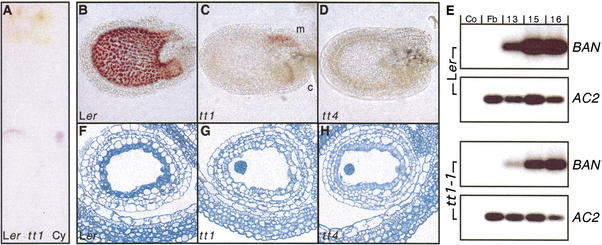
Seed-specific effects of the tt1 mutation. (A) Results of a TLC analysis of flavonoid aglycones accumulating in mature wild-type and tt1-1 mutant seeds. Cyanidin (Cy, red spot) serves as control. (B–D) Vanillin-stained 3-day-old Ler wild-type (B), tt1-1 (C), and tt4 (D) mutant seeds (longitudinal view). The red precipitate indicates presence of catechins. c, chalaza; m, micropyle. (E) BAN expression detected by quantitative RT–PCR after hybridization and autoradiography. Co, no template control; Fb, flower buds; 13, 15 and 16, stages of flower/silique development. Samples were derived from wild-type (Ler) or mutant (tt1-1) plants; the ACTIN2 transcript (AC2) was used as a control. The signals shown were obtained after 17 (BAN) and 13 cycles (ACTIN2). (F–H) Toluidine blue-stained transversal sections of 3-day-old wild-type (F), tt1-1 (G), and tt4 (H) mutant seeds.
To further characterize flavonoid biosynthesis in tt1-1, accumulation of catechins, the colorless precursors of condensed tannins, was monitored in seeds of all stages of embryo development. In the wild-type, vanillin staining revealed the onset of catechin accumulation at day two after pollination. Staining occurred throughout wild-type seed coats, but was restricted to the micropylar area and the chalazal bulb at the base of the seed in the tt1-1 mutant (Fig. 5B,C). Seeds of the tt4 mutant, defective in CHS, completely lack catechins (Fig. 5D). Catechin synthesis requires expression of BAN, which most probably encodes LAR. Therefore, BAN mRNA accumulation was monitored. Expression was detected in both young siliques of wild-type and tt1-1 mutant plants, but with a marked quantitative difference (Fig. 5E). The reduced signal from tt1-1 is consistent with BAN expression in the micropylar area and the chalazal bulb, but a lack of BAN gene activity in the remaining part of the seed.
Microscopy investigation of seed sections revealed the endothelium, which normally accumulates catechins, to be morphologically altered in the tt1-1 mutant compared with the wild-type. In the wild-type, this cell layer contained granules and stained dark green with Toluidine Blue, indicating the accumulation of phenolic compounds from early seed development on (Fig. 5F). In contrast, tt1 mutant seeds completely lack granules and green staining and have endothelium cells with a somewhat irregular shape (Fig. 5G), while tt4 mutant seeds still show light staining of the endothelium at young stages (Fig. 5H). These data show that the endothelium cells were directly affected by the tt1 mutation. In addition, the failure to accumulate catechins at day two after pollination was the earliest detectable difference between wild-type and tt1 mutants during development.
Subcellular localization and overexpression of TT1
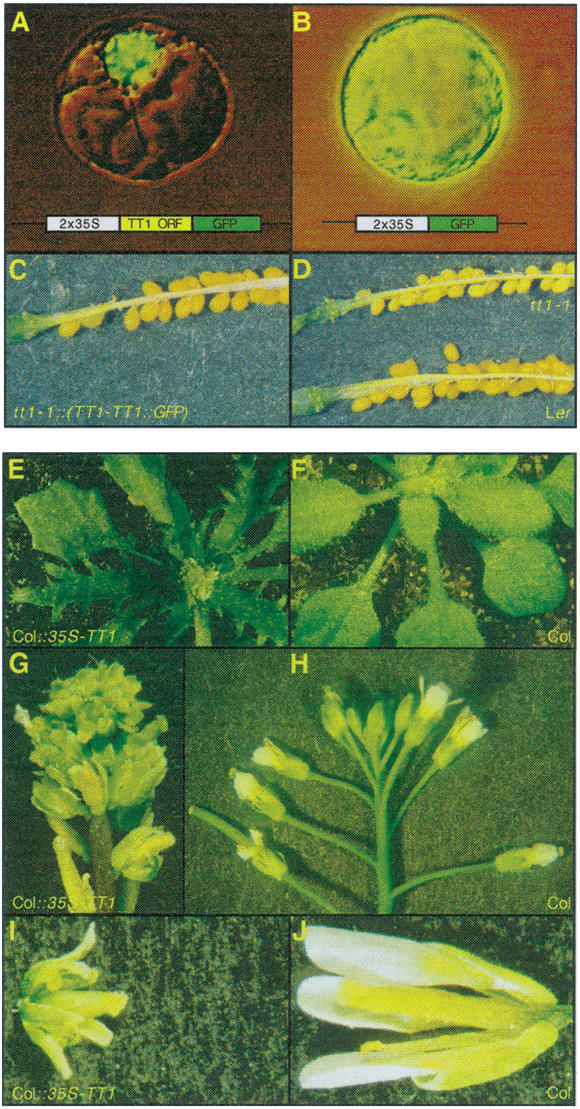
The subcellular localization of the TT1 protein was analyzed using protoplasts of suspension-cultured A. thaliana cells transfected with a construct expressing the complete TT1 protein fused to the green fluorescent protein (GFP). The TT1∷GFP fusion protein was detected exclusively in the nuclei of all protoplasts investigated (Fig. 6A,B), indicating that TT1 is a nuclear protein. In a complementation experiment, the TT1∷GFP fusion protein was shown to be functional in restoring the defect in pigment accumulation of tt1 seeds (Fig. 6C,D).
