Abstract
The ETS transcription factors perform distinct biological functions despite conserving a highly similar DNA-binding domain. One distinguishing property of a subset of ETS proteins is a conserved region of 80 amino acids termed the Pointed (PNT) domain. Using enzyme kinetics we determined that the Ets-1 PNT domain contains an ERK2 docking site. The docking site enhances the efficiency of phosphorylation of a mitogen-activated protein kinase (MAPK) site N-terminal to the PNT domain. The site enhances ERK2 binding rather than catalysis. Three hydrophobic residues are involved in docking, and the previously determined NMR structure indicates that these residues are clustered on the surface of the Ets-1 PNT domain. The docking site function is conserved in the PNT domain of the highly related Ets-2 but not in the ets family member GABPα. Ablation of the docking site in Ets-1 and Ets-2 prevented Ras pathway-mediated enhancement of the transactivation function of these proteins. This study provides structural insight into the function of a MAPK docking site and describes a unique activity for the PNT domain among a subset of ets family members.
Keywords: MAPK, Ets-1, docking site, phosphorylation
The ets gene family encodes evolutionarily related transcription factors that regulate multiple biological functions. Members of this family conserve an 85-amino-acid DNA-binding ETS domain that binds the core DNA sequence 5′-GGA(A/T)-3′ (Graves and Petersen 1998; Sharrocks 2001). The ets genes are found in all metazoans studied to date and number 8 in Drosophila melanogaster (Hsu and Schulz 2000), 10 in Caenorhabditis elegans (Hart et al. 2000), and 25 in humans. Strikingly, distinct ETS proteins can regulate diverse biological processes despite having the ability to bind highly similar DNA sequences. For instance, gene-targeting studies in mice indicate that different ETS proteins play critical roles in varied physiological processes such as hematopoiesis, apoptosis, and regulation of the extracellular matrix (Bartel et al. 2000). In addition, different ets genes are responsible for regulating embryogenesis and neural development in Drosophila (Hsu and Schulz 2000). This diversity of function raises the question of how specificity is generated among members of this gene family.
To regulate the transcription of unique genes, individual ETS proteins have evolved different molecular features. One feature conserved in ∼40% of ets family members is the 80-amino-acid Pointed (PNT) domain. Surprisingly, this domain can perform distinct functions among ETS proteins. For instance, the PNT domain of the ets family member TEL can serve as a transcriptional repression module (Fenrick et al. 1999; Wang and Hiebert 2001). The repression activity may be related to the ability of the PNT domain to form helical polymers (Kim et al. 2001). The TEL PNT domain is also fused to heterologous proteins as a result of translocations associated with leukemia (Golub et al. 1994, 1995, 1996; Lacronique et al. 1997). Oligomerization of the PNT domain is hypothesized to play an important role in the genesis of the associated leukemia. In contrast, the Ets-1 PNT domain enhances the activity of the Ets-1 transactivation domain but does not function in transcriptional repression (Schneikert et al. 1992). Also, unlike the TEL PNT domain, the Ets-1 PNT domain is monomeric (Slupsky et al. 1998). Biochemical reasons for differences in function are suggested by structural studies that indicate that the PNT domains of these two proteins have similar folds but divergent surface chemistry. Owing to the lack of conservation of surface residues, PNT domains of different ETS proteins may interact with distinct protein partners, thereby establishing unique biological functions within the family.
The specificity of ETS protein function also is enhanced by signal transduction pathways (Yordy and Muise-Helmericks 2000). For instance, the RAS/RAF/MEK/ERK pathway modulates the activities of vertebrate Ets-1 and Ets-2 and their apparent ortholog in Drosophila, Pnt-P2. These proteins are phosphorylated at a conserved mitogen-activated protein kinase (MAPK) phosphoacceptor site N-terminal to the PNT domain (Brunner et al. 1994; O'Neill et al. 1994; Yang et al. 1996). The MAPKs ERK1 and ERK2 phosphorylate Ets-1 and Ets-2 at this site, and the highly related Drosophila MAPK Rolled phosphorylates Pnt-P2. Phosphorylation of this site results in enhancement of the transactivation activity of these proteins by an undetermined mechanism (Brunner et al. 1994; O'Neill et al. 1994; Yang et al. 1996). The biological relevance of this phosphoacceptor is firmly established by genetic studies in Drosophila that show that MAPK phoshorylation of Pnt-P2 is required for R7 photoreceptor development (Brunner et al. 1994; O'Neill et al. 1994).
This report shows that the PNT domain regulates Ets-1 and Ets-2 phosphorylation by serving as an ERK2 docking site. Docking sites are defined as short sequence motifs that lie distal to the phosphoacceptor in the linear amino acid sequence and increase the efficiency of substrate phosphorylation (Holland and Cooper 1999; Sharrocks et al. 2000). In addition, these motifs can affect the accuracy and specificity of MAPK phosphorylation. Docking sites are thought to function by interacting with MAPKs outside the catalytic site, thereby increasing the concentration of the phosphoacceptor near the enzyme. A number of classes of proteins contain MAPK docking sites, including kinases, phosphatases, scaffold proteins, and transcription factors.
Despite numerous examples, little is known about the structural details of docking sites. To date, docking sites in transcription factors have been identified exclusively by deletion and site-directed mutagenesis of residues that are not defined by secondary or tertiary structural information. Lacking a structural context, these mutagenesis experiments could not identify residues that define the docking interfaces. Additionally, although docking sites often lie distant to the phosphoacceptor in the linear amino acid sequence, the relative positions of these elements in three-dimensional space is not known.
This report provides insights into the structural features of transcription factor docking sites as well as the specificity of ETS protein function. The previously determined NMR structure of the PNT domain allowed us to map a hydrophobic surface on the Ets-1 PNT domain required for efficient phosphorylation by ERK2. Kinetic data indicate that the residues in this surface mediate a binding event rather than an alteration of catalysis. Enzyme kinetics also indicate that the docking site is conserved in the PNT domain of Ets-2 but not in the ets family member GABPα. Functionally, the docking site is required for Ras-pathway-mediated enhancement of the transactivation activity of Ets-1 and Ets-2. In sum, this study reveals the structural context of a MAPK docking site in a transcription factor and highlights ERK2 docking as a distinguishing feature of a subset of PNT domains of ets family members.
Results
ERK1/2 binds an Ets-1(29–138) affinity column
The PNT domain is hypothesized to be a protein–protein interaction motif. To identify protein partners, an affinity chromatography experiment was performed using Ets-1(29–138) as a ligand, which includes the PNT domain (residues 54–132) and MAPK phosphoacceptor site (T38). A calf thymus nuclear extract was used as a source of potential partner proteins because of the abundance of Ets-1 in T lymphocytes. In the chromatography experiment, several proteins from the extract bound the Ets-1(29–138) column, and one protein was identified by immunoblotting to be the MAPK ERK1 and/or ERK2 (data not shown). The ERKs may have interacted with Ets-1 solely through the MAPK phosphoacceptor; however, we speculated that a docking site distal to the MAPK phosphorylation site also mediated the interaction.
