Abstract
The stem–loop binding protein (SLBP) is the posttranscriptional regulator of histone mRNA in metazoan cells. SLBP binds histone pre-mRNAs and facilitates 3′-end processing by promoting stable association of U7 snRNP with the pre-mRNA. To identify other factors involved in histone pre-mRNA processing, we used a modified yeast two-hybrid assay in which SLBP and its RNA target were coexpressed as bait. A novel zinc finger protein, hZFP100, which interacts with the SLBP/RNA complex but not with free SLBP, was cloned. The interaction requires regions of SLBP that are important for histone pre-mRNA processing. Antibodies to hZFP100 precipitate U7 snRNA, and expression of hZFP100 in Xenopus oocytes stimulates processing of histone pre-mRNA, showing that hZFP100 is a component of the processing machinery.
Keywords: Histone mRNA, two-hybrid, U7 snRNP, zinc finger, RNA binding
In metazoans replication-dependent histone mRNAs are not polyadenylated but instead terminate with a highly conserved stem–loop structure, consisting of 6 bp and a 4-nt loop (Marzluff 1992). The replication-dependent histone mRNAs are formed from longer pre-mRNA transcripts by endonucleolytic cleavage 4–5 nt downstream from the stem–loop (Gick et al. 1986; Dominski and Marzluff 1999). This 3′-end processing reaction requires at least two trans-acting factors: (1) the stem–loop binding protein, SLBP (Wang et al. 1996a), also known as the hairpin binding protein (Martin et al. 1997), which associates with the stem–loop structure (SL); and (2) U7 snRNP, which recognizes a purine-rich sequence, the histone downstream element (HDE), located ∼10 nt downstream from the cleavage site (Strub et al. 1984; Georgiev and Birnstiel 1985; Mowry and Steitz 1987a).
Human SLBP is a 31-kD protein containing 270 amino acids and is composed of three domains, the centrally located RNA-binding domain (RBD) and its flanking N-terminal and C-terminal domains (Wang et al. 1996a). The RBD, which is unique among all known RNA-binding proteins, mediates binding of SLBP to the stem–loop structure (Wang et al. 1996a; Martin et al. 2000; Dominski et al. 2001). The U7 snRNP is a minor snRNP, represented in an average mammalian cell by ∼103–104 particles (Birnstiel and Schaufele 1988; Smith et al. 1991; Grimm et al. 1993). The human U7 snRNP contains the 63-nt U7 snRNA (Mowry and Steitz 1987b) and associated proteins, including a set of Sm proteins and at least two additional U7-specific proteins (Smith et al. 1991; Stefanovic et al. 1995; Pillai et al. 2001). Binding of U7 snRNP to the pre-mRNA occurs primarily via base pairing between the HDE and the 5′ end of U7 snRNA (Schaufele et al. 1986; Bond et al. 1991). Additional factors involved in 3′-end processing of histone pre-mRNAs, including the factor(s) responsible for catalyzing the cleavage reaction, have not yet been identified. After the completion of 3′-end processing, SLBP remains associated with the terminal stem–loop and assists the mature histone mRNA to the cytoplasm, where it possibly regulates histone mRNA translation (Sun et al. 1992; Gallie et al. 1996) and stability (Pandey and Marzluff 1987). The efficiency of 3′-end processing of replication-dependent histone pre-mRNAs is cell cycle regulated and reaches a peak level during the S phase, followed by a rapid decline on completion of DNA replication (Stauber and Schümperli 1988; Harris et al. 1991). The high efficiency of 3′-end processing resumes at the end of G1 phase, prior to the next round of DNA replication. Restricting the 3′-end processing of histone pre-mRNA to the S phase is at least in part achieved by cell cycle regulation of SLBP, which accumulates to high levels only during S phase and is degraded through the proteasome pathway at the completion of S phase (Whitfield et al. 2000).
The HDE sequence varies considerably among the 60 different mammalian replication-dependent histone pre-mRNAs (Wang et al. 1996b). Because there is only a single U7 snRNA gene in mammalian cells (Gruber et al. 1991; Turner et al. 1996), binding of U7 snRNP to the pre-mRNA mediated by base-pairing does not result in formation of a sufficiently stable complex for efficient 3′-end processing of most histone pre-mRNAs. SLBP associated with the pre-mRNA stabilizes the binding of U7 snRNP, suggesting that SLBP bound to the pre-mRNA might interact with one of the U7 snRNP specific proteins (Dominski et al. 1999). In support of this, even a small increase in the distance between the stem–loop structure and the HDE results in a rapid decline in processing efficiency (Scharl and Steitz 1994, 1996), most likely by preventing productive interaction between SLBP and U7 snRNP, bound to their respective sites on the pre-mRNA (Dominski et al. 1999). This additional interaction is critical in processing of histone pre-mRNAs, for example, mouse histone H1t pre-mRNA, that contain an HDE capable of forming only a weak duplex with the U7 snRNA (Dominski et al. 1999). Conversely, pre-mRNA substrates such as mouse H2a-614 pre-mRNA, which contain an HDE that forms a nearly perfect duplex with U7 snRNA, can be processed in vitro with a modest efficiency even in the absence of SLBP (Melin et al. 1992; Streit et al. 1993; Spycher et al. 1994; Dominski et al. 1999).
To date, SLBP remains the only factor specifically involved in histone pre-mRNA 3′-end processing that is available for both biochemical and genetic studies. Here we describe an RNA-supplemented yeast two-hybrid screen used for the identification of human proteins interacting with the SLBP/SL complex. This modified version of the yeast two-hybrid system, in which SLBP fused to the GAL4 DNA-binding domain is coexpressed with the stem–loop RNA, allowed cloning of a new factor involved in histone pre-mRNA 3′-end processing. This factor, designated hZFP100, has an estimated molecular mass of 100 kD and contains 18 C2H2 zinc fingers. hZFP100 is associated with the U7 snRNP in nuclear extracts from HeLa cells. Moreover, when hZFP100 is expressed in Xenopus oocytes, it stimulates 3′-end processing of the histone pre-mRNA transcribed from the exogenous mouse H2a-614 histone gene. The RNA-supplemented yeast two-hybrid system may be a valuable general method for identifying proteins interacting with complexes of other known RNA-binding proteins and their respective RNA targets.
Results
Screening of a HeLa cDNA library for proteins interacting with the SLBP/SL RNA complex
SLBP stimulates 3′-end processing by stabilizing the association of U7 snRNP with the pre-mRNA substrate (Melin et al. 1992; Streit et al. 1993; Spycher et al. 1994; Dominski et al. 1999). This might be achieved by a direct interaction between SLBP tightly bound to the stem–loop structure and a protein stably or transiently associated with the U7 snRNP. The yeast two-hybrid system is routinely used for the cloning of proteins that interact in vivo with a previously cloned protein of interest (Chien et al. 1991). Here, we used a modification of this system for identifying a component of the U7 snRNP that interacts with SLBP during 3′-end processing of histone pre-mRNA.
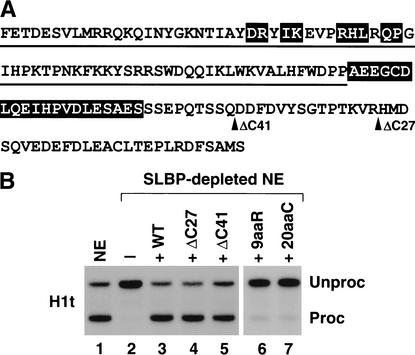
Because the C-terminal region of SLBP contains sequences that result in self-activation in the two-hybrid system (L.-X. Zheng and W.F.M., unpubl.), we tested deletions of the C terminus for their ability to process histone pre-mRNA and to self-activate the reporter genes in the two-hybrid system. The sequence of the RBD and the C-terminal domain of SLBP with the mutations used is shown in Figure 1A. A nuclear extract depleted of SLBP is inactive in H1t histone pre-mRNA processing, and the control level of processing efficiency can be restored by addition of recombinant SLBP expressed in baculovirus (Fig. 1B, lanes 1–3; Dominski et al. 1999, 2001). Deletion of 41 amino acids from the C terminus (ΔC41) reduced processing efficiency ∼25% (Fig. 1B, lane 5), whereas deletion of the last 27 amino acids (ΔC27) had no effect on processing efficiency (Fig. 1B, lane 4). In addition, the ΔC27 mutant SLBP no longer self-activated the HIS3 reporter gene and was consequently selected as bait for the yeast two-hybrid screen.
Figure 1.