Figure 6.
Analysis of the TT1 protein. (A–B) Subcellular localization of TT1. Pictures showing typical A. thaliana protoplasts transiently transfected with either p2x35S-TT1∷GFP or p2x35S-GFP. Schematic drawings of the constructs are inserted into the respective panel. (C–D) Complementation of tt1-1 by the TT1∷GFP fusion. Opened silique of a representative transgenic T1 plant (C) in comparison to mutant and wild-type (D). The genotype of the plants is indicated for the respective panels. (E–J) Morphology of TT1-OX plants (E,G,I) in comparison to wild-type (F,H,J). (E,F) Rosette leaves of mature plants at bolting. (G,H) Inflorescences of a TT1-OX and a Col wild-type plant. (I,J) Isolated flowers shortly after pollination.
To examine possible TT1 function(s) in organs where the protein is usually not expressed, we analyzed transgenic plants ectopically expressing TT1 under the control of the 35S CaMV promoter (TT1-OX). Most kanamycin-resistant T1 plants showed aberrant leaves. In contrast to the round shape of wild-type cotyledons and first leaves, cotyledons of TT1-OX plants were narrow and elongated, first rosette leaves were markedly angular, and older rosette leaves were deeply serrated and lancet-like (Fig. 6E,F). Flowering was delayed, elongation of inflorescence internodes was repressed, and flowers developed without pedicels (Fig. 6G,H). TT1–OX plants as well as their flowers were reduced in size in comparison with wild-type and showed reduced greenish petals (Fig. 6I,J). Most of the flowers were sterile. These severe phenotypic aberrations suggest that ectopic expression of the TT1 protein affects the regulation of a variety of processes in plant development and differentiation.
Identification and analysis of TT1-like sequences
Database searches revealed five putative proteins that showed more than 70% sequence similarity to TT1. For all of these genes the corresponding cDNAs were amplified by RT–PCR. The exon/intron structures of these genes were identical to that of TT1, with the splice site junction within the first zinc finger motif (cf. Fig. 3D). Additional TT1-like genes were shown to occur in various other plant species (Table 1).
Table 1.
Plant protein sequences containing two TFIIIA-type zinc finger motifs used for distance analysis
| Name
|
Organism
|
Gene ID
|
Accession
|
% id.
|
% sim.
|
No of res.
|
|---|---|---|---|---|---|---|
| AtTT1 | Arabidopsis thaliana | At1g34790 | AF190298a | 100 | 100 | 151 |
| AtWIP2 | Arabidopsis thaliana | At3g57670 | AF254447a | 82 | 87 | 151 |
| AtWIP3 | Arabidopsis thaliana | At1g08290 | AF254448a | 75 | 86 | 152 |
| AtWIP4 | Arabidopsis thaliana | At3g20880 | AJ311811a | 80 | 88 | 151 |
| AtWIP5 | Arabidopsis thaliana | At1g51220 | AJ311810a | 81 | 88 | 151 |
| AtWIP6 | Arabidopsis thaliana | At1g13290 | AJ311809a | 77 | 85 | 153 |
| GmWIP1 | Glycine max | n.a. | AJ311808a | 81 | 89 | 151 |
| MtWIP1 | Medicago truncatula | n.a. | BE203286 | 81 | 87 | 108 |
| GaWIP1 | Gossypium arboreum | n.a. | BE052647 | 76 | 80 | 62 |
| OsWIP1 | Oryza sativa | n.a. | AC090054 | 78 | 87 | 151 |
| TtWIP1 | Triticum turgidum | n.a. | BF293596 | 80 | 87 | 140 |
| HvWIP1 | Hordeum vulgare | n.a. | AJ234704 | 72 | 82 | 52 |
| StPCP1 | Solanum tuberosum | n.a. | X82328b | 34 | 46 | 144 |
| ZmID1 | Zea mays | n.a. | AF058757b | 32 | 42 | 168 |
| PhEPF1 | Petunia hybrida | n.a. | X60700b | n.d. | n.d. | 149 |
| TaWZF1 | Triticum aestivum | n.a. | D16416b | n.d. | n.d. | 137 |
| AtSTZ1 | Arabidopsis thaliana | At1g27730 | X95573b | n.d. | n.d. | 123 |
The length of the zinc finger domains considered to be comparable to the WIP domain are indicated. Values for percent identity and similarity when compared to TT1 are from BlastP. n.d., not determined; n.a., not available.