The Ets-1 PNT domain contains an ERK2 docking site
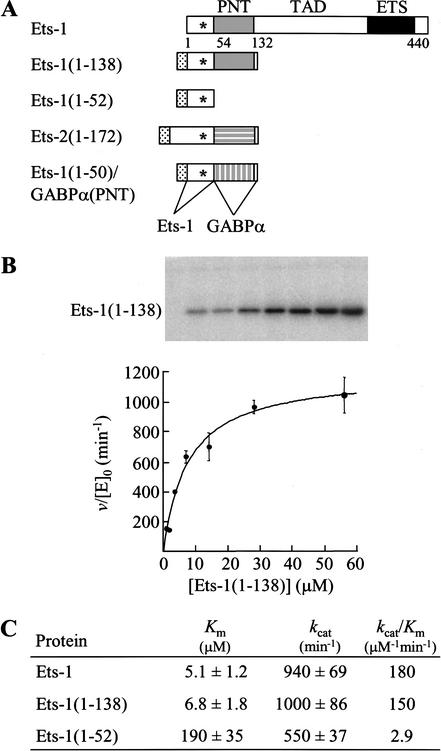
To localize a potential ERK2 docking site in Ets-1, full-length Ets-1 and two fragments of Ets-1 were generated for kinase assays (Fig. 1A). Full-length Ets-1 contains the MAPK phosphoacceptor site (T38), PNT domain, transactivation domain, and DNA-binding ETS domain (Fig. 1A). The two fragments were designed based on NMR solution structure data (Slupsky et al. 1998). The Ets-1(1–138) fragment retains the MAPK phosphorylation site that lies within disordered sequences and the globular five α-helix PNT domain (Fig. 1A). The Ets-1(1–52) fragment retains only the MAPK phosphorylation site (Fig. 1A) and presumably lacks secondary structural elements. Each fragment also contains a histidine tag to facilitate purification.
Figure 1.
An ERK2 docking site is localized to the PNT domain. (A) Schematic of Ets-1 and fragments of ETS proteins used in kinase assays. MAPK phosphorylation site (T38, asterisk) and histidine tag (stippled). (TAD) Transactivation domain. (B) Phosphorylation of Ets-1 by ERK2 follows Michaelis–Menten kinetics. Ets-1(1–138) was phosphorylated by ERK2 with [γ-32P]ATP under steady-state conditions as described in the Materials and Methods. PhosphorImage of a 15% SDS-polyacrylamide gel (top) displays reaction products. Initial reaction velocities, v, were divided by the ERK2 concentration, [E]0. Duplicate v/[E]0 values from a single experiment were averaged and are shown fit to the Michaelis–Menten equation v/[E]0 = kcat[S]/(Km + [S]), where [S] is the ETS protein concentration (bottom). Error bars indicate the range. (C) Kinetic parameters of Ets-1 and fragments phosphorylated by ERK2. Mean values for Km and kcat are reported ±S.E. and were determined as described in the Materials and Methods and B using data from two (Ets-1), five (Ets-1(1–138)), and three (Ets-1(1–52)) independent experiments. kcat/Km was determined by dividing individual values. A similar value for the 1–138 fragment (19 μM) was recently reported (Waas and Dalby 2001).
Kinase assays were performed in vitro under steady-state conditions with [γ-32P]ATP, and the initial rates of phosphorylation were fit to the Michaelis–Menten equation (Fig. 1B). The parameter Km, the apparent equilibrium dissociation constant of a kinase and its substrate, was used to compare the ability of ERK2 to bind ETS fragments. The Km of the Ets-1(1–138) fragment was nearly identical to that of full-length Ets-1 (6.8 ± 1.8 μM and 5.1 ± 1.2 μM, respectively; Fig. 1C). The fragment was phosphorylated on T38, as evidenced by the lack of phosphorylation of Ets-1(1–138;T38A) under assay conditions (data not shown). Additionally, the histidine tag did not affect the Km value (data not shown). These data suggest that the first 138 amino acids of Ets-1 are sufficient to achieve full binding activity with ERK2.
In contrast, Ets-1(1–52), which lacks the PNT domain and known secondary structural elements, had a Km ∼30-fold higher (190 ± 35 μM) than that of full-length Ets-1 and Ets-1(1–138) (Fig. 1C). The Km of this fragment is on the order of magnitude of previously characterized peptide substrates of ERK2 (∼300–500 μM; Gonzalez et al. 1991; Robinson et al. 1996). The kcat value showed only a slight twofold decrease, suggesting that removal of the PNT domain did not drastically affect enzyme catalysis. These data support the argument that the PNT domain contains an ERK2 docking site that enhances the binding affinity of the enzyme–substrate interaction.
L114, L116, and F120 form a potential docking interface
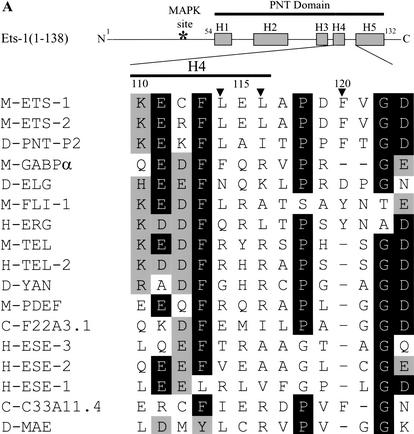
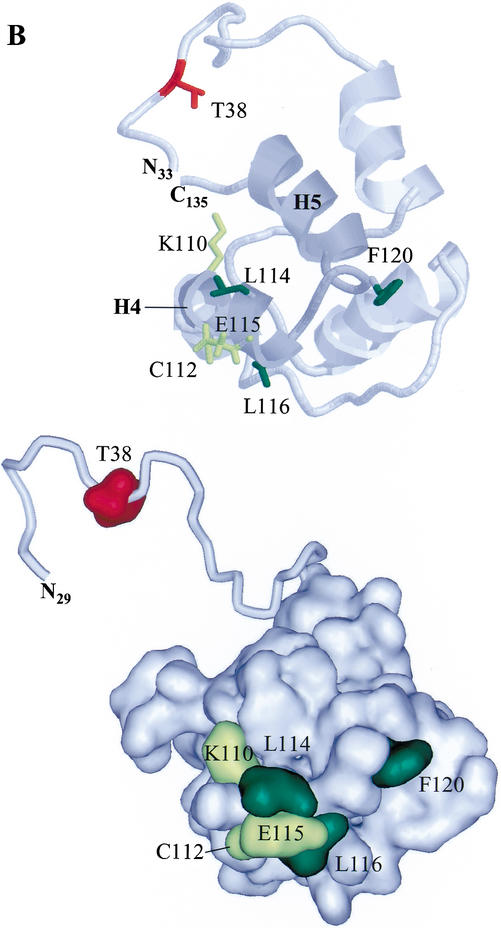
To define the docking site, the NMR solution structure of Ets-1(29–138) (Slupsky et al. 1998) was used to select surface residues of the PNT domain of Ets-1(1–138) for site-directed mutagenesis. The residues that were initially targeted were an LXL sequence (L114, L116) and a basic residue (K110) on the surface of helix H4 (Fig. 2A,B). These residues are in a sequence pattern described for other MAPK docking sites (Holland and Cooper 1999; Sharrocks et al. 2000). Mutation of L114 and L116 to alanines or arginines resulted in a modest fourfold increase in Km, whereas mutation of K110 to alanine did not result in a significant increase in Km (Table 1). These results suggest that the leucines, but not K110, play a role in ERK2 binding. However, the binding activity of these mutants was stronger than that of Ets-1(1–52) (Fig. 1C), which lacks the PNT domain, suggesting that other residues of the PNT domain also contribute to ERK2 binding. Mutation to alanine of the residues C112 and E115 (Fig. 2A), which lie near L114 and L116 on the surface of the PNT domain (Fig. 2B), did not alter the Km (Table 1). In contrast, mutation of the surface residue F120 (Fig. 2A,B) to alanine resulted in a striking 29-fold increase in Km (Table 1). This mutation, when combined with the L114R and L116R mutations, resulted in a fragment with a Km 40-fold higher than that of Ets-1(1–138) and similar to that of the fragment deleting the PNT domain, Ets-1(1–52) (Table 1). These data indicate that L114, L116, and in particular, F120, play an important role in Ets-1 docking with ERK2. The role of the phenylalanine also indicates that the site is distinct from other LXL-containing docking sites.