Human SLBP lacking the last 27 amino acids is fully active in 3′-end processing. (A) The sequence of the human SLBP RBD (underlined) and C-terminal domain is shown. The break points of the C-terminal deletions are indicated by arrows. The 9aaR and 20aaC SLBP mutants contain substitutions of the residues highlighted in the RBD and in the region C-terminal to the RBD, respectively. (B) Histone H1t pre-mRNA labeled at the 5′ end with 32PO4 was incubated at 32°C for 30 min in a nuclear extract prepared from mouse myeloma cells. The RNA was isolated, resolved by gel electrophoresis, and detected by autoradiography. Shown is the processing activity of (lane 1) control and (lane 2) SLBP-depleted nuclear extracts. (Lanes 3–7) The ability of 100 ng of different baculovirus-expressed SLBPs (indicated at the top of each lane) to restore histone pre-mRNA processing to the depleted nuclear extract. (WT) Wild type. Unproc and Proc indicate the input H1t pre-mRNA and mature product of 3′-end processing, respectively.
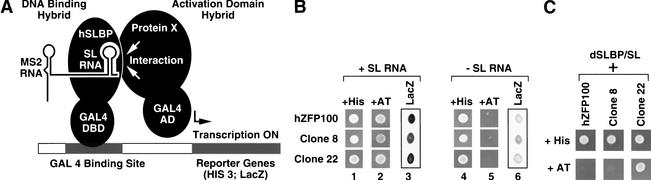
To select for proteins that bound to the SLBP/RNA complex rather than free SLBP, the yeast two-hybrid system was modified by introduction of a hybrid RNA containing the stem–loop structure (Fig. 2A; Wang et al. 1996a). The pGBT/SLBP/SLWT plasmid, which expresses both ΔC27 SLBP and an RNA containing the stem–loop, was introduced into the CG-1945 yeast reporter strain. This strain harboring the pGBT/SLBP/SLWT plasmid was used to screen a GAL4 AD/HeLa cDNA fusion library constructed in the pGAD GH vector (Clontech). We carried out four independent transformations with the cDNA library, each time systematically increasing the concentration of 3-AT in the medium (from 0 to 5.0 mM). The higher stringency of selection in the presence of 3-AT resulted in a drastic reduction in the number of colonies that grew on plates lacking histidine.
Figure 2.
The RNA-supplemented two-hybrid system. (A) The DNA-binding-domain hybrid is composed of the GAL4 DNA binding domain (DBD) fused to the ΔC27 mutant of human SLBP. The RNA component contains the RNase P leader at the 5′ end and two MS2-binding sites in the center, followed by the histone stem–loop. In the diagram only one MS2-binding site is depicted, and the RNase P leader is omitted. Protein X fused to the GAL4 activation domain (AD) interacts with the SLBP/RNA complex. The interaction brings the GAL4 AD into proximity to the promoter, resulting in activation of the HIS3 and LacZ reporter genes controlled by the GAL4-binding site. (B) Clones isolated in the RNA-supplemented two-hybrid system interact with the SLBP/SL complex but not with SLBP. Expression of two reporter genes, HIS3 and LacZ, was determined by testing the ability of yeast cells to grow on selective medium and by assaying β-galactosidase activity. To determine expression of the HIS3 gene, a suspension of yeast containing the library plasmid pGAD GH with the indicated cDNA insert, and the bait plasmid expressing SLBP with (left, lanes 1–3) or without stem–loop RNA (right, lanes 4–6), was spotted on medium with histidine (lanes 1,4) and on medium lacking histidine containing 10 mM 3-AT (lanes 2,5). hZFP100 and two other clones (8 and 22) isolated by the RNA-supplemented two-hybrid system are shown. Clone 8 is a putative 36-kD protein and clone 22 is ribosomal protein S15a. The ability of each clone to interact with the SLBP/RNA complex (lane 3) and free SLBP (lane 6) was confirmed by analysis of expression of the LacZ gene using a qualitative blue/white assay. After a 2-h incubation, cell suspensions were spotted on a white background and photographed. (C) The hSLBP was replaced in the bait construct shown in A with the Drosophila SLBP, and the ability of the three clones isolated in the initial screen to grow in the presence of 10 mM 3-AT was determined.
Only two unknown proteins were represented among >150 cDNA inserts analyzed in detail: a putative 36-kD protein (GenBank accession no. AF132946) with no similarity to any sequence available in the Swissprot database, and a 100-kD member of the zinc finger protein family, designated hZFP100 (see Fig. 4 below). The 36-kD protein was selected twice, in both cases only in the absence of 3-AT, but hZFP100 was repeatedly isolated in all four screens and represented ∼50% of the clones selected from the plates containing 2.5 mM 3-AT. Two ribosomal proteins, S15a and L22, were selected multiple times in the absence of 3-AT. Altogether, from more than 15 million transformants analyzed in multiple screens, hZFP100 was selected 25 times.
Figure 4.
hZFP100 is a novel 100-kD protein containing 18 C2H2 zinc fingers. (A) Amino acid sequence (1–871) of hZFP100 and nucleotide sequence of the 5′ and 3′ untranslated regions (UTRs). The nucleotide sequence in capital letters represents genomic sequence located immediately upstream of the first nucleotide of the cloned cDNA (in small letters), retrieved from the NCBI human genome database. The first in-frame Stop codon at the 5′ end of the clones is indicated with stars, and a presumptive TATAA box is boxed. The zinc fingers are underlined and numbered from 1 to 18, and the N-terminal region with a limited homology to KRAB is indicated with a thick line. The polyadenylation signal in the 3′ UTR is double underlined. (B) The 18 zinc fingers of hZFP100 are aligned with the conserved C and H residues involved in chelating the zinc ions and the conserved F and L residues indicated by arrows.
hZFP100 interacts with the SLBP/SL complex but not with SLBP alone
We tested whether hZFP100 interacts only with the SLBP/SL complex or whether the interaction can also occur in the absence of the stem–loop RNA, by transforming the hZFP100 into yeast expressing just the SLBP. The yeast cells expressing hZFP100, SLBP, and stem–loop RNA efficiently grew on medium containing 50 mM 3-AT (Fig. 2B, lane 2), but in the absence of the stem–loop RNA, growth was abolished by 2.5 mM 3-AT (Fig. 2B, lane 5), indicating that hZFP100 interacts very weakly, if at all, with free SLBP. The second reporter gene under control of the GAL4-binding site, LacZ, was also strongly activated by hZFP100 only in the presence of both SLBP and the histone stem–loop RNA (Fig. 2B, lane 3). In the absence of the RNA, the yeast cells remained white or turned only light blue (Fig. 2B, lane 6). A similar profile of activation of both reporter genes in the presence and absence of SL RNA was observed with two other clones: clone 8 (the unknown 36-kD protein) and clone 22 (ribosomal protein S15a; Fig. 2B).
The absolute requirement for the RNA component in the interaction between SLBP and hZFP100 could result from direct interaction between hZFP100 and the SLBP in the SLBP/RNA complex or from formation of a complex in which the two proteins bind independently to the bridging RNA containing the stem–loop. In the latter case, SLBP would recognize the stem–loop, whereas hZFP100 would interact with another portion of the RNA. Formation of such a complex is utilized in the three-hybrid system, designed for cloning RNA-binding proteins, and does not require any direct contact between the two proteins (SenGupta et al. 1996). We tested this possibility by replacing human SLBP used as bait in the screening procedure with the recently cloned Drosophila homolog dSLBP, which is similar to the human protein only in the RBD (Sullivan et al. 2001). The RBD of dSLBP cannot substitute for the RBD of human SLBP in histone 3′-end processing (Z. Dominski and W.F. Marzluff, unpubl.). Expression of hZFP100 in yeast expressing dSLBP and the stem–loop RNA did not allow growth of the yeast in the presence of 10 mM and higher concentrations of 3-AT (Fig. 2C, bottom, lane 1). In contrast, expression of clone 22 did support growth (Fig. 2C, bottom, lane 3) suggesting that it may form a bridging RNA complex (and not interact directly with SLBP) or interact with some region of SLBP common to the Drosophila and human proteins. hZFP100 also interacted very weakly and nonspecifically with the RNA bait alone in the standard three-hybrid assay (data not shown). These results strongly support the argument that hZFP100 directly interacts with SLBP associated with the stem–loop RNA.