This work.
TT1 and the TT1-like proteins from A. thaliana are most divergent in their N-terminal parts. Only one moderately conserved motif was detected by MEME analysis in this part of the proteins (consensus sequence V[D,A,T]L[H,Q][I,L]G[L,P,F]P). The highly conserved zinc finger domain is located at the C-termini of TT1 and the related proteins. As illustrated in Figure 7A, all of these proteins possess putative zinc binding as well as hydrophobic residues at the positions assumed to be required for maintaining a finger structure. Furthermore, the residues proposed to contact DNA in a zinc finger and the two putative nuclear localization signals are absolutely conserved.
Figure 7.
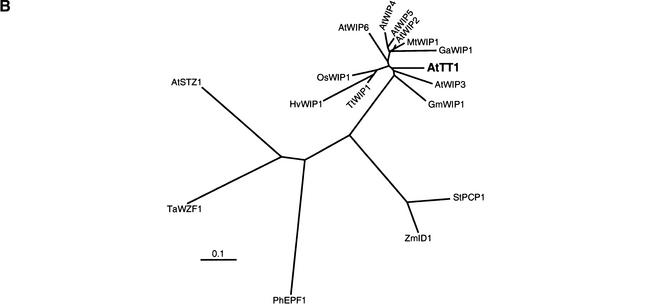
Relationship among various WIP proteins. (A) Amino acid sequence comparison of WIP domains. Part of the TT1 protein (Ler sequence) compared to the animal TFIIIA (Klug and Schwabe 1995) and the plant EPF (Takatsuji 1998) consensus sequences and aligned to partial TT1-like sequences. Only domains containing the sequence Trp-Ile-Pro (WIP, bold characters) are shown. Asterisks denote putative DNA binding residues. Black boxes indicate zinc binding residues. Conserved hydrophobic residues (h) are shaded. Amino acids conserved among all sequences are boxed. Dashes indicate gaps inserted to improve the alignment. Relative numbering without reference to a known start methionine is given in italics. (B) Distance analysis of several plant protein sequences containing two TFIIIA-type zinc finger motifs. For tree construction, only sequences of the zinc finger domains comparable to the WIP domain were used. See Table 1 for details on the factors included in the clustering. The scale bar indicates the distance in numbers of amino acid substitutions per site.
We compared the deduced amino acid sequences of the newly identified genes with established zinc finger sequences of animal origin and with those of other plant zinc finger proteins. All of the TT1-like proteins contained two zinc finger motifs of the TFIIIA-type. The only proteins known to contain a similar combination of zinc fingers are ID1 from Z. mays and PCP1 from potato (Solanum tuberosum). The zinc finger domains of TT1 and of the TT1-like proteins displayed more than 70% sequence identity when compared with one another, but only about 40% identity when compared with ZmID1 or StPCP1 (Table 1).
Sequence relationships among various plant zinc finger domains containing two TFIIIA-type motifs are illustrated in Figure 7B. Mono- and dicotyledonous TT1-like protein sequences are closely related, although they form separate branches in the clustering. All TT1-like sequences constitute a subclass which is not closely related to ZmID1 and StPCP1. With the exception of the presence of zinc fingers as such, they are unrelated to all other plant zinc finger proteins described to date.
Discussion
The TT1 locus encodes a zinc finger protein
The results presented above indicate that the TT1 gene encodes a nuclear protein that is conserved throughout the plant kingdom and defines a new subfamily of plant zinc finger proteins. Zinc fingers are protein motifs that form structures stabilized by zinc ions. The classical C2H2 motif, containing one pair each of cysteine and histidine residues that tetrahedrally bind a zinc ion, was first described for the Xenopus laevis general transcription factor TFIIIA (Miller et al. 1985). C2H2 zinc fingers represent one of the most common DNA binding motifs found in eukaryotic transcription factors (Wolfe et al. 1999). Direct sequence-specific interaction of the α-helix of a zinc finger with the major groove of DNA was first shown for Zif268 from mice (Pavletich and Pabo 1991). Zinc finger motifs have also been implicated in RNA binding and in mediating protein/protein interactions (for review, see Mackay and Crossley 1998). Animal factors of the C2H2 family play central roles in general housekeeping processes as well as in various tissue-specific differentiation processes. A prominent example is the erythroid Krüppel-like factor (EKLF), which is involved in erythroid cell differentiation and regulation of globin gene expression (Perkins 1999).
