Abstract
Budding yeast centromeres are comprised of ∼125-bp DNA sequences that direct formation of the kinetochore, a specialized chromatin structure that mediates spindle attachment to chromosomes. We report here a novel role for the histone deposition complex chromatin assembly factor I (CAF-I) in building centromeric chromatin. The contribution of CAF-I to kinetochore function overlaps that of the Hir proteins, which have also been implicated in nucleosome formation and heterochromatic gene silencing. cacΔ hirΔ double mutant cells lacking both CAF-I and Hir proteins are delayed in anaphase entry in a spindle assembly checkpoint-dependent manner. Further, cacΔ and hirΔ deletions together cause increased rates of chromosome missegregation, genetic synergies with mutations in kinetochore protein genes, and alterations in centromeric chromatin structure. Finally, CAF-I subunits and Hir1 are enriched at centromeres, indicating that these proteins make a direct contribution to centromeric chromatin structures.
Keywords: Centromere, kinetochore, histone, yeast, checkpoint, chromatin
High-fidelity chromosome segregation during cell division is required to ensure that daughter cells each receive a complete copy of the genome. Chromosome segregation is a closely monitored process (Amon 1999; Hoyt 2000); loss of proteins that monitor segregation is linked to loss of growth control in human cells (Cahill et al. 1998) and inviability in nematodes (Kitagawa and Rose 1999). Proper segregation of eukaryotic chromosomes requires centromeres, chromosomal domains that are the sites of microtubule attachment during mitosis and meiosis. Kinetochores are protein complexes that assemble onto centromeric loci and mediate spindle attachment to chromosomes. Each time a chromosome is replicated, this kinetochore protein complex must be assembled onto both daughter chromosomes to achieve bipolar attachment of sister chromatids. How centromeres direct kinetochore assembly and propagation is poorly understood, but is thought to occur by a chromatin-mediated epigenetic mechanism (for reviews, see Karpen and Allshire 1997; Wiens and Sorger 1998; Maggert and Karpen 2000; Sullivan 2001).
In the budding yeast Saccharomyces cerevisiae, centromere DNA sequences are 125 bp in length and contain three conserved elements (CDEI, II, III) that are present at all centromeres (Hyman and Sorger 1995). Despite these limited primary DNA sequence determinants, a specialized chromatin structure extends for several kilobases on either side of the core centromere region (Bloom and Carbon 1982). This specialized structure is apparently required for proper centromere function: mutations within histone proteins (Smith et al. 1996; Pinto and Winston 2000) or nucleosome remodeling enzymes (Tsuchiya et al. 1998) cause aberrations in this structure and reduce the fidelity of chromosome segregation.
In contrast to the limited primary sequence requirements for centromere function in S. cerevisiae, centromeres of multicellular organisms and fission yeast extend for many kilobases and are largely comprised of repetitive DNA (Chikashige et al. 1989; Hahnenberger et al. 1991; Sun et al. 1997). Despite these differences, specialized chromatin structures are important for centromere function in all organisms. Position-dependent gene silencing occurs within centric heterochromatin in fission yeast and Drosophila (Reuter and Spierer 1992; Grewal 2000). Silencing at fission yeast centromeres depends on many of the same proteins required for silencing the cryptic mating loci and telomere-proximal genes (Allshire et al. 1995). Mutations and drug treatments that reduce silencing at centromeric regions also decrease the fidelity of chromosome segregation (Allshire et al. 1995; Ekwall et al. 1997, 1999), thus demonstrating that heterochromatin structure contributes to centromere function.
The fundamental repeated structure of chromosomes is the nucleosome. Nucleosomes are assembled by ordered deposition of histones, mediated by histone-binding assembly factors. For example, chromatin assembly factor I (CAF-I) is a nuclear heterotrimeric protein complex that is conserved throughout the eukaryotes (Kaufman et al. 1995, 1997; Kamakaka et al. 1996; Kaya et al. 2001; Quivy et al. 2001). CAF-I performs the first step of nucleosome formation, delivery of histones H3/H4 to newly replicated DNA (Smith and Stillman 1991; Verreault et al. 1996). Consistent with the biochemical analyses of CAF-I, budding yeast cells lacking any or all of the three genes encoding CAF-I subunits (CAC1, CAC2, and CAC3) display reduced position-dependent gene silencing at telomeres (Enomoto et al. 1997; Kaufman et al. 1997; Monson et al. 1997), the silent mating loci (Enomoto and Berman 1998; Kaufman et al. 1998), and at ribosomal DNA (Smith et al. 1999). Gene silencing at these loci in yeast is a chromatin-mediated process analogous to heterochromatic silencing in multicellular organisms: mutation of histone termini (Kayne et al. 1988; Megee et al. 1990), histone-binding heterochromatin proteins (Hecht et al. 1995), or other genes that modulate the posttranslational modification of histone termini (Rundlett et al. 1996) affect the magnitude of this silencing.