Mapping the regions of SLBP interacting with hZFP100
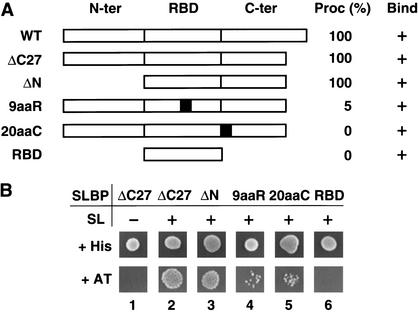
We have previously established that the entire N-terminal domain of SLBP consisting of 127 amino acids is dispensable for the activity of SLBP in 3′-end processing in vitro. The region absolutely essential for processing of H1t pre-mRNA is the RBD plus the first 20 amino acids immediately following the RBD (Dominski et al. 1999). To determine which regions of SLBP mediate its interaction with hZFP100, we replaced the ΔC27 SLBP bait with various mutant SLBPs (Fig. 3A) and tested expression of both HIS3 (Fig. 3B) and LacZ (data not shown) reporter genes. Removal of the 127-amino-acid N-terminal domain from SLBP (ΔN) had no effect on the interaction with hZFP100 (Fig. 3B, bottom, lane 3), whereas deletion of the entire C terminus, leaving only the RBD, eliminated expression of the HIS3 reporter gene (Fig. 3B, bottom, lane 6).
Figure 3.
hZFP100 does not interact with mutant SLBPs defective in histone pre-mRNA processing. (A) Diagram of the wild-type (WT) SLBP and five mutants containing specific alterations within different parts of the protein: N-terminal and C-terminal flanking domains (N-ter and C-ter, respectively) and the central RBD. The gray boxes in the 9aaR and 20aaC mutants represent the 9-amino-acid substitution in the RBD and the 20-amino-acid substitution in the C terminus shown in Figure 1A, respectively. Both mutations abolish in vitro processing of the H1t pre-mRNA without affecting binding to the stem–loop structure. The ability of each mutant SLBP to restore 3′-end processing to the SLBP-depleted nuclear extract (expressed as a percentage of the wild-type SLBP) and to bind the stem–loop RNA is shown to the right. (B) The effect of the SLBP mutations shown in A on the interaction with hZFP100 was determined by the ability of yeast cells to grow on medium lacking histidine and containing 10 mM 3-AT (bottom row). (Lanes 1,2) The strength of the interaction between SLBP ΔC27 and hZFP100 without and with coexpression of the SL RNA, respectively. SLBPs lacking the N terminus (ΔN, lane 3) and the processing- defective SLBPs 9aaR and 20aaC, and the SLBP RBD (lanes 4–6) were also assayed for the ability to interact with hZFP100 in the presence of the stem–loop RNA.
The RBD itself plays a dual role in processing: first, it mediates binding of SLBP to the stem–loop structure in the pre-mRNA; and second, it provides a number of residues involved in an additional interaction within the processing complex (Ingledue et al. 2000; Dominski et al. 2001). We have previously described mutations in the RBD (Dominski et al. 2001) and in the 20 amino acids C-terminal to the RBD (Dominski et al. 1999) that affect processing and recruitment of U7 snRNP without affecting RNA binding. Two of these mutants, 9aaR and 20aaC (shown in Fig. 1A,B, lanes 6,7), were used to identify clones that interact with regions of SLBP required for processing. When tested for their ability to interact with hZFP100, each of these mutants greatly reduced expression of the HIS3 gene (Fig. 3B, bottom, lanes 4,5) and had a similar effect on expression of the LacZ gene (data not shown). Clone 8, which was similar to hZFP100 in all the other tests for interaction with SLBP, did not interact with 20aaC SLBP, but, unlike hZFP100, did interact with 9aaR (data not shown). We conclude that the interaction of hZFP100 with the SLBP/SL complex depends on regions of SLBP that play an important role in 3′-end processing of histone pre-mRNA.
hZFP100 contains 18 zinc fingers of the C2H2 type
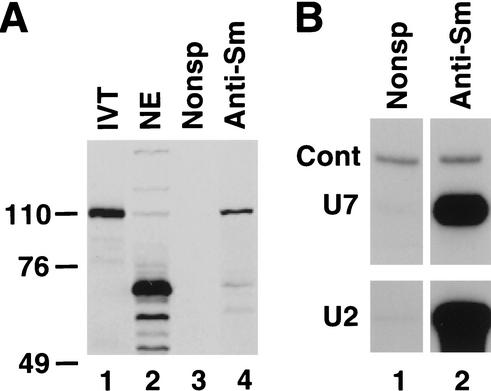
The 3.8-kb cDNA insert encoding hZFP100 predicts an 871-amino-acid protein with a calculated molecular mass of 101 kD. All yeast transformants expressing hZFP100 contained the identical cDNA insert (GenBank accession no. AF454744), indicating that they originated from the same clone. Only two partial ESTs (GenBank accession nos. AB032967 and AL080143) for hZFP100 were present in the human EST database, showing that the mRNA encoding hZFP100 is in very low abundance in the cell. The gene encoding hZFP100 is located in position 19q13.33 on Chromosome 19 and has not previously been annotated. The predicted gene structure has 4 exons, with the last exon encoding 80% of the protein. The sequence of the remainder of the 3′ UTR not present in the two-hybrid clone, consisting of 392 nucleotides, was retrieved from the database of human ESTs (GenBank accession no. AL080143). It contains a noncanonical polyadenylation signal AUUAAA, located 11 nt upstream from the site of the poly(A) addition (double underlined in Fig. 4A). The clone we isolated contained 192 nt of 5′ UTR that was in frame with hZFP100. Because the 5′ UTR did not contain any Stop codon in frame with the open reading frame (as expected for all inserts selected in the two-hybrid system), it was possible that the available cDNA represented only a partial hZFP100 mRNA lacking the bona fide initiation codon and that the actual protein is significantly larger than initially estimated. Inspection of the genomic sequence immediately preceding the sequence of the cDNA reveals a TATAA sequence (boxed in Fig. 4A) ∼25 nt upstream of the 5′ end of the clone and an adjacent in-frame Stop codon (starred in Fig. 4A). No ATG codons or obvious 3′ splice junctions were present in this region. We conclude from this analysis that the available cDNA represents close to the full-length hZFP100 mRNA, and the ATG codon at position 193 of the cDNA represents the start of translation. In support of this interpretation, the sizes of the hZFP100 produced by in vitro translation and of the protein precipitated by the anti-Sm antibody are identical (see Fig. 7A below).
Figure 7.
The anti-Sm antibody precipitates hZFP100 from nuclear extracts. (A) Anti-Sm antibody (lane 4) or a control antibody (lane 3) was bound to protein G agarose beads and incubated with 2 mg of protein from a nuclear extract from HeLa cells. The bound proteins were resolved by gel electrophoresis, together with a reticulocyte lysate programmed with hZFP100 DNA (lane 1) and 50 μg of the nuclear extract (lane 2). The proteins were transferred to nitrocellulose and hZFP100 was detected by Western blotting. (B) A portion of the same immunoprecipitates was used for preparation of RNA and detection of U7 snRNA and U2 snRNA by Northern blotting.
Inspection of the 871-amino-acid protein identified 18 zinc fingers of the C2H2 type fully conforming to the known consensus sequence (Fig. 4B). Each C2H2 zinc finger contains 21 amino acids with two conserved cysteines and histidines that coordinate binding of a zinc ion, as well as conserved phenylalanine and leucine residues at positions 8 and 14, respectively (Berg and Shi 1996). The first zinc finger is located between amino acids 211 and 231 and is separated from the next one by 90 amino acids. Zinc fingers 2–4, 5–8, 9–10, and 11–18 are clustered together, and each finger in the cluster is separated by 7 amino acids. Three of these interfinger spacers, referred to as H/C links, located in the C-terminal portion of hZFP100, contain the highly conserved sequence TGEKPY, found in nearly half of the known C2H2 zinc finger proteins that bind DNA (Laity et al. 2001). In addition to 18 typical zinc fingers, hZFP100 contains a number of degenerate zinc fingers with either deletions or substitutions of single conserved amino acids. At the N terminus of hZFP100 there is a weakly conserved element, identified in a number of proteins related to the Krüppel transcription factor, termed Krüppel-associated box, or KRAB (Laity et al. 2001). The possible KRAB element found in hZFP100 is shorter than the classical KRAB by ∼30 amino acids and is only 43% identical with KRAB.
Zinc fingers 2–8 interact with the SLBP/SL complex
We prepared a series of deletions to delineate the region of hZFP100 that interacts with the SLBP/SL complex. Deleting 15 (ΔC15) and 256 (ΔC256) amino acids from the C terminus of hZFP100, encompassing one and eight zinc fingers, respectively, did not significantly reduce the ability of yeast cells to grow on selective medium containing 10 mM 3-AT (Fig. 5A, bottom, lanes 4,5). Deletion of an additional 104 amino acids encompassing three zinc fingers (ΔC360) weakened the interaction between hZFP100 and the SLBP/SL complex, whereas removal of the next 73 amino acids (ΔC439) virtually abolished the interaction (Fig. 5A, bottom, lanes 6,7). No change in the strength of interaction with the SLBP/SL complex was observed after deleting the 64 amino acids encoded by the 5′ UTR of hZFP100 (Δ5′UTR; Fig. 5A, bottom, lane 2) or the first 168 amino acids of hZFP100 (ΔN168; Fig. 5A, bottom, lane 3). These experiments narrowed down the region of hZFP100 interacting with the complex of SLBP and SL RNA to 448 amino acids (168–615).