Approximately 30 C2H2 zinc finger proteins have been described in plants (Takatsuji 1999), but only few of them have been characterized genetically. SUPERMAN (AtSUP) contributes to the maintenance of floral whorl boundaries (Sakai et al. 1995), and AtFIS2 controls fertilization-independent seed development in A. thaliana (Luo et al. 1999). The StPCP1 gene, a homolog of COUCH POTATO from Drosophila melanogaster, was identified by its ability to confer growth on sucrose as the sole carbon source upon a sucrose uptake-deficient yeast strain (Kühn and Frommer 1995), and INDETERMINATE1 (ZmID1) is a transcriptional regulator of floral transition (Colasanti et al. 1998). Sequence-specific DNA binding has been shown for the zinc finger protein PhZPT2-1 from Petunia hybrida that interacts with the promoter of the 5-enolpyrovylshikimate-3-phosphate synthase (EPSPS) gene involved in the biosynthesis of aromatic amino acids (Takatsuji et al. 1994). These data suggest that in plants, similar to the situation in animals, many zinc finger proteins act as transcription factors with regulatory functions in development and differentiation.
The predicted TT1 protein contains two motifs that perfectly match established zinc finger consensus sequences. The N-terminal motif belongs to the C2H2 class, whereas the more C-terminal motif has an imperfect C2HC configuration. However, based on the conserved spacing and the presence of hydrophobic key residues required for maintaining the finger structure, the second finger also appears to be of the TFIIIA-type (Fig. 7A). Additional cysteine and histidine residues in the carboxy terminus of the protein show less conserved spacing but might lead to the formation of two additional fingers. The TT1 protein was localized to the nucleus and contains two motifs that fit the consensus sequence of SV40-type nuclear localization signals. The ability of the TT1–GFP fusion protein to complement the tt1 mutation illustrates the biological relevance of this finding. Moreover, TT1 contains an hDLNh-motif also found in several other unrelated plant zinc finger proteins (Meissner and Michael 1997; Takatsuji 1998), as well as motifs that may be implicated in phosphorylation, transcriptional activation, and protein/protein interaction (Fig. 3B). Therefore, TT1 is likely to act as a transcriptional regulator rather than an enzyme involved in seed pigment formation. In some way similar to the animal zinc finger factor EKLF, TT1 might function in the differentiation of specific cell types. While most known regulatory proteins controlling flavonoid biosynthesis in various plant species contain MYB or bHLH domains, TT1 is the first zinc finger factor required for cell-specific flavonoid accumulation.
Involvement of TT1 in seed coat pigmentation
The mutant line K60 used to clone TT1 contains an insertion of the 8-kb En-1 element in the first exon of the gene, which most probably renders tt1-2∷En a null allele. Sequencing of the tt1-1 allele revealed three nucleotide mismatches that lead to a truncated protein lacking the zinc finger domain. Since no phenotypic difference was observed between the tt1-1 and tt1-2∷En alleles, tt1-1 apparently also represents a null allele. Normal activity of the TT1 promoter in tt1-1 indicates that the effect of the tt1 mutation results from a lack of TT1 protein, not from a changed TT1 expression pattern.
All available data indicate that the tt1 mutation has no effect on flavonoid biosynthesis in vegetative plant parts, but specifically affects the pigmentation of a single cell layer of the seed coat. Consistently, those steps of flavonoid biosynthesis that are considered “early” according to the expression patterns of the respective genes during seedling development (CHS, CFI, F3H, F3′H, and FLS; Kubasek et al. 1992) are not altered in tt1 mutant plants at the RNA level, and flavonoid glycoside product accumulation patterns were normal. Developing tt1 seeds, however, differed from wild-type in their ability to accumulate catechins, the biosynthetic precursors of condensed tannins. Catechin accumulation is generally restricted to the endothelium layer of the seed coat (Devic et al. 1999), but occurs also in the chalaza and at the micropylar area of the seed. The accumulation of catechins appeared to be normal in these tissues in tt1 (Fig. 5). This observation is in agreement with the results of studies using tt1/tt12 double mutants (Debeaujon et al. 2001), which indicated that pigmentation of chalaza and seed body are under different genetic control. In addition, it confirms that the tt1 mutation does not affect the ability of the plants to synthesize catechins as such. Because of the observed catechin accumulation, some expression of BAN (cf. Fig. 1) was to be expected in tt1-1. The reduction of BAN expression in the mutant compared to the wild-type (Fig. 5E) can be attributed to the absence of BAN gene activity in the endothelium of tt1 seed. In this respect, TT1 differs clearly from the pathway-specific factor TT8 (Nesi et al. 2000) which controls BAN gene activity in general and which prevents pigmentation of chalaza and the micropylar area when mutated (tt8; Debeaujon et al. 2001).
The endothelium cells of the tt1 mutant show characteristics distinct from cells present at the same location in the wild-type and the tt4 mutant. These differences could account for the reduced size and altered dormancy of tt1-1 reported previously (Debeaujon et al. 2000). Based on our data, we hypothesize that the lack of TT1 function in young endothelium cells would cause altered differentiation resulting in incompetence for pigment synthesis and hence the observed yellow appearance of mature tt1 mutant seeds. This hypothesis explains the cell-type specificity of the tt1 phenotype, and also resolves the discrepancy between normal flavonoid biosynthesis throughout tt1 mutant plants and in distinct cells of developing seeds but absence of catechins from the endothelium cell layer.