The histone regulatory (HIR) genes HIR1, HIR2, HIR3, and HPC2 (Osley and Lycan 1987; Xu et al. 1992) encode proteins implicated in a nucleosome assembly pathway that functionally overlaps CAF-I (Sharp et al. 2001). Mutations in HIR genes have minor effects on silencing at telomeres and the silent mating loci (Kaufman et al. 1998), and growth of S. cerevisiae cells is not affected when either CAC genes encoding subunits of CAF-I or HIR genes are mutated. However, cacΔ hirΔ double mutant cells display synergistic reduction of position-dependent gene silencing at both telomeres and the silent mating loci, and exhibit slow growth (Kaufman et al. 1998; Qian et al. 1998). These phenotypes occur regardless of which of the three cacΔ or four hirΔ gene deletions are combined (Kaufman et al. 1998; Qian et al. 1998) Thus, Hir proteins become important for normal growth and silencing in the absence of CAF-I, indicating functional overlap between these two groups of proteins.
In this work, we show that histone-binding and -deposition proteins responsible for heterochromatin formation are also required for proper centromere function in budding yeast. We discovered this because budding yeast cells lacking both CAF-I and Hir proteins experienced a delay specifically during the G2/M phase of the cell cycle that resulted in part from activation of the spindle assembly checkpoint. cacΔ hirΔ double mutant cells displayed rates of chromosome loss and nondisjunction approximately two orders of magnitude greater than wild-type cells or cacΔ or hirΔ single mutant cells. cacΔ and hirΔ deletions together showed strong genetic interactions with conditional alleles of genes encoding kinetochore proteins. We show localization of CAF-I subunits and Hir1 protein to centromeric chromatin; in the absence of these proteins, centromeric chromatin structure is altered. Together, these data show that CAF-I and Hir proteins have overlapping roles in maintaining a centromeric chromatin structure compatible with kinetochore function.
Results
G2/M delay and mitotic checkpoint activation in cacΔ hirΔ cells
SIR (silent information regulator) genes are required for formation of silent chromatin structures at telomeres and cryptic mating (HM) loci, but are dispensable for growth (Loo and Rine 1995). CAC and HIR genes also contribute to the stability of heterochromatic gene silencing, yet in contrast to sirΔ mutants, cacΔ hirΔ double mutants display marked growth defects (Kaufman et al. 1998; Qian et al. 1998). Therefore, CAF-I and Hir proteins have roles outside of known regions of silent chromatin. We investigated the growth defect of cacΔ hirΔ cells to gain insight into these other functions.
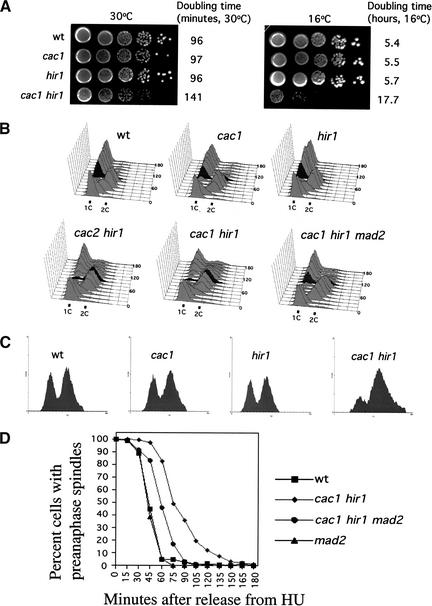
The slow-growth phenotype of cacΔ hirΔ cells is apparent in early cell divisions after completion of meiosis (Kaufman et al. 1998). This defect persists throughout subsequent mitotic divisions, resulting in the formation of small colonies at both physiological (30°C) and cold (16°C) temperatures (Fig. 1A). At 30°C, cac1Δ hir1Δ cells displayed a doubling time in liquid culture ∼45 min longer than that observed for wild-type cells or cac1Δ or hir1Δ single mutants. At 16°C, this effect was greatly exacerbated; cac1Δ hir1Δ cells required nearly 18 h to complete a cell cycle, compared with 5.4 h for wild-type cells.