Figure 5.
hZFP100 interacts with the SLBP/SL RNA complex through zinc fingers 2–8. (A) Yeast expressing the SLBP/SL complex were transformed with the various deletion mutants of hZFP100 indicated at the top of each column. The ability of the mutants to interact with the SLBP/SL complex was determined by plating yeast cells on medium with 10 mM 3-AT (bottom row). (Top row) The control rate of yeast growth on medium containing histidine (no selection applied). Wild type (WT) represents the original hZFP100 clone isolated from the screen. This clone encodes the 871-amino acid hZFP100 extended at the N terminus by an additional 64 amino acids generated by translation of the 5′ UTR. The Δ5′UTR clone encodes the full-length hZFP100. (B) The amino acid sequence of hZFP100 from 312 to 628 is shown with the zinc fingers underlined. The solid arrowheads indicate break points of the C-terminal deletions. (C) The minimal region of hZFP100 required for efficient interaction with the SLBP/SL complex. Four overlapping portions of the cDNA for hZFP100 were amplified by PCR and cloned in-frame downstream of the region encoding the GAL4 AD. The lengths of the truncated proteins and the regions they span in the full-length hZFP100 are indicated to the right. Each of the four regions of hZFP100 were tested for interaction with the SLBP/SL complex, as in A.
To further define the region required for interaction with SLBP, a series of overlapping subclones was constructed and analyzed. Subclones containing residues 312–628 (zinc fingers 2–10) and 312–552 (zinc fingers 2–8) interacted strongly with the SLBP/SL complex (Fig. 5C, rows 1,2). The subclone 416–628, which removed zinc fingers 2–4, had significantly reduced binding (Fig. 5C, row 3). Because a protein containing amino acids 1–511 also bound weakly (ΔC360; Fig. 5A, bottom, lane 6), this defines a C-terminal boundary of the region required for binding between amino acids 511 and 552. A protein (amino acids 416–552) containing just zinc fingers 5–8 did not interact with SLBP (Fig. 5C, row 4). From analysis of both the deletion mutants and the minigenes, we conclude that the critical region of hZFP100 interacting with the SLBP/SL complex is located in the central part of the protein, between amino acids 312 and 552, and contains zinc fingers 2–8.
Immunoprecipitation of U7 snRNP by the hZFP100 antibody
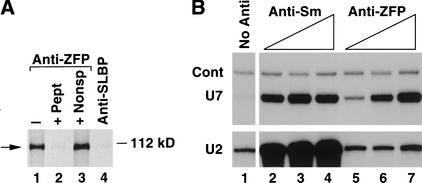
The antibody raised against a peptide containing the C-terminal 18 amino acids of hZFP100 immunoprecipitated the 35S-labeled protein expressed in the rabbit reticulocyte extract (Fig. 6A, lane 1). The efficiency of precipitation of hZFP100 from the reticulocyte lysate varied between 30% and 50%, depending on the batch of affinity-purified antibody used. Binding of the antibody to the zinc finger protein was completely inhibited by the peptide used for immunization but not by a nonspecific peptide (Fig. 6A, lanes 2 and 3, respectively). Unrelated antibodies, including the anti-SLBP antibody (Fig. 6A, lane 4), did not precipitate the hZFP100.
Figure 6.
The anti-hZFP100 antibody precipitates U7 snRNA. (A) hZFP100 was synthesized in a rabbit reticulocyte lysate in the presence of 35S methionine. The lysate was treated with anti-hZFP100 in the absence (lane 1) or presence (lane 2) of the antigenic peptide, in the presence of a nonspecific peptide (lane 3), or with anti-SLBP (lane 4). The proteins were resolved by electrophoresis in a 12% SDS-polyacrylamide gel and detected by autoradiography. (B) A nuclear extract from HeLa cells was incubated with increasing amounts of anti-Sm (5–25 μg; lanes 2–4) or anti-hZFP100 (1–5 μg; lanes 5–7) antibody. RNA was prepared from the immunoprecipitates and analyzed by Northern blotting for U7 snRNA (top) or U2 snRNA (bottom). Lane 1 shows the snRNA bound to the beads in the absence of antibody. Cont represents a synthetic RNA containing the U7 snRNA sequence (Dominski et al. 1999), used as a control for precipitation and hybridization.
To determine whether hZFP100 is associated with the U7 snRNP we tested the ability of the antibody to hZFP100 to precipitate U7 snRNA. We incubated a constant amount of a nuclear extract from HeLa cells with increasing amounts of the anti-hZFP100, prepared RNA from the immunoprecipitates, and assayed for the presence of U7 snRNA by Northern blotting. As a control the immunoprecipitates were analyzed for the presence of U2 and U1 (data not shown). Increasing amounts of the anti-hZFP100 precipitated increasing amounts of U7 snRNA (Fig. 6B, top, lanes 5–7), and 20%–25% of the U7 snRNA in the nuclear extract was precipitated by the highest concentration of the antibody. Other preparations of affinity-purified anti-hZFP100 were able to remove as much as 40% of U7 snRNA from the nuclear extract. Further increasing the concentration of the antibody in the nuclear extract or repeated incubation of the supernatant with a second aliquot of antibody did not result in precipitation of additional U7 snRNA (data not shown). Precipitation of the U7 snRNP by the anti-hZFP100 was blocked by the antigenic peptide (data not shown). Under the same conditions, U1 snRNA (data not shown) and U2 snRNA were detected only at background levels (Fig. 6B, bottom, lanes 5–7), similar to the amount of these snRNAs bound to the beads in the absence of antibody (Fig. 6B, bottom, lane 1).
The mouse monoclonal antibody Y-12 recognizes an epitope on the Sm proteins that are present in the nucleoplasmic snRNPs, including U7 snRNP (Strub and Birnstiel 1986; Mowry and Steitz 1987b). We tested this antibody in parallel with the anti-hZFP100 for its ability to precipitate U7 snRNA. The anti-Sm antibody precipitated similar amounts of the U7 snRNP as the anti-hZFP100 antibody (Fig. 6B, top, lanes 2–4). It also precipitated the U2 snRNA (Fig. 6B, bottom, lanes 2–4) and U1 snRNA (data not shown). The intensity of the band corresponding to the synthetic U7 snRNA (Cont), added to the immunoprecipitates to control for RNA recovery, was similar in all samples.
Anti-Sm immunoprecipitate is enriched in hZFP100
The anti-hZFP100 readily detected the in vitro synthesized hZFP100 in a Western blot assay (Fig. 7A, lane 1). The same preparation of the antibody barely detected hZFP100 in a HeLa nuclear extract (Fig. 7A, lane 2), indicating that there are very small amounts of the protein in the extract. In addition to detecting hZFP100, the anti-hZFP100 also detected a number of relatively abundant proteins of lower molecular weight (Fig. 7A, lane 2). To determine whether hZFP100 was present in snRNPs, the nuclear extract was precipitated with the anti-Sm antibody. A small fraction of the anti-Sm immunoprecipitate was analyzed for the presence of U2 and U7 snRNA by Northern blotting (Fig. 7B, lane 2), and the remainder was analyzed for hZFP100 by Western blotting. Immunoprecipitation of the nuclear extract with the anti-Sm antibody greatly enriched the amount of hZFP100 that was detected by Western blotting (Fig. 7A, lane 4), but not the cross-reacting proteins. The hZFP100 was not detected in immunoprecipitates obtained with a control monoclonal antibody (Fig. 7A, lane 3) or with several other antibodies (data not shown). Thus, the Sm-antibody effectively precipitates both U7 snRNA and the hZFP100, consistent with the concept that hZFP100 is associated with the U7 snRNP.