The various phenotypic alterations observed in TT1-OX plants (Fig. 6) support a role of TT1 in development. The overall phenotype of TT1-OX plants might result to a large extent from interference of the ectopically expressed protein with unrelated processes that are normally controlled by TT1-like genes which are expressed in many parts of the plant (M. Sagasser and B. Weisshaar, unpubl.).
TT1 defines a novel class of plant zinc finger proteins
C2H2 zinc finger proteins from animals generally contain multiple fingers separated by short invariant spacers of 6–8 amino acids (Klug and Schwabe 1995). Most plant proteins of this type contain fewer fingers (1–4; Takatsuji 1998) that are separated by comparatively long spacers of high variability in sequence and length (e.g., 19–65 amino acids in P. hybrida EPFs; Kubo et al. 1998). TT1 contains two zinc fingers with a wide spacing of 63 amino acids and thus displays features typical of plant zinc finger proteins.
Another characteristic feature of most plant C2H2 zinc fingers is the presence of a stretch of six amino acids (QALGGH) at the N terminus of the helical region. This sequence was found in nearly all plant C2H2 zinc finger proteins described to date, classified as EPF-type proteins (Takatsuji 1999). However, because most of the corresponding genes were isolated by similarity-based cloning methods (Meissner and Michael 1997), it is reasonable to assume a bias towards the detection of closely related genes. The only plant C2H2 zinc finger proteins without the QALGGH motif described to date have been identified by sequence-independent genetic methods: AtFIS2, containing only one zinc finger, and ZmID1 and StPCP1, both of which contain two fingers.
TT1 represents a novel type of plant zinc finger protein without appreciable similarity to any known protein of bacterial, fungal, or animal origin and with moderate similarity to ZmID1 and StPCP1. However, five sequences with high similarity to TT1 are present in the A. thaliana genome. TT1-like sequences were also detected in a number of very distantly related species (Table 1). We therefore propose a new class of plant C2H2 zinc finger proteins that is characterized by the presence of a novel type of domain containing one C2H2 and one C2HC zinc finger motif but not the QALGGH sequence. We designate this protein region the WIP domain according to the first three amino acid residues of the consensus sequence (Fig. 7A).
Regions outside the WIP domain exhibit little sequence conservation and may therefore determine distinct functional specificities among WIP proteins. On the other hand, the high degree of sequence identity within the WIP domains suggests that this domain represents a structural element involved in a common function. Similar domain conservation patterns are also observed in many families of plant transcription factors (Riechmann et al. 2000). The WIP domain may function in the interaction with other factors and/or in recognizing DNA. Conservation of both the spacing between the zinc fingers in the WIP domain and the putative DNA binding residues (asterisks in Fig. 7A) within the WIP domain zinc fingers might indicate recognition of similar target sequences.
Plant genomes contain numerous genes encoding potential WIP domain transcription factors. The evidence of WIP factor genes in cereals suggests a common evolutionary origin prior to the separation of mono- and dicotyledonous plants, and may indicate a universal occurrence of these factors throughout the plant kingdom. Different subtypes of zinc finger proteins might be similarly central to differentiation and development in plants and animals. Our results regarding TT1 and other WIP factors will facilitate further studies of the involvement of zinc finger proteins in these processes in plants.
Materials and methods
Materials
Seeds of tt1-1 were obtained from the Nottingham Arabidopsis Stock Center (NASC, stock no. NW82). Plants were grown as described (Wisman et al. 1998). Filters representing the IGF-BAC library and selected BAC clones were obtained from the RZPD (Berlin, Germany). The cDNA encoding GmWIP1 was produced in the public soybean EST project (R. Shoemaker et al., GenBank) and obtained from Genome Systems (St. Louis, MO).
Molecular biology techniques
Sequences of plasmid DNA and PCR fragments were determined by the MPIZ DNA core facility ADIS using BigDye terminator chemistry on Applied Biosystems 377 sequencers. Oligonucleotides were purchased from MWG (Ebersberg, Germany). BAC DNA was column-purified using premixed reagents (QIAGEN) following the manufacturer's recommendations.
Biochemical analyses and cytological staining procedures
TLC was performed as described (Focks et al. 1999). Six milligrams of seeds were extracted, and 10% of the extract was loaded on a TLC plate. Cyanidin was obtained from Apin Chemical (Oxon, UK). Vanillin staining was performed as described by Devic et al. (1999). Tissue fixation, embedding, sectioning, and Toluidine Blue staining were carried out according to published protocols (Windsor et al. 2000).