Figure 2.
An ERK2 docking site lies on the surface of the Ets-1 PNT domain. (A) Position of docking residues in the secondary structure of Ets-1(1–138). Rectangles indicate α-helices, and the asterisk indicates the position of the MAPK phosphorylation site (T38) within the flexible N-terminal region of Ets-1 (top; Slupsky et al. 1998). Sequence lineup (bottom) of helix H4 and the loop between helices H4 and H5 of the PNT domains of ets family members as well as Drosophila Mae, which lacks an ETS domain. Residues conserved in >50% of proteins are shaded in black; conservative substitutions found in >50% of proteins are shaded in gray. (M) Murine; (H) human; (D) Drosophila; (C) C. elegans. Numbering is for murine Ets-1. Arrowheads mark residues involved in ERK2 docking in Ets-1 and Ets-2 (Tables 1, 2). (B) Positions of docking residues in the NMR solution structure of Ets-1(29–138) (Slupsky et al. 1998). Ribbon display (top) and surface rendering (bottom) show α-helices (H1–H5). Helices H4 and H5, and the connecting loop are shaded in dark gray (top). Mutagenized residues are highlighted: residues that gave negligible increases in Km when mutated (yellow); residues that gave fourfold or higher increases in Km when mutated (green). The disordered N-terminal region containing the phosphoacceptor (T38; red) is shown in ribbon display and in different orientations to highlight its flexibility (top and bottom). The PNT domain also shows strong structural similarity to the SAM domain in helices H2–H5 (Stapleton et al. 1999). The figure was generated using RasMol version 2.6 and WebLab ViewerPro 4.0 (Accelrys, San Diego, CA).
Table 1.
Kinetic analysis of Ets-1(1–138)
| Protein [Ets-1(1–138)]
|
|
Kma (μM)
|
Fold increase in Kmb
|
kcata (min−1)
|
kcat/Kmc (μM−1 min−1)
|
|---|---|---|---|---|---|
| Ets-1(1–138) | (5)d | 6.8 ± 1.8e | — | 1000 ± 86 | 150 |
| C112A | (2) | 7.3 ± 1.1 | 1.1 ± 0.33 | 970 ± 47 | 130 |
| E115A | (2) | 8.6 ± 2.6 | 1.2 ± 0.48 | 1100 ± 120 | 130 |
| K110A | (2) | 11 ± 1.6 | 1.6 ± 0.48 | 1100 ± 60 | 100 |
| L114A;L116A | (3) | 27 ± 12 | 4.0 ± 2.1 | 1100 ± 150 | 41 |
| L114R;L116R | (3) | 29 ± 8 | 4.3 ± 1.6 | 1200 ± 92 | 41 |
| F120A | (4) | 200 ± 24 | 29 ± 8.4 | 860 ± 43 | 4.3 |
| L114R;L116R;F120A | (2) | 270 ± 63 | 40 ± 14 | 850 ± 64 | 3.1 |
Value ± S.E. determined by nonlinear least squares fit to the Michaelis–Menten equation.
Relative to the Km of Ets-1(1–138).
Determined by dividing individual values.
Number of independent experiments performed in duplicate.
Similar value obtained by Waas and Dalby (2001)
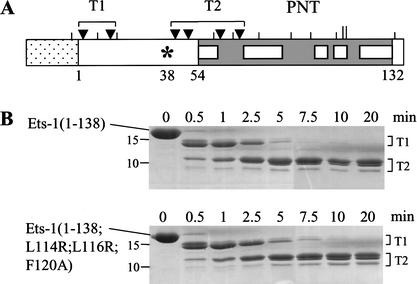
The location of L114, L116, and F120 on the surface of the Ets-1 PNT domain (Fig. 2B) suggests that these residues form an interface that contacts ERK2. Using partial trypsin proteolysis, we addressed an alternative possibility that single amino acid substitutions could affect docking by disrupting the structure of the PNT domain. Potential trypsin proteolysis sites exist throughout Ets-1(1–138) (Fig. 3A). However, the wild-type PNT domain was refractory to cleavage (Fig. 3A), presumably owing to the highly folded structure of the domain (Fig. 2B; Slupsky et al. 1998). Thus, mutations that severely disrupt the structure of the PNT domain are predicted to alter the pattern of protease sensitivity. The kinetics and pattern of partial trypsin proteolysis of Ets-1(1–138;L114R;L116R;F120A) were nearly identical to those of Ets-1(1–138) (Fig. 3B), suggesting that the mutations did not disrupt the fold or flexibility of the PNT domain. These data, combined with the NMR-based structure and kinase assays, suggest that L114, L116, and F120 comprise an interface that docks with ERK2.
Figure 3.
Trypsin sensitivity of Ets-1(1–138) fragments is unaffected by mutation of the docking site. (A) Schematic of Ets-1(1–138) depicting potential trypsin cleavage sites. Histidine tag (stippled), PNT domain with α-helices (gray with white boxes), potential trypsin cleavage sites (vertical lines). Ets-1(1–138;L114R;L116R;F120A) has two additional potential cleavage sites (L114R and L116R, longer lines). Arrowheads indicate residues, clustered as T1 and T2, that were cleaved by partial proteolysis of Ets-1(1–138) as determined by N-terminal sequencing. (B) Partial trypsin digest of Ets-1(1–138) (top) and Ets-1(1–138;L114R;L116R;F120A) (bottom). Proteins were incubated with trypsin for indicated times then electrophoresed on an 18% SDS-polyacrylamide gel. Digital image shows Coomassie Blue-stained gels with size standards noted (in kilodaltons). Clusters of cleavage sites (T1 and T2) in A are the proposed N termini of the indicated protein bands.
The docking site is conserved in the PNT domain of Ets-2
Ets-1 and Ets-2 are closely related proteins that conserve both the MAPK phosphorylation site and the PNT domain. The amino acid sequences spanning these functional elements are 70% identical, suggesting that Ets-1 and Ets-2 have similar three-dimensional structures in these regions. Notably, the residues comprising the ERK2 docking site in Ets-1 are conserved in Ets-2 (L148, L150, and F154; Fig. 2A), predicting that the PNT domain of Ets-2 also contains an ERK2 docking site.
To test for a functional ERK2 docking site, kinase assays were performed with a histidine-tagged fragment of Ets-2 encompassing amino acids 1–172 that includes the MAPK site (T72) and PNT domain (Fig. 1A). The Km and kcat/Km (a measure of a protein's ability to serve as a substrate) of this protein (Table 2) were similar to the values for Ets-1(1–138) (Table 1). Mutation of the proposed docking residues L148 and L150 to arginines resulted in a fivefold increase in Km (Table 2), akin to the effect of mutating L114 and L116 in Ets-1 (Table 1). Strikingly, mutation of the other proposed docking residue, F154, to alanine caused a 75-fold increase in Km (Table 2), and combination of the L148R, L150R, and F154A mutations resulted in a 96-fold increase in Km (Table 2). These values were accompanied by minimal changes in kcat, indicating disruption of a binding event. Indistinguishable trypsin proteolysis patterns were observed for Ets-2(1–172) and Ets-2(1–172;L148R;L150R;F154A) (data not shown), suggesting that the mutations do not significantly alter the structure of the Ets-2 PNT domain. These data show that the ERK2 docking site is conserved in Ets-2.
Table 2.