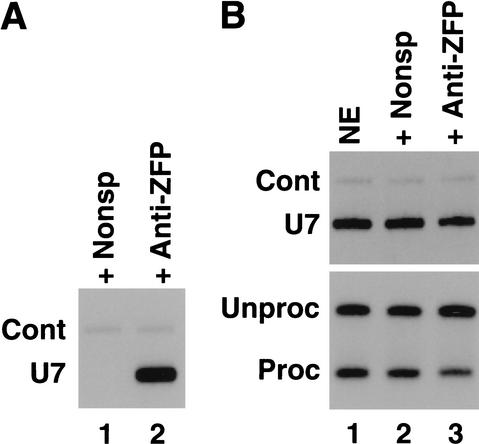
Because hZFP100 is associated with some of the U7 snRNPs, we sought to determine whether it might be involved in histone pre-mRNA processing, either as a component of an active U7 snRNP or possibly as an inhibitor of U7 snRNP. We attempted to deplete hZFP100 from a HeLa nuclear extract active in histone pre-mRNA processing with the anti-hZFP100 antibody. As found previously, significant amounts of U7 snRNA were immunoprecipitated by the anti-hZFP100 antibody (Fig. 8A). Analysis of the U7 snRNA remaining in the extract showed that ∼40% of the U7 snRNA had been removed from the extract by the antibody (Fig. 8B, top), resulting in a twofold reduction in processing efficiency (Fig. 8B, bottom). Treatment of the nuclear extract with a nonspecific antibody did not result in the removal of U7 snRNA and consequently did not significantly affect processing (Fig. 8B, lane 2, top and bottom, respectively). These results suggest that hZFP100 plays a positive role in histone pre-mRNA processing as part of the active U7 snRNP.
Figure 8.
Treatment of nuclear extracts with anti-hZFP100 reduces in vitro processing efficiency. A nuclear extract from HeLa cells was incubated with a control antibody or the anti-hZFP100 antibody, and the antibody complexes were adsorbed on protein A agarose beads. (A) RNA immunoprecipitated by either a control antibody (lane 1) or anti-hZFP100 (lane 2) was analyzed for the presence of U7 snRNA as described in Figure 7. (B) Equal amounts of the nuclear extract (lane 1) and the extracts immunodepleted with the control antibody (lane 2) or the anti-hZFP100 antibody (lane 3) were analyzed for the presence of U7 snRNA (top), and the ability to process a radiolabeled synthetic histone H2a-614 pre-mRNA (bottom). The processed and unprocessed RNAs were resolved on an 8.5% polyacrylamide gel and detected by autoradiography as in Figure 1B.
hZFP100 stimulates 3′-end processing of histone mRNA in Xenopus oocytes
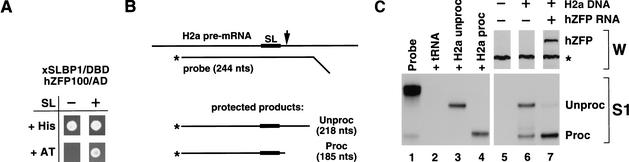
Because we were not able to completely abolish the in vitro processing activity by immunodepletion with the hZFP100 antibody, we took a different approach to show the importance of hZFP100 in histone pre-mRNA processing. We used an in vivo system based on injection of histone genes into Xenopus oocytes and measuring the efficiency of histone pre-mRNA processing. Histone pre-mRNAs transcribed from injected mouse histone genes are processed in Xenopus oocytes (Williams et al. 1994; Wang et al. 1999). Moreover, Xenopus SLBP1 (xSLBP1) is as effective as human SLBP in processing histone H2a-614 pre-mRNA in mammalian nuclear extracts (Ingledue et al. 2000), which shows that components of the processing machinery in the two organisms are exchangeable. As a prerequisite for performing the in vivo studies, we established that hZFP100 interacts with the complex formed between the SL RNA and xSLBP1, using the yeast two-hybrid system (Fig. 9A, lane 2).
Figure 9.
hZFP100 stimulates processing of histone pre-mRNA in Xenopus oocytes. (A) hZFP100 interacts with the complex formed between the SL RNA and Xenopus SLBP1 (xSLBP1). Yeast expressing the GAL4 DBD fused with xSLBP1 in the absence or presence of the stem–loop RNA were transformed with hZFP100 and tested for the ability to grow in the presence of 10 mM 3-AT. (B) A diagram of the S1 nuclease protection assay. The protected fragments Unproc and Proc correspond to mouse H2a-614 pre-mRNA and mRNA, respectively. (C) Control oocytes (lane 5), oocytes injected with the H2a-614 gene (lane 6), or oocytes injected with the hZFP100 mRNA followed 12 h later by injection with the H2a-614 gene (lane 7) were analyzed for hZFP100 by Western blotting (top). The star indicates a Xenopus protein that cross-reacts with the anti-hZFP100 antibody. RNA was prepared from the oocytes, and the amount of processed and unprocessed histone mRNA was measured by an S1 nuclease protection assay. Lanes 3 and 4 show the product protected by a synthetic H2a-614 pre-mRNA and H2a-614 mRNA, respectively. The position of the undigested probe is shown in lane 1 and the probe analyzed with yeast tRNA in lane 2.
To determine whether expression of hZFP100 could affect processing of histone pre-mRNA, a synthetic mRNA encoding hZFP100 was injected into the cytoplasm of Xenopus oocytes, and 12 h later the mouse histone H2a-614 gene was injected into the nucleus. Both the pre-mRNA and the final cleavage product can be readily and specifically detected in preparations of total oocyte RNA by an S1 nuclease protection assay using a probe complementary to the mouse gene (Fig. 9B; Ingledue et al. 2000). The size of S1 nuclease protection products corresponding to H2a-614 pre-mRNA (unprocessed) and mRNA (processed) was precisely determined by using in the assay two synthetic RNAs terminating at the appropriate sites (Fig. 9C, bottom, lanes 3 and 4, respectively). Twelve hours after injection of capped and polyadenylated hZFP100 mRNA into the cytoplasm of stage VI oocytes, hZFP100 was detected by Western blotting using the anti-hZFP100 antibody (Fig. 9C, top, lane 7). hZFP100 was present in both the nucleus and the cytoplasm of the oocyte (data not shown). In the oocytes injected with the mouse H2a-614 gene and not expressing hZFP100, only 30% of the H2a-614 pre-mRNA was processed (Fig. 9C, bottom, lane 6). The efficiency of 3′-end processing in the oocytes expressing hZFP100 was >95% (Fig. 9C, bottom, lane 7), showing that hZFP100 is involved in histone pre-mRNA processing, most likely by associating with U7 snRNP and interacting with the SLBP/pre-mRNA complex.
Discussion
Previously we have identified two regions of SLBP essential for 3′-end processing of the H1t pre-mRNA; the RBD located in the center of the protein and the first 20 amino acids of the C-terminal domain. In addition to mediating binding of SLBP to the stem–loop in the pre-mRNA, the RBD also directly participates in recruitment of U7 snRNP to its binding site in the pre-mRNA, the HDE (Dominski et al. 1999, 2001). This most likely occurs through the interaction between SLBP bound to the pre-mRNA and a protein of the U7 snRNP or another factor that would bridge SLBP and U7snRNP. Here we have used a novel variation of the yeast two-hybrid system designed to clone proteins that interact with the SLBP/stem–loop-RNA complex. A single low-abundance zinc finger protein, hZFP100, that is associated with U7 snRNP and can stimulate histone pre-mRNA processing, was isolated.
The RNA-supplemented two-hybrid system can be used to clone proteins interacting with protein–RNA complexes
In our modified version of the yeast two-hybrid system, an RNA containing the stem–loop structure was expressed from the RNA polymerase III type promoter of the yeast RNase P gene, that is, as originally used in the three-hybrid system (SenGupta et al. 1996) to clone SLBP (Wang et al. 1996a). The vast majority of the clones obtained in this screen did not interact with SLBP in the absence of the SL RNA, suggesting that the RNA is expressed in excess over SLBP in the yeast, and that all the SLBP is present in a complex with the RNA. Interestingly, several clones we isolated in a two-hybrid screen using only SLBP as bait in the absence of the RNA (L.-X. Zheng and W.F. Marzluff, unpubl.) interacted more weakly with the SLBP/RNA complex than with the free SLBP. These clones were not obtained in the modified two-hybrid screen, further showing that all the SLBP in the yeast is associated with the stem–loop RNA.