GUS activity was histochemically localized using 2 mM 5-bromo-4-chloro-3-indoyl glucuronide, 2 mM K3Fe6(CN)6, 50 mM NaCl in 0.1 M Tris/HCl (pH 7). Plant material was incubated in 90% acetone for 30 min at 4°C, washed with buffer, vacuum infiltrated with the substrate solution for 30 min, and incubated at 37°C. In transformants, blue staining became visible after at least 24 h incubation. No staining was observed in wild-type plants. For photography, chlorophyll was removed by repeated ethanol washes and the material was transferred to 50% glycerol prior to mounting. For sectioning, the material was fixed (2.5% glutaraldehyde, 50 mM NaCl, 0.1 M Tris/HCl at pH 7) for 24 h at 4°C and embedded in 6% agarose. Sections (20–30 μm) were obtained using a vibratome (Leica Nussloch, Germany).
Isolation of TT1
The En-1 mutagenized population was described previously (Wisman et al. 1998). For cosegregation analysis, genomic DNA from a family of 53 6K60 plants that segregated for the tt1 phenotype was prepared using the Nucleon Phytopure kit (Amersham Pharmacia Biotech). The DNA was digested with EcoRV and subjected to DNA gel blot analysis using an En-1-specific probe representing bp 7631 to 8141 of GenBank accession no. M25427. A 0.9-kb genomic DNA fragment that cosegregated with the tt1 phenotype was considered to represent an insertion of the En-1 element into the TT1 gene (i.e., the Col allele tt1-2∷En). Flanking regions of En-1 insertions were amplified by rapid amplification of genomic ends (RAGE) essentially as described (Cormack and Somssich 1997). A PCR fragment that hybridized to the cosegregating 0.9-kb genomic DNA fragment was inserted into pCR-TOPO2.1 (Invitrogen) and used to screen the IGF BAC library (Mozo et al. 1998). One of five BACs hybridizing to the tt1-2∷En probe (F3N5) was selected for further analysis.
Cleaved amplified polymorphic sequence marker analysis
The selected flanking DNA fragment was mapped using Lister and Dean (1993) recombinant inbred lines (RILs). PCR conditions were as described (Wisman et al. 1998); primers were oSA117 (5′-CATACCTGAAGATTGTTGTAGCG-3′) and oSA126 (5′-GTACGTAAGTCACACAATATGATTC-3′); the amplified fragment of 840 bp contains one (Col) or two (Ler) MaeIII restriction sites. Of the 100-line RIL mapping set (stock no. NW1899), 94 were used for mapping.
Isolation of the TT1 cDNA
An internal fragment of 806 bp of the TT1 cDNA covering the intron position was amplified by RT–PCR using primers oSA105 (5′-CACCACCACTATACGAGATATCCTC-3′) and oSA130 (5′-GTCCTTAAGAGAACGTTTGTG-3′) derived from the predicted exon sequences, and the Titan-one-tube RT–PCR-System (Roche). RNA from immature Col siliques was chosen as a template as TT1 was expected to act during seed development. The TT1 transcriptional start site as well as 5′ and 3′ sequences of the TT1 cDNA were determined using premixed reagents according to the manufacturer's recommendations (5′/3′ RACE kit; Roche). Similar experiments were performed with RNA from Ler and tt1-1.
Sequencing strategy and sequence analysis
A 12-kb SpeI fragment of BAC F3N5 that hybridized to the tt1-2∷En probe was cloned into pBluescript II SK(+) (Stratagene) to give pSK-F3N5-S/S12. Templates for sequencing of pSK-F3N5-S/S12 were produced using the genome priming system GPS1 (NEB, Schwalbach, Germany). Sequence assembly was performed using Sequencher version 3.1 (Gene Codes Corporation, Ann Arbor, MI) and Genetics Computer Group software (v. 10.0, Genetics Computer Group, Madison, WI). Exon and gene predictions were done with NetPlantGene2. For distance analysis, amino acid sequences were aligned using ClustalW. The consensus tree presented was drawn with TreeView (Page 1996). Conserved motifs were detected by MEME analysis (Bailey and Elkan 1994).
Expression analysis
RNA from various plant organs was extracted as described (Feldbrügge et al. 1994). Oligo(dT)-primed first strand cDNA was synthesized with Omniscript RT (QIAGEN). PCR with primers oSA105 and oSA130 was performed using Advantage cDNA polymerase (Clontech) and 20 ng cDNA as template. Amplification of the ACTIN2 control transcript (An et al. 1996) was performed using primers Actin2-1 (5′-TCCCTCAATCTC ATCTTCTTCC-3′) and Actin2-2 (5′-GACCTGCCTCATCA TACTCG-3′). Quantitative RT–PCR was performed essentially as described (Nesi et al. 2000). The BAN fragment was amplified using primers oSA214 (5′-GCTATAAGTGCCGGAATCACG-3′) and oSA215 (5′-CATGGACCAGACTCTTACACAC-3′).