Kinetic analysis of Ets-2(1–172)
| Protein [Ets-2(1–172)]
|
Kma (μM)
|
Fold increase in Kmb
|
kcata (min−1)
|
kcat/Kmc (μM−1 min−1)
|
|---|---|---|---|---|
| Ets-2(1–172) | 2.8 ± 0.41 | — | 420 ± 19 | 150 |
| L148R;L150R | 15 ± 3.1 | 5.4 ± 1.4 | 720 ± 46 | 48 |
| F154A | 210 ± 37 | 75 ± 17 | 560 ± 38 | 2.7 |
| L148R;L150R; F154A | 270 ± 51 | 96 ± 23 | 530 ± 40 | 2.0 |
Value ± S.E. determined by nonlinear least squares fit of data from two independent experiments performed in duplicate to the Michaelis–Menten equation.
Relative to the Km of Ets-2(1–172).
Determined by dividing individual values.
The Drosophila protein Pnt-P2 is encoded by a gene that is an apparent ortholog of the genes encoding Ets-1 and Ets-2. Pnt-P2 conserves the Ras/MAPK-targeted phosphoacceptor found N-terminal to the PNT domain in Ets-1 and Ets-2 (Brunner et al. 1994; O'Neill et al. 1994). Additionally, the PNT domain of Pnt-P2 contains sequence closely resembling the ERK2 docking site of Ets-1 and Ets-2 (L228, I230, and F234; Fig. 2A). It is likely this sequence functions as a docking site for the Drosophila MAPK related to ERK2, Rolled.
The docking site is not conserved in the PNT domain of GABPα
The juxtaposition of the MAPK phosphorylation site and the PNT domain is not conserved in ETS proteins other than vertebrate Ets-1 and Ets-2 and Drosophila Pnt-P2. This suggests that the other PNT domains may not serve as ERK2 docking sites. Consistent with this hypothesis, the residues involved in docking are well-conserved in five vertebrate forms of Ets-1 (rat, mouse, chicken, frog, human) and Ets-2 (mouse, chicken, frog, human, cow) but not in the PNT domains of other ets family members (Fig. 2A). In GABPα, for instance, the two leucines of the docking site in Ets-1 are replaced by a phenylalanine and an arginine, and the loop between helix H4 and helix H5 is shortened and lacks the phenylalanine (Fig. 2A). Therefore, although GABPα is phosphorylated by MAPKs at other sites (Fromm and Burden 2001), we predicted that the PNT domain of GABPα would not contain an ERK2 docking site.
To test this prediction, the N-terminal extension of Ets-1 (residues 1–50) was fused to the PNT domain of GABPα to generate a chimeric substrate (Fig. 1A). In kinetic assays with ERK2, the Ets-1/GABPα chimera had a 56-fold higher Km (380 ± 81 μM) than that of Ets-1(1–138) (Table 1). Furthermore, kcat/Km was similar for the chimera (2.1 μM−1 min−1), Ets-1(1–52), and Ets1(1–138;L114R;L116R;F120A) (Fig. 1C; Table 1). These data indicate that the PNT domain of GABPα does not efficiently dock with ERK2 and supports the hypothesis that the LXLXXXF motif is critical for docking.
To more rigorously establish a role for the LXLXXXF sequence in docking, these residues were introduced into the GABPα PNT domain and tested for ERK2 binding by kinetic analysis. Leucine residues were inserted at the appropriate positions in the predicted helix H4 of the GABPα PNT domain, and a phenylalanine was introduced in the loop between helices H4 and H5 (Fig. 2A). A valine was also introduced immediately C-terminal of the phenylalanine to conserve the spacing of the loop (Fig. 2A). The Ets-1/GABPα chimera with the introduced docking residues had a 2.4-fold lower Km (160 ± 35 μM) than the chimera with the wild-type GABPα PNT domain. We conclude that introduction of these residues generated a docking site, albeit compromised relative to the site in Ets-1 and Ets-2. It is possible that the side chains of the introduced docking residues may not be oriented in the GABPα PNT domain in an optimal conformation for ERK2 interaction. Alternatively, additional residues may contribute to full docking site function in Ets-1 and Ets-2.
The docking site is required for Ras/MAPKpathway-mediated enhancement of transactivation by Ets-1 and Ets-2
Phosphorylation of Ets-1 and Ets-2 in vivo by the RAS/RAF/MEK/ERK pathway stimulates transactivation of genes regulated by Ras-responsive elements (RREs) (Yang et al. 1996). Enhancement of transactivation by Ets-1 and Ets-2 is reduced by ablating the MAPK phosphoacceptor site (Yang et al. 1996). Therefore, mutations in the docking site that decrease levels of phosphorylation are predicted to reduce Ras/MAPK-pathway-mediated enhancement of the transactivation activity of Ets-1 and Ets-2.
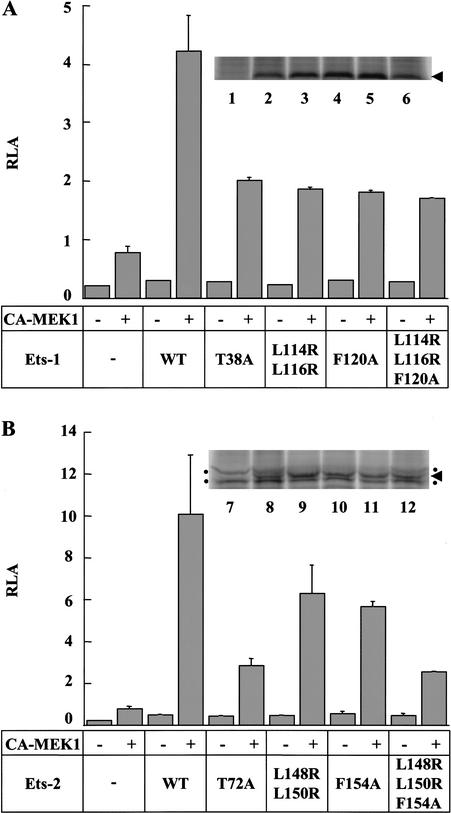
To test the in vivo function of the docking site, a transient expression reporter assay was established in NIH3T3 cells. The reporter was constructed by inserting the RRE from the promoter of the matrix metalloproteinase-9 gene, which has a composite ETS/AP-1 binding site (Watabe et al. 1998), upstream of the firefly luciferase gene. Plasmids encoding full-length Ets-1, Ets-2, or the phosphoacceptor mutants Ets-1(T38A) and Ets-2(T72A) were introduced into cells with or without a plasmid encoding a constitutively active form of MEK1 (CA-MEK1; Mansour et al. 1994). MEK1 specifically phosphorylates and activates ERK1 and ERK2, which are endogenous in NIH3T3 cells, but not other subfamilies of MAPKs (Chang and Karin 2001). The results for the wild-type and phosphoacceptor mutants are consistent with previous studies (Fig. 4A,B; Yang et al. 1996) that show cooperation between ETS proteins and the Ras signaling pathway in the stimulation of transcription and the requirement of T38 and T72 of Ets-1 and Ets-2, respectively, for enhancement of transactivation.
Figure 4.