Using the modified two-hybrid screen, a novel zinc finger protein, hZFP100, was cloned that interacted only with the SLBP/RNA complex and did not interact with SLBP mutants that are inactive in histone pre-mRNA processing. Other proteins isolated in this screen could be involved in subsequent steps of histone mRNA metabolism (transport, translation, or mRNA stability). A similar RNA-supplemented two-hybrid system has recently been used for analyzing a ternary complex formed between Nanos, Pumilio, and its RNA target that is involved in translational control of hunchback mRNA (Sonoda and Wharton 1999). Therefore, this approach extends the repertoire of the yeast systems for analyzing macromolecular interactions and may prove to be a general method for cloning and studying proteins that interact with protein/RNA complexes.
hZFP100 is a member of the large family of proteins containing C2H2-type zinc fingers, first discovered in the Xenopus transcription factor TFIIIA (Pelham and Brown 1980). It is estimated that zinc finger proteins may comprise 1% of all proteins encoded by the human genome (Hoovers et al. 1992), and they represent 0.7% of the proteins encoded in the Caenorhabditis elegans genome (Clarke and Berg 1998). Although best known as sequence-specific DNA-binding motifs in many transcription factors, the C2H2 zinc fingers also can mediate binding of proteins to single- (Draper 1995) and double-stranded RNAs (Finerty and Bass 1997, 1999), RNA–DNA hybrids (Shi and Berg 1995), and to other zinc finger proteins (Mackay and Crossley 1998). Zinc finger proteins of the C2H2 type have been implicated in many aspects of cellular RNA metabolism, including pre-mRNA splicing (Larsson et al. 1995; Davies et al. 1998) and RNA storage (Joho et al. 1990). Some zinc finger proteins contain as many as 37 zinc fingers (Andreazzoli et al. 1993), and are localized exclusively in the cytoplasm. Clearly, the zinc finger is a protein motif that has evolved to participate in many protein/ligand interactions and may play roles in multiple aspects of mRNA metabolism.
hZFP100 is associated with U7 snRNP
hZFP100 is present in very low concentrations, as expected for a protein that may be a component of the U7 snRNP, which is present in only about 1000 molecules per cell. Consistent with this interpretation, only two partial ESTs were present in the human EST database as of June 2001, and the gene was not annotated in the initial analysis of the human genome. Other critical components of the histone pre-mRNA processing machinery are also likely to be low-abundance proteins. In contrast, SLBP is required stoichiometrically for pre-mRNA processing and is much more abundant than U7 snRNP.
Our results indicate that hZFP100 plays an important role in histone pre-mRNA processing by interacting with the SLBP/RNA complex. Some of the amino acids in the RBD of SLBP critical for histone pre-mRNA processing and interaction with hZFP100 are adjacent to amino acids critical for RNA binding (Dominski et al. 2001). Therefore, it is possible that hZFP100 interacts with both the SLBP and with the stem–loop in the pre-mRNA. This interaction may position the U7 snRNP to facilitate base-pairing between the U7 snRNA and the HDE. Given the intimate contact of hZFP100 with the SLBP/RNA complex, it is possible that this interaction is important for determining the position of the cleavage site used to form histone mRNA. Steitz and co-workers have shown that there is a stringent distance requirement between the HDE and the stem–loop (Scharl and Steitz 1994, 1996; Cho et al. 1995), consistent with a close contact between the U7snRNP and the SLBP/RNA complex. It is possible that hZFP100 may also make nonspecific contacts with the RNA between the stem–loop and the cleavage site, stiffening this region and in effect serving as the molecular ruler postulated by Scharl and Steitz (1994, 1996).
Recently another U7 snRNP protein was cloned by isolation of the U7 snRNP from HeLa nuclear extracts (Pillai et al. 2001). This protein, Lsm10, is a novel Sm protein in the U7 snRNP that replaces SmD1 found in other snRNPs (Pillai et al. 2001). It is likely that this protein helps recognize the novel Sm site in U7 snRNA that is critical for proper function of the U7 snRNP (Grimm et al. 1993; Stefanovic et al. 1995). There was no evidence of higher molecular weight polypeptides of the size of hZFP100 in these preparations, but the U7 snRNP prepared in this way was not active in processing (Smith et al. 1991), suggesting that some components had been lost during purification. It is therefore possible that hZFP100 is a component of the active U7 snRNP and is relatively loosely bound to the U7 snRNP particle.
Other possible functions of hZFP100 in expression of replication-dependent histone genes
Formation of the 3′ end of polyadenylated mRNAs (Connelly and Manley 1988; Proudfoot 1989) is coupled to transcription termination. We previously showed that deletion of either the stem–loop or the HDE, required for 3′-end processing of replication-dependent histone pre-mRNAs, abolishes transcriptional termination, indicating that 3′ processing of replication-dependent histone pre-mRNAs is also mechanistically coupled to transcription termination (Chodchoy et al. 1987). hZFP100 has some characteristics of a DNA-binding zinc finger protein including three C-terminal C2H2 zinc fingers linked by a seven-amino-acid sequence TGEKPY that is highly conserved in the sequence of Krüppel-like transcription factors. In addition, the first 200 amino acids of hZFP100 activated expression of reporter genes when fused to the GAL4 DNA-binding domain (Z. Dominski, unpubl.). This raises the possibility that hZFP100 might possibly play a role in initiation or termination of transcription of the histone genes coupling these processes to 3′-end processing of the nascent pre-mRNA. It is not uncommon for C2H2 zinc finger proteins to play a role in both transcription regulation and RNA metabolism (Ladomery 1997), and the dual function is best illustrated by known activities of a prototype zinc finger protein, the Xenopus TFIIIA (el Baradi and Pieler 1991; Theunissen et al. 1992). In addition to regulating transcription of the 5S rRNA genes, this factor associates with the 5S rRNA, thus facilitating its nuclear export and storage in the cytoplasm (Guddat et al. 1990).
Conclusions
hZFP100 is a novel protein associated with U7 snRNP and involved in histone pre-mRNA processing. Like many other potential proteins that affect the metabolism of a subset of mRNAs, it is a very low abundance protein. It is likely that there are additional low-abundance proteins involved in both histone pre-mRNA processing as well as in controlling histone mRNA metabolism, and we are currently trying to identify additional components in these processes.
Materials and methods
Construction of clones
C-Terminal deletion mutants of SLBP, ΔC27, and ΔC41 were constructed by PCR amplification of the SLBP cDNA encoding amino acids 1–243 and 1–229, respectively (Fig. 1A). The pGBT/SLBP/SLWT plasmid used as the bait to isolate proteins interacting with the SLBP/RNA complex was constructed in two steps. First, SLBP ΔC27 was subcloned into a pGBT9 derivative containing NcoI and SacI sites in the multiple cloning site (kindly provided by Yue Xiong, UNC), resulting in the pGBT/SLBP plasmid. This plasmid, expressing SLBP ΔC27 fused in frame to the GAL4 DNA-binding domain (DBD), was cleaved at the unique AatII site and blunt-end-ligated to a 534-nt SpeI fragment of the polIII/SLWT plasmid (Wang et al. 1996a). The SpeI fragment encompasses the entire transcription unit, which allows efficient expression of a nuclear RNA containing the histone stem–loop (SL) structure from the RNA Pol III specific promoter of the yeast RNase P gene. The resulting pGBT/SLBP/SLWT plasmid is thus capable of expressing both components of the SLBP/SL complex. The deletions and modifications of SLBP, Xenopus SLBP1 (xSLBP1), and hZFP100 were constructed by standard technology. All clones were sequenced in the Sequencing Facility at UNC, Chapel Hill. Details of each construct are available on request.
The RNA-supplemented two-hybrid screen
Yeast transformation, minipreparation of the yeast DNA, and rescue of the library plasmid were carried out according to standard yeast protocols, as described elsewhere (Dominski and Marzluff 2001). The RNA-supplemented two-hybrid screen was performed in the yeast strain CG-1945 transformed with the pGBT/SLBP/SLWT bait plasmid (TRP1 selection). The yeast cells transformed with the HeLa cDNA library (LEU2 selection) were plated on synthetic defined (SD) medium lacking tryptophan, leucine, and histidine, supplemented with increasing concentrations of 3-aminotriazole (3-AT). For analysis of the interaction between selected library clones and various SLBP variants in the presence and absence of the SL RNA, the yeast transformants were initially grown on plates containing histidine and subsequently scraped and resuspended in sterile water to a density of ∼106 cells/mL. The cell suspension (5–10 μL) was spotted on selective SD medium lacking histidine, supplemented with various concentrations of 3-AT. The same amount of the cell suspension was also applied to medium containing histidine to control for growth under nonselective conditions. After incubation at 27°C for 2 d, the plates were photographed using a video camera, and the images were processed by NIH Image software (National Institutes of Health).
Liquid assay for β-galactosidase activity
Expression of the LacZ reporter gene was determined by qualitative analysis of β-galactosidase activity according to a modified version of the liquid assay, as previously described (Dominski and Marzluff 2001). For illustrations, a portion of the cell suspension was spotted on a white membrane, dried, and photographed using a video camera. The images were processed by NIH Image software (National Institutes of Health).
Expression and purification of recombinant SLBP
The wild-type and deletion mutants of SLBP were expressed in Sf9 insect cells using the baculovirus expression system, and the His-tagged proteins were purified as previously described (Dominski et al. 1999, 2001).