GFP fusion
To study the localization of TT1 within the cell, the TT1 ORF was inserted into the NcoI site of pAVA393 (von Arnim et al. 1998; pers. comm.) to yield a translational TT1∷GFPm5 fusion protein expressed under the control of the double 35S CaMV promoter (construct p2x35S-TT1∷GFP). The control construct pAVA393 is referred to as p2x35S-GFP. Procedures for transient transfection of cultured A. thaliana cells (At7) were as described (Hartmann et al. 1998). Incubation was at 26°C for 20 h in the dark. GFP fluorescence was detected using a Zeiss (Oberkochen, Germany) Axiophot microscope and digitized with a CCD camera.
Construction of transgenes
All constructs for plant transformation were based on the binary vector pGPTV-TATA, a derivative of pGPTV (Becker et al. 1992) containing the TATA-GUS cassette of pBT10 (Sprenger-Haussels and Weisshaar 2000). Plants were transformed by in planta infiltration (Bechtold et al. 1993) using Agrobacterium tumefaciens strain GV3101 (Koncz and Schell 1986). For complementation of the tt1-1 mutant, the insert of pSK-F3N5-S/S12 was released with SpeI and ligated into pGPTV-TATA to yield pGPTV-F3N5-S/S12. tt1-1 plants transformed with pGPTV-TATA served as controls. Three independent T1 plants were analyzed for complementation of the tt1 phenotype; all of them did produce brown seeds. Presence of the transgene (Col) in the genomes of transformants (Ler) was verified using the MaeIII polymorphism.
For overexpression, the TT1 ORF was amplified with primers introducing a NcoI site at the first ATG and a SacI site at the STOP codon. The ORF was inserted into pBT8-35S (Sprenger-Haussels and Weisshaar 2000) downstream of the cauliflower mosaic virus (CaMV) 35S promoter. The 35S promoter-TT1 ORF cassette was constructed into pGPTV-TATA by replacing TATA-GUS sequences, and transformed into A. thaliana accession Col. All T1 plants displaying a strong TT1-OX phenotype (about 70% of the kanamycin-resistant T1 plants) were sterile.
To construct a TT1 promoter GUS fusion, a 1.1-kb region 5′ of the TT1 ORF was PCR-amplified using oSA150 (5′-ATAAC TAGTATATTAGAAGTAATACTTG-3′) containing a SpeI site and a complementary primer introducing an NcoI site at the putative ATG start codon. The PCR product was ligated into pBT10, and the TT1-promoter-GUS fragment was constructed into pGPTV-TATA to yield pTT1-GUS, which was transformed into A. thaliana accession Col. GUS activity was tested in T1 and T2 plants.
The T-DNA from which TT1∷GFP was expressed under the control of the TT1 promoter was created by replacing the 2x35S promoter of p2x35S-TT1∷GFP by the 1.1kb TT1 promoter fragment. The resulting hybrid gene was transferred into the binary vector and transformed into tt1-1 plants.
Acknowledgments
We thank Ellen Wisman and her coworkers at MPIZ for providing lines from the En-1 mutagenized population and for helpful discussions, as well as Ute Tartler for technical assistance. We thank Ila Rouhara, Rainer Franzen, Rolf-Dieter Hirtz, and Elmon Schmelzer for help with histology and microscopy, and Jürgen Prell for his advice on the vibratome. We thank Maarten Koornneef, Imre Somssich, and Paul Rushton for critically reading the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
While this paper was under review, Nesi et al. (Plant Cell 13: 2099–2114) published the isolation of the TT2 gene, which encodes the R2R3–MYB factor AtMYB123.
Footnotes
E-MAIL weisshaa@mpiz-koeln.mpg.de; FAX 49-221-506-2851.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.212702.