Docking site mutants of Ets-1 and Ets-2 affect Ras-pathway-induced transcription. (A) Reporter assay with Ets-1 docking site mutants. NIH3T3 cells were transfected with a firefly luciferase gene reporter under the control of a MMP-9 Ras-responsive element (RRE) (2.5 μg) and the pRL-null internal control plasmid (0.5 μg). As indicated, a plasmid encoding a constitutively active form of MEK1 (CA-MEK1; 0.1 μg) and a plasmid encoding wild-type or mutant Ets-1 (0.1 μg) were cotransfected. Firefly luciferase activity was normalized to Renilla luciferase activity (relative luciferase activity, RLA). Bars depict the mean ± S.D. of a representative experiment with each assay performed in duplicate (Ets-1 and mutants thereof transfected without CA-MEK1 were performed once). The experiment was repeated four times with similar results. Inset shows protein expression levels as detected by immunoprecipitation from metabolically labeled, transfected cells: (lane 1) mock-transfected; (lane 2) FLAG-Ets-1; (lane 3) FLAG-Ets-1(T38A); (lane 4) FLAG-Ets-1(L114R;L116R); (lane 5) FLAG-Ets-1(F120A); (lane 6) FLAG-Ets-1(L114R;L116R;F120A). (B) Reporter assay with Ets-2 docking site mutants. Transfection as in A with plasmids encoding wild-type or mutant Ets-2 (0.1 μg). Inset shows protein expression levels as determined above; dots indicate background bands; arrowhead, ETS protein. (Lane 7) Mock-transfected; (lane 8) FLAG-Ets-2; (lane 9) FLAG-Ets-2(T72A); (lane 10) FLAG-Ets-2(L148R;L150R); (lane 11) FLAG-Ets-2(F154A); (lane 12) FLAG-Ets-2(L148R;L150R;F154A).
To assess the role of the docking site in transactivation, constructs expressing docking site mutants of full-length Ets-1 or Ets-2 were used in the transient expression assay. Strikingly, MEK1 stimulation of the transactivation activity of Ets-1(L114R;L116R), Ets-1(F120A), and Ets-1(L114R;L116R;F120A) was abrogated to the same degree as that of the phosphoacceptor mutant (Fig. 4A). The Ets-2 docking site mutants differed slightly in that they showed a graded ability to transactivate the reporter (Fig. 4B). The ability to transactivate was consistent with the graded pattern of increase in Km observed for proteins with the same mutations in kinase assays (Fig. 4B; Table 2). A parallel set of experiments performed with FLAG-epitope tagged ETS proteins displayed similar results except that both FLAG-tagged Ets-1 and Ets-2 docking site mutants showed a graded ability to transactivate the reporter (data not shown), similar to untagged Ets-2 mutants (Fig. 4B). Controls showed that protein levels in transfected NIH3T3 cells were similar for wild-type and mutant ETS proteins (Fig. 4A,B). The simplest interpretation of these data is that mutation of the docking site reduces phosphorylation of Ets-1 and Ets-2 at T38 or T72, respectively, resulting in lower levels of transactivation. We conclude that the docking site plays an important role in Ras pathway-dependent enhancement of transactivation by Ets-1 and Ets-2.
Discussion
Structural and mechanistic insights into ERK2 substrate docking
This report identifies three hydrophobic residues in the PNT domain that enhance phosphorylation of Ets-1 by ERK2. Several lines of evidence suggest that these residues form a hydrophobic ERK2 interaction surface, despite residing ∼80 amino acids C-terminal of the ERK2 phosphoacceptor site. First, in phosphorylation assays with ERK2, mutation of L114, L116, and F120 caused a 40-fold increase in Km. This increase was not accompanied by large changes in kcat, indicating that the mutations disrupted ERK2 binding rather than catalysis. Second, in the NMR solution structure, L114, L116, and F120 are clustered on the surface of the Ets-1 PNT domain, where they could simultaneously contact ERK2. Third, the triple-point mutant and the PNT-domain-deletion mutant had similar kcat/Km values, indicating an essential role for L114, L116, and F120 in ERK2 docking. Finally, Ras/MAPK-mediated enhancement of the transactivation activity of Ets-1 was reduced by mutation of the hydrophobic residues, showing that the docking site functions in vivo within the context of the full-length protein. Docking sites exist in many other proteins; however, this hydrophobic surface of the Ets-1 PNT domain is the first kinase docking interface in a transcription factor to be defined by high-resolution structural data.
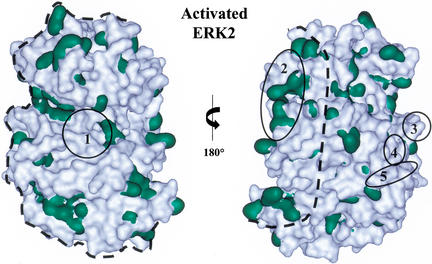
Using the available structures, we sought to model the interaction of dual-phosphorylated ERK2 (Canagarajah et al. 1997) with the phosphoacceptor and docking site of Ets-1 (Slupsky et al. 1998). Because of the highly flexible nature of the extension containing the phosphoacceptor in Ets-1, one specific interaction could not be modeled. Instead, we estimated the potential surfaces of ERK2 with which the Ets-1 docking site might interact, assuming that the phosphoacceptor and the docking site simultaneously contact ERK2. To delimit the range of potential interaction surfaces for the docking site, the phosphoacceptor was tethered in the ERK2 catalytic site, and the PNT domain was rotated around the surface of ERK2 (Fig. 5).
Figure 5.
Potential docking interfaces of ERK2. Potential docking interfaces of ERK2 as determined by molecular modeling (area within dashed line) on two views of the activated ERK2 structure (Canagarajah et al. 1997). Dual-phosphorylated ERK2 can dimerize with a Kd of 7.5 nM (Khokhlatchev et al. 1998); however, monomeric ERK2 was used for modeling because most ERK2 was estimated to be monomeric at the concentrations (0.1 nM) used in the kinase assays. Hydrophobic residues (Val, Ile, Leu, Trp, Phe, Ala, and Met) are shown in green. Surfaces involved in the catalytic site (1) and ERK2 dimer interface (2) are noted. The ED (TT) site (3), CD domain (4), and surface containing Y314 and Y315 (5) are implicated to dock with other proteins (Tanoue et al. 2000, 2001; Xu et al. 2001). The figure was generated using WebLab ViewerPro 4.0 (Accelrys, San Diego, CA).
Because the Ets-1 docking site is hydrophobic, it is assumed to interact with a hydrophobic surface of ERK2. Several hydrophobic surfaces lie within the determined range (Fig. 5). Interestingly, surfaces of ERK2 proposed to mediate interactions with other docking sites in phosphatases, upstream and downstream kinases, and the transcription factor Elk-1 do not lie within our estimated range (Fig. 5; Tanoue et al. 2000, 2001; Xu et al. 2001). Because known sites are not within our modeled range, we propose that the Ets-1 PNT domain interacts with a surface that has not been previously described as a docking interface on ERK2.
Kinetics and thermodynamics of docking
Direct binding of the Ets-1 PNT domain to ERK2 was not detected in several protein interaction assays (data not shown). Despite having micromolar Km values, ERK2 interactions with Ets-1 that precede phosphorylation or occur in the absence of ATP may have high dissociation rate constants, making them difficult to detect by conventional interaction assays. However, the interaction with ERK2 was detected on the affinity column using Ets-1(29–138) as a ligand (data not shown). The high concentration of ligand on the column matrix may have favored detection of this binding event. We do not suspect auxillary proteins are responsible for the interaction because purified, recombinant ERK2 also bound the column.
Despite an inability to detect direct binding, kinetic assays indicate that the docking site plays an important role in the efficiency of phosphorylation. This observation is supported by examining the thermodynamic contribution of the docking site to binding ERK2 during the phosphorylation reaction. Calculated based on kcat/Km values, the interaction of ERK2 with the Ets-1(1–138) fragment had a Gibbs standard free energy (ΔG°) of ∼−7.1 kcal/mole. In contrast, the ΔG° of the Ets-1 triple-mutant interaction with ERK2 was ∼−4.9 kcal/mole. Thus, the free energy contribution to binding (ΔΔG°) of the docking site was ∼2.2 kcal/mole, a significant portion (∼30%) of the overall binding energy, illustrating the importance of the docking site for Ets-1 phosphorylation by ERK2.