In vitro synthesis and labeling of RNA
In vitro 3′-end processing was carried out using 86-nt pre-mRNAs containing the stem–loop structure and the U7 binding site from either the mouse H2a-614 or H1t pre-mRNAs. The sequences of both RNAs and methods for their synthesis and labeling have been described previously (Dominski et al. 1995, 1999; Marzluff et al. 1997). Capped and polyadenylated mRNA encoding hZFP100 used for oocyte injection was synthesized by SP6 transcription of the pSP64T/hZFP100 plasmid linearized with PvuII, as described (Wang et al. 1999). The identity of the mouse histone H2a-614 mRNA and pre-mRNA expressed in Xenopus oocytes was confirmed by using a synthetic RNA ending either precisely at the cleavage site or 104 nt further downstream, respectively. The two RNAs were generated by T7 transcription of PCR amplification products containing the T7 promoter and the mouse histone H2a-614 gene.
Preparation of nuclear extracts and 3′-end processing
Preparation of nuclear extracts from both mouse myeloma and human HeLa cells, immunodepletion of SLBP and 3′-end processing of histone pre-mRNA were performed as previously described (Dominski et al. 1995, 1999; Marzluff et al. 1997). HeLa cells were provided by the National Cell Culture Center (Minneapolis, MN).
Generation and affinity purification of anti-hZFP100 antibodies
Rabbits were injected at Pocono Farms (Candenensis, PA) with a KLH-conjugated C-terminal peptide corresponding to the last 18 amino acids of hZFP100. The serum was purified using the C-terminal peptide coupled to an agarose support using the SulfoLink kit (Pierce). The affinity-purified polyclonal antibody (average protein concentration of 0.5 μg/μL) was eluted from the column with 100 mM Glycine-HCl (pH 2.5) and immediately neutralized.
In vitro synthesis of hZFP100
hZFP100 was synthesized in the rabbit reticulocyte extract using the coupled transcription and translation system (TnT) from Promega (Madison, WI) according to the manufacturer's protocol. Each reaction (50 μL total volume) contained 2.5 μg of supercoiled pSP64T/hZFP100 DNA and was carried out in the absence of labeled amino acids. When required, the TnT reaction was supplemented with 35S-methionine (NEN), and unlabeled methionine was omitted.
Western blotting
Proteins were resolved by electrophoresis on 12% SDS polyacrylamide gels, transferred to nitrocellulose, and analyzed by Western blotting as previously described (Whitfield et al. 2000).
Immunoprecipitation of processing complexes
Processing complexes containing the U7 snRNP were formed on H2a-614 pre-mRNA in a HeLa nuclear extract and immunoprecipitated as described (Dominski et al. 1999, 2001). Each reaction contained 25 ng of the pre-mRNA substrate, 75 μL of the nuclear extract, and 20 mM EDTA (pH 8) in a total volume of 100 μL. The samples were incubated at 22°C for 5 min to allow formation of the processing complexes, supplemented with 5 μg of affinity-purified antibody directed against either SLBP or hZFP100, and incubated at 4°C with rotation for 1 h. Antibody–antigen complexes were adsorbed on 20 μL of protein A agarose beads (GIBCO BRL), and RNA was prepared from the immunoprecipitate. The RNA was resolved on an 8.5% polyacrylamide/7M urea gel and transferred to Hybond-N+ synthetic membrane (Amersham), and the U7 snRNA was detected by hybridization with a riboprobe complementary to human U7 snRNA. Prior to phenol extraction, 0.5 ng of a synthetic mouse U7 snRNA was added to each sample to monitor the efficiency of RNA recovery and hybridization with the anti-U7 probe. This RNA hybridizes with the human U7 snRNA probe, and contains additional sequences on both ends of the U7 snRNA sequence, thus migrating significantly slower than the endogenous human U7 snRNA (Dominski et al. 1999, 2001). The specificity of U7 snRNA precipitation was determined by reblotting the same membrane with a probe complementary to human U1 snRNA or U2 snRNA (Bond 1988).
Immunoprecipitation of U7 snRNP
The U7 snRNP was directly immunoprecipitated from 100 μL of HeLa nuclear extract by addition of either 25 μg of the anti-Sm monoclonal antibody (Lerner and Steitz 1979) or 5 μg of anti-hZFP100 antibody. The samples containing the nuclear extract and the antibody were rotated at 4°C for 1 h. The immunocomplexes were absorbed on protein A agarose beads and further processed as described above for analysis of U7 snRNA.
Detection of hZFP100 in anti-Sm precipitate
For immunoprecipitation of anti-Sm reactive snRNPs, a 500-μL aliquot of mouse ascitic fluid containing the monoclonal anti-Sm antibody Y-12 (Lerner et al. 1981) was rotated at 4°C for 2 h with 50 μL of protein G agarose beads (GIBCO BRL) in the presence of 100 mM Tris-HCl (pH 7). The beads were collected, washed several times with buffer D (20 mM HEPES-KOH at pH 7.9, 100 mM KCl, 0.5 mM DTT, 0.2 mM EDTA at pH 8.0, 20% glycerol), and mixed with 250 μL of HeLa nuclear extract. After rotation at 4°C for 2 h, the supernatant was removed and the beads were washed several times with buffer D/0.1% NP-40 and tested for the presence of both U7 snRNA by Northern blotting (20% of the beads) and hZFP100 by Western blotting (80% of the beads).
Injection of Xenopus oocytes
Stage V–VI oocytes were injected in the cytoplasm with 30 nL of a solution containing 1.5 ng of the in vitro synthesized capped and polyadenylated mRNA encoding hZFP100. Twelve hours after injection of the synthetic mRNA, the nucleus of each healthy oocyte was injected with 15 nL of a solution containing blue dextran and 750 pg of plasmid DNA containing the mouse H2a-614 gene. The oocytes were incubated for an additional 12 h, and the total RNA for each experiment was isolated from a pool of 30 oocytes. The histone mRNAs were detected by S1 nuclease mapping as previously described (Ingledue et al. 2000).
Acknowledgments
This work was supported by NIH grant GM29832 to W.F.M. Z.D. was a recipient of a Faculty Development Award from the University of North Carolina and a grant from the North Carolina Biotechnology Center. We thank Matt Purdy for technical assistance. We thank Joan Steitz (Yale University, New Haven) for a gift of Y-12 anti-Sm antibodies and hybridomas.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL marzluff@med.unc.edu; FAX (919) 966-6821.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.932302.
References
- Andreazzoli M, De Lucchini S, Costa M, Barsacchi G. RNA binding properties and evolutionary conservation of the Xenopus multifinger protein Xfin. Nucleic Acids Res. 1993;21:4218–4225. doi: 10.1093/nar/21.18.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, Shi YG. The galvanization of biology: A growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- Birnstiel ML, Schaufele FJ. Structure and function of minor snRNPs. In: Birnstiel ML, editor. Structure and function of major and minor small ribonucleoprotein particles. Berlin, Germany: Springer-Verlag; 1988. pp. 155–182. [Google Scholar]
- Bond U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988;7:3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond UM, Yario TA, Steitz JA. Multiple processing-defective mutations in a mammalian histone premessenger RNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes & Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DC, Scharl EC, Steitz JA. Decreasing the distance between the two conserved sequence elements of histone pre-messenger RNA interferes with 3′ processing in vitro. RNA. 1995;1:905–914. [PMC free article] [PubMed] [Google Scholar]
- Chodchoy N, Levine BJ, Sprecher C, Skoultchi AI, Marzluff WF. Expression of mouse histone genes: Transcription into 3′ intergenic DNA and cryptic processing sites downstream from the 3′ end of the H3 gene. Mol Cell Biol. 1987;7:1039–1047. doi: 10.1128/mcb.7.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke ND, Berg JM. Zinc fingers in Caenorhabditis elegans: Finding families and probing pathways. Science. 1998;282:2018–2022. doi: 10.1126/science.282.5396.2018. [DOI] [PubMed] [Google Scholar]
- Connelly S, Manley J. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes & Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Davies RC, Calvio C, Bratt E, Larsson SH, Lamond AI, Hastie ND. WT1 interacts with the splicing factor U2AF65 in an isoform-dependent manner and can be incorporated into spliceosomes. Genes & Dev. 1998;12:3217–3225. doi: 10.1101/gad.12.20.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- ————— Three-hybrid screens for RNA-binding proteins: Proteins binding the 3′ end of histone mRNA. Meth Mol Biol. 2001;177:291–318. doi: 10.1385/1-59259-210-4:291. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Sumerel J, Hanson RJ, Whitfield ML, Marzluff WF. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA. 1995;1:915–923. [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Zheng L-X, Sanchez R, Marzluff WF. The stem–loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol Cell Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Erkmann JA, Greenland JA, Marzluff WF. Mutations in the RNA binding domain of stem–loop binding protein define separable requirements for RNA binding and histone pre-mRNA processing. Mol Cell Biol. 2001;21:2008–2017. doi: 10.1128/MCB.21.6.2008-2017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE. Protein–RNA recognition. Annu Rev Biochem. 1995;64:593–620. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]