References
- An YQ, Mcdowell JM, Huang S, Mckinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Mol Biol Genet. 1993;316:1194–1199. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Chapple C, Shirley B, Zook M, Hammerschmidt R, Somerville SC. Arabidopsis (eds. E.M. Meyerowitz and C.R. Somerville), pp. 989–1030. Cold Spring Harbor Laboratory Press, New York, NY. 1994. Secondary metabolism in Arabidopsis. [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf generated signal required for the transition to flowering in maize. Cell. 1998;93:593–603. doi: 10.1016/s0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- Cormack RS, Somssich IE. Rapid amplification of genomic ends (RAGE) as a simple method to clone flanking genomic DNA. Gene. 1997;194:273–276. doi: 10.1016/s0378-1119(97)00205-9. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Leon-Kloosterziel K, Koornneef M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 2000;122:403–414. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell. 2001;13:853–872. doi: 10.1105/tpc.13.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M. The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J. 1999;19:387–398. doi: 10.1046/j.1365-313x.1999.00529.x. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Feldbrügge M, Sprenger M, Dinkelbach M, Yazaki K, Harter K, Weisshaar B. Functional analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell. 1994;6:1607–1621. doi: 10.1105/tpc.6.11.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Sagasser M, Weisshaar B, Benning C. Characterization of tt15, a novel transparent testa mutant of Arabidopsis thaliana (L.) Heynh. Planta. 1999;208:352–357. doi: 10.1007/s004250050569. [DOI] [PubMed] [Google Scholar]
- Franzmann LH, Yoon ES, Meinke DW. Saturating the genetic map of Arabidopsis thaliana with embryonic mutations. Plant J. 1995;7:341–350. [Google Scholar]
- Hartmann U, Valentine WJ, Christie JM, Hays J, Jenkins GI, Weisshaar B. Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol Biol. 1998;36:741–754. doi: 10.1023/a:1005921914384. [DOI] [PubMed] [Google Scholar]
- Klug A, Schwabe JWR. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Koornneef M. Mutations affecting the testa colour in Arabidopsis. Arab Inf Serv. 1990;27:1–4. [Google Scholar]
- Kubasek WL, Shirley BW, Mckillop A, Goodman HM, Briggs W, Ausubel FM. Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell. 1992;4:1229–1236. doi: 10.1105/tpc.4.10.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo H, Peeters AJM, Aarts MGM, Pereira A, Koornneef M. ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell. 1999;11:1217–1226. doi: 10.1105/tpc.11.7.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, Nishino T, Takatsuji H. Cys2/His2 zinc-finger protein family of petunia: Evolution and general mechanism of target-sequence recognition. Nucleic Acids Res. 1998;26:608–616. doi: 10.1093/nar/26.2.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C, Frommer WB. A novel zinc finger protein encoded by a couch potato homologue from Solanum tuberosum enables a sucrose transport-deficient yeast strain to grow on sucrose. Mol Gen Genet. 1995;247:759–763. doi: 10.1007/BF00290408. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- Meissner R, Michael AJ. Isolation and characterization of a diverse family of Arabidopsis two and three-fingered C2H2 zinc finger protein genes and cDNAs. Plant Mol Biol. 1997;33:615–624. doi: 10.1023/a:1005746803089. [DOI] [PubMed] [Google Scholar]
- Miller J, Mclachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R. How genes paint flowers and seeds. Trends Pl Sci. 1998;3:212–217. [Google Scholar]
- Mozo T, Fischer S, Shizuya H, Altmann T. Construction and characterization of the IGF Arabidopsis BAC library. Mol Gen Genet. 1998;258:562–570. doi: 10.1007/s004380050769. [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: Crystal structure of a Zif268 DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Pelletier MK, Burbulis IE, Shirley BW. Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and endproducts in Arabidopsis seedlings. Plant Mol Biol. 1999;40:45–54. doi: 10.1023/a:1026414301100. [DOI] [PubMed] [Google Scholar]
- Perkins A. Erythroid Kruppel-like factor: From fishing expedition to gourmet meal. Int J Biochem Cell Biol. 1999;31:1175–1192. doi: 10.1016/s1357-2725(99)00083-7. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature. 1995;378:199–203. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B. Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol Chem. 2000;381:749–753. doi: 10.1515/BC.2000.095. [DOI] [PubMed] [Google Scholar]
- Shirley BW. Flavonoids in seeds and grains: Physiological function, agronomic importance and the genetics of biosynthesis. Seed Sci Res. 1998;8:415–422. [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995;8:659–671. doi: 10.1046/j.1365-313x.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- Smyth D, Bowman J, Meyerowitz E. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger-Haussels M, Weisshaar B. Transactivation properties of parsley proline rich bZIP transcription factors. Plant J. 2000;22:1–8. doi: 10.1046/j.1365-313x.2000.00687.x. [DOI] [PubMed] [Google Scholar]
- Takatsuji H. Zinc-finger transcription factors in plants. Cell Mol Life Sci. 1998;54:582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H. Zinc-finger proteins: The classical zinc finger emerges in conteporary plant science. Plant Mol Biol. 1999;39:1073–1078. doi: 10.1023/a:1006184519697. [DOI] [PubMed] [Google Scholar]
- Takatsuji H, Nakamura N, Katsumoto Y. A new family of zinc finger proteins in petunia: Structure, DNA sequence recognition, and floral organ-specific expression. Plant Cell. 1994;6:947–958. doi: 10.1105/tpc.6.7.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng X-W, Stacey MG. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene. 1998;221:35–43. doi: 10.1016/s0378-1119(98)00433-8. [DOI] [PubMed] [Google Scholar]
- Weisshaar B, Jenkins GI. Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol. 1998;1:251–257. doi: 10.1016/s1369-5266(98)80113-1. [DOI] [PubMed] [Google Scholar]
- Windsor J, Symonds V, Mendenhall J, Lloyd A. Arabidopsis seed coat development: Morphological differentiation of the outer integument. Plant J. 2000;22:483–493. doi: 10.1046/j.1365-313x.2000.00756.x. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid Biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate new phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci. 1998;95:12432–12437. doi: 10.1073/pnas.95.21.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 1999;3:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]