Comparisons to other docking sites
The ERK2 docking site in the Ets-1 and Ets-2 PNT domain (LXLXXXF) differs in sequence composition and position with respect to the phosphoacceptor compared with other MAPK docking sites. The critical role of the C-terminal phenylalanine in the Ets-1 and Ets-2 docking site distinguishes it from LXL docking motifs in the bZIP transcription factor c-Jun (Kallunki et al. 1996) and the ETS proteins Elk-1 and SAP-1 (Yang et al. 1998a,b; Galanis et al. 2001). Additionally, the Ets-1 docking site is ∼80 residues C-terminal of a single phosphoacceptor, whereas the LXL docking motifs of c-Jun, Elk-1, and SAP-1 lie N-terminal to multiple phosphoacceptors (Kallunki et al. 1996; Yang et al. 1998a,b; Galanis et al. 2001). The Ets-1 and Ets-2 docking site also is positioned differently than the FXF docking motif found in Elk-1 and SAP-1, as this site lies immediately C-terminal of a cluster of phosphoacceptors (Jacobs et al. 1999; Fantz et al. 2001; Galanis et al. 2001). Determination of the structural features of these docking sites would facilitate further functional and mechanistic comparisons.
The solution structure of the LXL-motif-containing ERK2 binding domain in the phosphatase MKP-3 was recently solved (Farooq et al. 2001). There are several differences in the structure and function of this domain compared to the PNT domain. First, the secondary structural elements and overall fold of this domain are distinct from the PNT domain. Second, the residues important for docking lie within different structural elements from those in the PNT domain. Third, the MKP-3 docking site is proposed to interact with ERK2 outside the range of interfaces determined in our Ets-1 modeling experiment. Therefore, the MKP-3 LXL motif appears to function in a structurally and mechanistically distinct fashion from that of the LXL motif in the Ets-1 PNT domain.
MAPK substrate specificity
The docking function of the Ets-1 and Ets-2 PNT domain is expected to play a role in the specificity of MAPK phosphorylation. Besides ERKs, there are two other main classes of MAPKs, the p38 and JNK subfamilies. Preliminary in vitro experiments indicate that Ets-1 is a substrate for the MAPK p38, but the phosphoacceptor is not T38 (data not shown). More importantly, mutation of the ERK2 docking site does not affect the efficiency of p38 phosphorylation. A JNK MAPK also phosphorylates Ets-2 at T72 (Smith et al. 2000); however, no kinetic analysis was performed. Further studies are required to understand the role of the docking site in determining the specificity of MAPK phosphorylation of Ets-1 and Ets-2.
Specificity of function among the ets family of transcription factors
Members of the ets family of transcription factors conserve the highly similar DNA-binding ETS domain, yet regulate diverse sets of genes (Graves and Petersen 1998). There are many molecular features that contribute to the unique functions of ETS proteins. For instance, the PNT domain is conserved among ∼40% of ETS proteins, but can perform distinct functions in different proteins. The PNT domain has roles in homotypic oligomerization in TEL (Jousset et al. 1997; Kim et al. 2001); heterotypic oligomerization in TEL, Yan, Mae, and Pnt-P2 (Potter et al. 2000; Baker et al. 2001); interaction with the SUMO conjugating enzyme Ubc9 in TEL (Chakrabarti et al. 1999, 2000); and binding transcriptional corepressors in TEL (Fenrick et al. 1999; Wang and Hiebert 2001). This study provides data that indicate a novel role of the PNT domain as an ERK2 docking site. This docking function is apparent in the PNT domains of Ets-1 and Ets-2, and presumably Drosophila Pnt-P2. However, the docking sequences are not conserved in other PNT domains, and the PNT domain of GABPα does not efficiently dock with ERK2 in kinase assays. We speculate that an ERK docking site evolved within an ancestral ortholog of the genes encoding Ets-1, Ets-2, and Pnt-P2. This site enhances phosphorylation of the N-terminal phosphoacceptor site, thereby lending a unique function to 2 of 25 ETS proteins in the human genome and 1 of 10 in the Drosophila genome.
The PNT domains of other proteins also are implicated in Ras/MAPK signaling. The PNT domain of the Drosophila protein Mae oligomerizes with the PNT domain of Yan and assists phosphorylation of Yan by the MAPK Rolled (Baker et al. 2001). It was speculated that the PNT domain of Mae interacts with Rolled; however, the Mae PNT domain lacks the docking site found in the PNT domain of Ets-1 and Ets-2 (Fig. 2A). This suggests that among ETS proteins the PNT domain may be able to regulate MAPK phosphorylation by multiple mechanisms.
In conclusion, our findings show how a structural domain, through changes in its surface residues, can establish varied functions among the ets gene family members. This diversification has facilitated the utility of different family members in regulating the complicated arrays of genomic information in higher eukaryotes. Therefore, this study sheds new light on the role of families of DNA-binding proteins in the metazoa.
Materials and methods
Plasmid construction
Bacterial expression plasmids encoding histidine-tagged murine Ets-1(1–138) (described previously by Slupsky et al. 1998), Ets-1(1–52), Ets-2(1–172), and Ets-1(1–50)/GABPα(167–254) were generated by PCR-amplification of the appropriate cDNA and subcloning into the NdeI and HindIII sites of pET28a(+) (Novagen). The mammalian expression plasmid pEVRFO-Ets-1 was described previously (Nelsen et al. 1993). The mammalian expression plasmid encoding Ets-2 was generated by subcloning the murine ets-2 open reading frame (ORF) into the BamHI site of the pEVRFO plasmid (Matthias et al. 1989). Site-directed mutagenesis was conducted using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Mammalian expression plasmids encoding FLAG-epitope-tagged ETS proteins were generated by PCR-amplification of the ORFs of pEVRFO plasmids encoding wild-type or mutant Ets-1 and Ets-2 followed by subcloning into the PstI and ApaI sites of pCMV-Tag2a (Stratagene). The luciferase reporter was constructed by inserting the Ras-responsive element spanning −527 to −544 of the matrix metalloproteinase (MMP)-9 gene (Watabe et al. 1998) into the MluI and NheI sites of the pGL3-basic plasmid (Promega) containing 72 bp of the rat prolactin minimal promoter (Ingraham et al. 1988).
Expression and purification of ETS proteins
Full-length murine Ets-1 was expressed and purified as previously described (Cowley and Graves 2000) with the addition of a final purification step. Following S-Sepharose chromatography, Ets-1 was dialyzed in Buffer A (25 mM Tris-HCl at pH 7.9, 10% [v/v] glycerol, 0.1 mM ethylenediaminetetraacetic acid [EDTA], 150 mM KCl, and 1 mM dithiothreitol [DTT]), and purified on a Sephacryl-200 size exclusion column (Amersham-Pharmacia) in the same buffer. Recombinant histidine-tagged ETS proteins were expressed in Escherichia coli BL21(DE3) cells. Proteins were purified at 4°C on a 5-mL HiTrap chelating column (Amersham-Pharmacia) and a 16/60 Superdex-75 size exclusion column (Amersham-Pharmacia) and were stored in Buffer A at −80°C in single use aliquots. The concentration of native proteins was determined by absorbance at 280 nm with calculated extinction coefficients (for Ets-1(1–138) and mutants thereof, ɛ280 = 23,240 M−1 cm−1, except for Ets-1(1–138;C112A), ɛ280 = 23,120 M−1 cm−1; for Ets-2(1–172) and mutants thereof, ɛ280 = 28,930 M−1 cm−1; Gill and von Hippel 1989). The concentration of Ets-1(1–52) was determined with Bradford reagent (Bio-Rad). Mass spectrometry and N-terminal sequencing of several mutants indicated that the N-terminal methionine of the histidine tag was cleaved (data not shown).