- el Baradi T, Pieler T. Zinc finger proteins: What we know and what we would like to know. Mech Dev. 1991;35:155–169. doi: 10.1016/0925-4773(91)90015-x. [DOI] [PubMed] [Google Scholar]
- Finerty PJ, Jr, Bass BL. A Xenopus zinc finger protein that specifically binds dsRNA and RNA–DNA hybrids. J Mol Biol. 1997;271:195–208. doi: 10.1006/jmbi.1997.1177. [DOI] [PubMed] [Google Scholar]
- ————— Subsets of the zinc finger motifs in dsRBP-ZFa can bind double-stranded RNA. Biochemistry. 1999;38:4001–4007. doi: 10.1021/bi9825293. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Lewis NJ, Marzluff WF. The histone 3′-terminal stem–loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 1996;24:1954–1962. doi: 10.1093/nar/24.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev O, Birnstiel ML. The conserved CAAGAAAGA spacer sequence is an essential element for the formation of 3′ termini of the sea urchin H3 histone mRNA by RNA processing. EMBO J. 1985;4:481–489. doi: 10.1002/j.1460-2075.1985.tb03654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O, Krämer A, Keller W, Birnstiel ML. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986;5:1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Stefanovic B, Schümperli D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 1993;12:1229–1238. doi: 10.1002/j.1460-2075.1993.tb05764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A, Soldati D, Burri M, Schümperli D. Isolation of an active gene and of two pseudogenes for mouse U7 small nuclear RNA. Biochim Biophys Acta Gene Struct Expr. 1991;1088:151–154. doi: 10.1016/0167-4781(91)90167-k. [DOI] [PubMed] [Google Scholar]
- Guddat U, Bakken AH, Pieler T. Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in Xenopus oocytes. Cell. 1990;60:619–628. doi: 10.1016/0092-8674(90)90665-2. [DOI] [PubMed] [Google Scholar]
- Harris ME, Böhni R, Schneiderman MH, Ramamurthy L, Schümperli D, Marzluff WF. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoovers JM, Mannens M, John R, Bliek J, van Heyningen V, Porteous DJ, Leschot NJ, Westerveld A, Little PF. High-resolution localization of 69 potential human zinc finger protein genes: A number are clustered. Genomics. 1992;12:254–263. doi: 10.1016/0888-7543(92)90372-y. [DOI] [PubMed] [Google Scholar]
- Ingledue TC, Dominski Z, Sanchez R, Erkmann JA, Marzluff WF. Dual role for the RNA binding domain of Xenopus laevis SLBP1 in histone pre-mRNA processing. RNA. 2000;6:1635–1648. doi: 10.1017/s1355838200000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joho KE, Darby MK, Crawford ET, Brown DD. A finger protein structurally similar to TFIIIA that binds exclusively to 5S RNA in Xenopus. Cell. 1990;61:293–300. doi: 10.1016/0092-8674(90)90809-s. [DOI] [PubMed] [Google Scholar]
- Ladomery M. Multifunctional proteins suggest connections between transcriptional and post-transcriptional processes. Bioessays. 1997;19:903–909. doi: 10.1002/bies.950191010. [DOI] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: New insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Larsson SH, Charlieu JP, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie ND. Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- Martin F, Schaller A, Eglite S, Schümperli D, Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Michel F, Zenklusen D, Müller B, Schümperli D. Positive and negative mutant selection in the human histone hairpin-binding protein using the yeast three-hybrid system. Nucleic Acids Res. 2000;28:1594–1603. doi: 10.1093/nar/28.7.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF. Histone 3′ ends: Essential and regulatory functions. Gene Expr. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Whitfield ML, Dominski Z, Wang Z-F. Identification of the protein that interacts with the 3′ end of histone mRNA. In: Richter JD, editor. mRNA formation and function. New York: Academic Press; 1997. pp. 163–193. [Google Scholar]
- Melin L, Soldati D, Mital R, Streit A, Schümperli D. Biochemical demonstration of complex formation of histone pre- mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 1992;11:691–697. doi: 10.1002/j.1460-2075.1992.tb05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry KL, Steitz JA. Both conserved signals on mammalian histone pre-mRNAs associate with small nuclear ribonucleoproteins during 3′ end formation in vitro. Mol Cell Biol. 1987a;7:1663–1672. doi: 10.1128/mcb.7.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science. 1987b;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- Pandey NB, Marzluff WF. The stem–loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB, Brown DD. A specific transcription factor that can bind to either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci. 1980;77:4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Will CL, Lührmann R, Schümperli D, Müller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989;14:105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- Scharl EC, Steitz JA. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 1994;13:2432–2440. doi: 10.1002/j.1460-2075.1994.tb06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Length suppression in histone messenger RNA 3′-end maturation: Processing defects of insertion mutant premessenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc Natl Acad Sci. 1996;93:14659–14664. doi: 10.1073/pnas.93.25.14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F, Gilmartin GM, Bannwarth W, Birnstiel ML. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature. 1986;323:777–781. doi: 10.1038/323777a0. [DOI] [PubMed] [Google Scholar]
- SenGupta DJ, Zhang BL, Kraemer B, Prochart P, Fields S, Wickens M. A three-hybrid system to detect RNA–protein interactions in vivo. Proc Natl Acad Sci. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Berg JM. Specific DNA–RNA hybrid binding by zinc finger proteins. Science. 1995;268:282–284. doi: 10.1126/science.7536342. [DOI] [PubMed] [Google Scholar]
- Smith HO, Tabiti K, Schaffner G, Soldati D, Albrecht U, Birnstiel ML. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2‘-O-methyl oligoribonucleotides. Proc Natl Acad Sci. 1991;88:9784–9788. doi: 10.1073/pnas.88.21.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes & Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C, Streit A, Stefanovic B, Albrecht D, Koning THW, Schümperli D. 3′ end processing of mouse histone pre-mRNA: Evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 1994;22:4023–4030. doi: 10.1093/nar/22.20.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber C, Schümperli D. 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 1988;16:9399–9413. doi: 10.1093/nar/16.20.9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic B, Hackl W, Lührmann R, Schümperli D. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 1995;23:3141–3151. doi: 10.1093/nar/23.16.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A, Koning TW, Soldati D, Melin L, Schümperli D. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 1993;21:1569–1575. doi: 10.1093/nar/21.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K, Birnstiel ML. Genetic complementation in the Xenopus oocyte: Co-expression of sea urchin histone and U7 RNAs restores 3′ processing of H3 pre-mRNA in the oocyte. EMBO J. 1986;5:1675–1682. doi: 10.1002/j.1460-2075.1986.tb04411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K, Galli G, Busslinger M, Birnstiel ML. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 1984;3:2801–2807. doi: 10.1002/j.1460-2075.1984.tb02212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes & Dev. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J-H, Pilch DR, Marzluff WF. The histone mRNA 3′ end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res. 1992;20:6057–6066. doi: 10.1093/nar/20.22.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen O, Rudt F, Guddat U, Mentzel H, Pieler T. RNA and DNA binding zinc fingers in Xenopus TFIIIA. Cell. 1992;71:679–690. doi: 10.1016/0092-8674(92)90601-8. [DOI] [PubMed] [Google Scholar]
- Turner PC, Whalen A, Schümperli D, Matera AG. The Bona Fide mouse U7 snRNA gene maps to a different chromosome than two U7 pseudogenes. Genomics. 1996;31:250–252. doi: 10.1006/geno.1996.0042. [DOI] [PubMed] [Google Scholar]
- Wang Z-F, Whitfield ML, Ingledue TI, Dominski Z, Marzluff WF. The protein which binds the 3′ end of histone mRNA: A novel RNA- binding protein required for histone pre-mRNA processing. Genes & Dev. 1996a;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- Wang Z-F, Tisovec R, Debry RW, Frey MR, Matera AG, Marzluff WF. Characterization of the 55 kilobase mouse histone gene cluster on chromosome 3. Genome Res. 1996b;6:702–714. doi: 10.1101/gr.6.8.702. [DOI] [PubMed] [Google Scholar]
- Wang Z-F, Ingledue TC, Dominski Z, Sanchez R, Marzluff WF. Two Xenopus proteins that bind the 3′ end of histone mRNA: Implications for translational control of histone synthesis during oogenesis. Mol Cell Biol. 1999;19:835–845. doi: 10.1128/mcb.19.1.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Zheng L-X, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AS, Ingledue TC, Kay BK, Marzluff WF. Changes in the stem–loop at the 3′ terminus of histone mRNA affects its nucleocytoplasmic transport and cytoplasmic regulation. Nucleic Acids Res. 1994;22:4660–4666. doi: 10.1093/nar/22.22.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]