Expression and purification of activated ERK2
Histidine-tagged rat ERK2 was expressed in bacteria and phosphorylated in bacteria by a coexpressed, constitutively active mutant of human MEK1 (generous gift of Melanie Cobb; Khokhlatchev et al. 1997). Activated ERK2 was purified on a 1-mL HiTrap chelating column (Amersham-Pharmacia) and a HR5/5 Mono Q column. The most active fractions, as assessed by SDS-PAGE and immunoblotting with an antibody that recognizes dual-phosphorylated ERK2 (New England BioLabs), were dialyzed in 25 mM Tris-HCl (pH 7.5), 20% glycerol, 0.1 mM EDTA, 50 mM KCl, and 1 mM DTT, then diluted to 100 nM as determined by Bradford assay and stored at −80°C in single use aliquots.
Kinase assays
Kinase reactions were performed in buffer containing 25 mM Tris-HCl (pH 7.9), 87.5 mM KCl, 5% glycerol, 10 mM magnesium acetate, 0.5 mg/mL bovine serum albumin, 10 mM DTT, 2 mM ATP (Amersham-Pharmacia), 25 mCi/mmole [γ-32P]ATP (ICN), and 0.1 nM ERK2 in a total volume of 20 μL. ATP was in excess (>10× the estimated Km(ATP)). Reactions were performed at 30°C and were initiated by the addition of ATP. After 15 min, reactions were stopped by the addition of 3× SDS sample buffer and electrophoresed on an SDS-polyacrylamide gel. Gels were stained with Coomassie Blue, dried on Whatman paper, and exposed to a phosphor screen. Radioactivity was detected by PhosphorImaging (Storm 860, Molecular Dynamics) and quantified relative to an ATP standard curve using ImageQuant software (v. 5.0, Molecular Dynamics). Less than 15% of ETS protein was phosphorylated, and phosphate incorporation was linear during the experiment. Initial reaction velocities (v) were determined by dividing product concentration by the time of the reaction. Km, the Michaelis constant, and kcat, the turnover number, were determined by fitting v/[E]0 (where [E]0 is the total ERK2 concentration) from two to five independent experiments performed in duplicate to the Michaelis–Menten equation {v/[E]0 = kcat[S]/(Km + [S])}, where [S] is the ETS protein concentration, by nonlinear least squares analysis (Kaleidagraph v. 3.0).
Transient transfection and luciferase assay
NIH3T3 cells (generous gift of Peggy Farnham, University of Wisconsin, Madison) were grown in DMEM (Dulbecco modified Eagle medium) with 10% bovine calf serum (BCS). Cells (1.5 × 105) were plated in 35-mm tissue culture dishes and were transfected with calcium phosphate precipitates of plasmids encoding wild-type or mutant versions of Ets-1 and Ets-2, a plasmid encoding a constitutively active version of MEK1 (ΔN3;S218E;S222D; Mansour et al. 1994), an internal control plasmid pRL-null (Promega), and the MMP-9 luciferase reporter. Twenty to twenty-four hours after transfection the media was changed to DMEM containing 0.5% BCS and the cells were incubated for an additional 20–24 h. Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) on a Dynex Technologies, MLX Microtiter Plate Luminometer. Firefly luciferase activity was normalized to Renilla luciferase activity. Each transfection was performed in duplicate and the relative luciferase activity (RLA) was reported. The experiment was performed twice with ETS protein lacking FLAG tags and twice with FLAG-tagged ETS proteins; presence of the FLAG tag did not significantly affect results.
To detect expression of ETS proteins, 5 × 105 NIH3T3 cells plated in 60-mm tissue culture dishes were transfected with calcium phosphate precipitates of 20 μg of expression plasmid encoding wild-type or mutant FLAG-epitope-tagged ETS protein. Twenty hours following transfection, cells were metabolically labeled for 15 h in 1.5 mL of DMEM (minus methionine) containing 0.5% BCS, 2 mM glutamine, and 150 μCi of [35S]methionine (Amersham Pharmacia). Cells were lysed in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton-X 100, and the complete, miniprotease inhibitor cocktail (Roche). FLAG-tagged ETS proteins were immunoprecipitated with ANTI-FLAG M2 Affinity Gel (Sigma) in Tris-buffered saline. Proteins were eluted by the addition of 3× SDS sample buffer and heating at >95°C for 10 min then electrophoresed on a 10% SDS-polyacrylamide gel. Radioactive proteins on dried gels were detected by PhosphorImaging and quantified relative to background bands using ImageQuant software.
Partial trypsin proteolysis sensitivity assays
Ets-1(1–138) or Ets-1(1–138;L114R;L116R;F120A) (110 μg each) was incubated with 5.5 μg of trypsin in a total reaction volume of 55 μL at 22.5°C. Aliquots of 5 μL (10 μg of total ETS protein) were taken at various time intervals, added to 3× SDS sample buffer, and heated at >95°C for 5 min to stop the proteolysis reaction. Proteolyzed fragments were electrophoresed on an 18% SDS-polyacrylamide gel and were detected by Coomassie Blue staining. To identify sites of proteolysis, a similar assay was performed, and the pool of fragments was transferred to PVDF for N-terminal sequencing.
Acknowledgments
We thank Charles Foulds for the MMP-9 reporter plasmid, those cited within the text for reagents, and Frank Whitby for assistance with molecular modeling. We also thank members of the Graves lab for advice and Janet Lindsley, Lawrence McIntosh, and David Virshup for critical reading of the manuscript. The National Institutes of Health supported this work (GM38663 to B.J.G., predoctoral fellowship CA09602 to J.J.S., and CA24014 to Huntsman Cancer Center for support of core facilities). Support from the Huntsman Cancer Foundation is also gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL barbara.graves@hci.utah.edu; FAX (801) 585-1980.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.950902.
References
- Baker DA, Mille-Baker B, Wainwright SM, Ish-Horowicz D, Dibb NJ. Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila. Nature. 2001;411:330–334. doi: 10.1038/35077122. [DOI] [PubMed] [Google Scholar]
- Bartel FO, Higuchi T, Spyropoulos DD. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19:6443–6654. doi: 10.1038/sj.onc.1204038. [DOI] [PubMed] [Google Scholar]
- Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- Chakrabarti SR, Sood R, Ganguly S, Bohlander S, Shen Z, Nucifora G. Modulation of TEL transcription activity by interaction with the ubiquitin-conjugating enzyme UBC9. Proc Natl Acad Sci. 1999;96:7467–7472. doi: 10.1073/pnas.96.13.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti SR, Sood R, Nandi S, Nucifora G. Posttranslational modification of TEL and TEL/AML1 by SUMO-1 and cell-cycle-dependent assembly into nuclear bodies. Proc Natl Acad Sci. 2000;97:13281–13285. doi: 10.1073/pnas.240315897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Cowley DO, Graves BJ. Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes & Dev. 2000;14:366–376. [PMC free article] [PubMed] [Google Scholar]
- Fantz DA, Jacobs D, Glossip D, Kornfeld K. Docking sites on substrate proteins direct extracellular signal-regulated kinase to phosphorylate specific residues. J Biol Chem. 2001;276:27256–27265. doi: 10.1074/jbc.M102512200. [DOI] [PubMed] [Google Scholar]
- Farooq A, Chaturvedi G, Mujtaba S, Plotnikova O, Zeng L, Dhalluin C, Ashton R, Zhou MM. Solution structure of ERK2 binding domain of MAPK phosphatase MKP-3: Structural insights into MKP-3 activation by ERK2. Mol Cell. 2001;7:387–399. doi: 10.1016/s1097-2765(01)00186-1. [DOI] [PubMed] [Google Scholar]
- Fenrick R, Amann JM, Lutterbach B, Wang L, Westendorf JJ, Downing JR, Hiebert SW. Both TEL and AML-1 contribute repression domains to the t(12;21) fusion protein. Mol Cell Biol. 1999;19:6566–6574. doi: 10.1128/mcb.19.10.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm L, Burden SJ. Neuregulin-1-stimulated phosphorylation of GABP in skeletal muscle cells. Biochemistry. 2001;40:5306–5312. doi: 10.1021/bi002649m. [DOI] [PubMed] [Google Scholar]
- Galanis A, Yang SH, Sharrocks AD. Selective targeting of MAPKs to the ETS domain transcription factor SAP-1. J Biol Chem. 2001;276:965–973. doi: 10.1074/jbc.M007697200. [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Golub TR, Barker GF, Lovett M, Gilliland DG. Fusion of PDGF receptor β to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- Golub TR, Barker GF, Bohlander SK, Hiebert SW, Ward DC, Bray-Ward P, Morgan E, Raimondi SC, Rowley JD, Gilliland DG. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Goga A, Barker GF, Afar DEH, McLaughlin J, Bohlander SK, Rowley JD, Witte ON, Gilliland DG. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol Cell Biol. 1996;16:4107–4116. doi: 10.1128/mcb.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez FA, Raden DL, Davis RJ. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
- Graves BJ, Petersen JM. Specificity within the ets family of transcription factors. In: Woude GV, Klein G, editors. Advances in cancer research. San Diego, CA: Academic Press; 1998. pp. 1–55. [DOI] [PubMed] [Google Scholar]
- Hart AH, Reventar R, Bernstein A. Genetic analysis of ETS genes in C. elegans. Oncogene. 2000;19:6400–6408. doi: 10.1038/sj.onc.1204040. [DOI] [PubMed] [Google Scholar]
- Holland PM, Cooper JA. Protein modification: Docking sites for kinases. Curr Biol. 1999;9:329–331. doi: 10.1016/s0960-9822(99)80205-x. [DOI] [PubMed] [Google Scholar]
- Hsu T, Schulz RA. Sequence and functional properties of Ets genes in the model organism Drosophila. Oncogene. 2000;19:6409–6416. doi: 10.1038/sj.onc.1204033. [DOI] [PubMed] [Google Scholar]
- Ingraham HA, Chen RP, Mangalam HJ, Elsholtz HP, Flynn SE, Lin CR, Simmons DM, Swanson L, Rosenfeld MG. A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell. 1988;55:519–529. doi: 10.1016/0092-8674(88)90038-4. [DOI] [PubMed] [Google Scholar]
- Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes & Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- Jousset C, Carron C, Boureux A, Quang CT, Oury C, Dusanter-Fourt I, Charon M, Levin J, Bernard O, Ghysdael J. A domain of TEL conserved in a subset of ETS proteins defines a specific oligomerization interface essential to the mitogenic properties of the TEL-PDGFRβ oncoprotein. EMBO J. 1997;16:69–82. doi: 10.1093/emboj/16.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- Khokhlatchev A, Xu S, English J, Wu P, Schaefer E, Cobb MH. Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. Efficient synthesis of active protein kinases. J Biol Chem. 1997;272:11057–11062. doi: 10.1074/jbc.272.17.11057. [DOI] [PubMed] [Google Scholar]
- Khokhlatchev AV, Canagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb MH. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, Faham S, Bowie JU. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20:4173–4182. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, et al. A TEL–JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, Vande Woude GF, Ahn NG. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- Matthias P, Muller MM, Schreiber E, Rusconi S, Schaffner W. Eukaryotic expression vectors for the analysis of mutant proteins. Nucleic Acids Res. 1989;17:6418. doi: 10.1093/nar/17.15.6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen B, Gang T, Erman B, Gregoire J, Maki R, Graves B, Sen R. Regulation of lymphoid-specific immunoglobulin μ heavy chain gene enhancer by ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- O'Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Potter MD, Buijs A, Kreider B, van Rompaey L, Grosveld GC. Identification and characterization of a new human ETS-family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood. 2000;95:3341–3348. [PubMed] [Google Scholar]
- Robinson MJ, Harkins PC, Zhang J, Baer R, Haycock JW, Cobb MH, Goldsmith EJ. Mutation of position 52 in ERK2 creates a nonproductive binding mode for adenosine 5′-triphosphate. Biochemistry. 1996;35:5641–5646. doi: 10.1021/bi952723e. [DOI] [PubMed] [Google Scholar]
- Schneikert J, Lutz Y, Wasylyk B. Two independent activation domains in c-Ets-1 and c-Ets-2 located in non-conserved sequences of the ets gene family. Oncogene. 1992;7:249–256. [PubMed] [Google Scholar]
- Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD, Yang SH, Galanis A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- Slupsky CM, Gentile LN, Donaldson LW, Mackereth CD, Seidel JJ, Graves BJ, McIntosh LP. Structure of the Ets-1 pointed domain and mitogen-activated protein kinase phosphorylation site. Proc Natl Acad Sci. 1998;95:12129–12134. doi: 10.1073/pnas.95.21.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Schaffner AE, Hofmeister JK, Hartman M, Wei G, Forsthoefel D, Hume DA, Ostrowski MC. ets-2 is a target for an Akt (protein kinase B)/jun N-terminal kinase signaling pathway in macrophages of motheaten-viable mutant mice. Mol Cell Biol. 2000;20:8026–8034. doi: 10.1128/mcb.20.21.8026-8034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton D, Balan I, Pawson T, Sicheri F. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat Struct Biol. 1999;6:44–49. doi: 10.1038/4917. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Maeda R, Adachi M, Nishida E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waas WF, Dalby KN. Purification of a model substrate for transcription factor phosphorylation by erk2. Protein Expr Purif. 2001;23:191–197. doi: 10.1006/prep.2001.1491. [DOI] [PubMed] [Google Scholar]
- Wang L, Hiebert SW. TEL contacts multiple co-repressors and specifically associates with histone deacetylase-3. Oncogene. 2001;20:3716–3725. doi: 10.1038/sj.onc.1204479. [DOI] [PubMed] [Google Scholar]
- Watabe T, Yoshida K, Shindoh M, Kaya M, Fujikawa K, Sato H, Seiki M, Ishii S, Fujinaga K. The Ets-1 and Ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. Int J Cancer. 1998;77:128–137. doi: 10.1002/(sici)1097-0215(19980703)77:1<128::aid-ijc20>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Xu B, Stippec S, Robinson FL, Cobb MH. Hydrophobic as well as charged residues in both MEK1 and ERK2 are important for their proper docking. J Biol Chem. 2001;276:26509–26515. doi: 10.1074/jbc.M102769200. [DOI] [PubMed] [Google Scholar]
- Yang B-S, Hauser CA, Henkel G, Colman MS, Van Beveren C, Stacey KJ, Hume DA, Maki RA, Ostrowski MC. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets-1 and c-Ets-2. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Whitmarsh AJ, Davis RJ, Sharrocks AD. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998a;17:1740–1749. doi: 10.1093/emboj/17.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol Cell Biol. 1998b;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy JS, Muise-Helmericks RC. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]