Figure 1.
cacΔ hirΔ cells activate the MAD2-dependent spindle assembly checkpoint. (A) Slow growth of cac1Δ hir1Δ cells. Yeast strains PKY346 (wt), PKY1100 (cac1Δ), PKY1154 (hir1Δ), and PKY1168 (cac1Δ hir1Δ) were grown to log phase at 30°C, serially diluted, and grown on YPAD agar at 30°C for 2 d or 16°C for 12 d. Cell doubling time in liquid media for each strain is indicated to the right. (B) cacΔ hirΔ cells experience a MAD2-dependent G2/M phase delay. Yeast cells were grown to log phase at 30°C and arrested in α-factor (10 μg/mL) for 3 h. Cells were washed and released into pheromone-free media at 30°C. Aliquots of cultures were removed at 15-min intervals for a period of 3 h. α-Factor was added back to cultures at 55 min after the initial release to capture cells in the subsequent G1 phase of the cell cycle. DNA content was measured by FACS. Selected time points are marked for clarity. Strains are as in A, with the addition of PKY1340 (cac2Δ hir1Δ) and PKY1257 (cac1Δ hir1Δ mad2Δ). (C) Pronounced G2/M delay in cac1Δ hir1Δ cells at 16°C. Strains are as in A. Yeast cells were grown to log phase at 30°C and synchronized by arrest in 0.2 M hydroxyurea (HU) for 2 h. Cells were washed and released into 16°C media for 24 h. DNA content was measured by FACS. (D) Kinetics of spindle elongation. Yeast cells were grown to log phase at 30°C, arrested in α-factor for 3 h, washed, and released into media containing 0.2 M HU for 2 h at 30°C. Cells were then released from the HU block at 30°C. α-Factor was added back to cultures at 30 min after HU release to capture cells in the subsequent G1 phase of the cell cycle. Aliquots of cultures were taken at 15-min intervals for 3 h after release from HU and processed for tubulin (YOL134) and DNA (DAPI) staining of cells. Approximately 200 cells at each time point were then scored for spindle length and nuclear morphology.Shown at each time point are the percentage of cells in the population with short, bipolar spindles associated with an undivided nucleus at the bud neck. Yeast strains used were PKY346, wt; PKY1168, cac1Δ hir1Δ; PKY1257, cac1Δ hir1Δ mad2Δ; PKY1265, mad2Δ.
To determine whether the slow growth of cacΔ hirΔ cells reflected a delay in any particular stage in the cell cycle, we performed flow cytometric analysis of cells grown at either 30°C or 16°C (Fig. 1B,C). Haploid cells grown at 30°C were arrested in G1 by treatment with the mating pheromone α-factor, and released into pheromone-free media for a single synchronous cell cycle. The cell cycle profiles of cac1Δ and hir1Δ cells were very similar to wild-type cells. In these strains, the majority of cells completed mitosis 90 min after α-factor release (marked in dark gray) as indicated by the return to 1C DNA content. In contrast, in cac1Δ hir1Δ and cac2Δ hir1Δ double mutant cells, mitotic exit was not completed until 135–150 min after α-factor release, indicating an ∼45-min elongation of the G2/M stage of the cell cycle. Consistent with the results at 30°C, asynchronous cultures of cac1Δ hir1Δ cells grown at 16°C displayed a greater percentage of cells with 2C DNA content compared to wild-type, cac1Δ, or hir1Δ cells (Fig. 1C). Therefore, a G2/M delay occurred at both growth temperatures analyzed. Furthermore, microscopic examination of cells grown at either 30°C or 16°C showed that cac1Δ hir1Δ cells accumulated with a single nucleus and short bipolar spindle near the bud neck, a morphology consistent with a delay prior to anaphase (Fig. 1D; data not shown). These mitotic delay phenotypes were observed for all cacΔ hirΔ double mutant combinations tested. We conclude that cells require CAF-1 and Hir protein function to traverse mitosis in an efficient manner.
Checkpoints are surveillance mechanisms that impede cell cycle progression in the presence of damaged molecules or insufficient preparation for subsequent events (Hartwell and Weinert 1989). At the metaphase-to-anaphase transition, two prominent checkpoints monitor chromosomal events. The MEC1-dependent DNA damage checkpoint monitors the presence of chromosomal lesions such as DNA strand breaks or stalled replication forks (Weinert et al. 1994), and the MAD2-dependent spindle assembly checkpoint monitors the presence of unattached kinetochores (Amon 1999). We hypothesized that one of these checkpoints was involved in mediating the cell cycle delay of cac1Δ hir1Δ cells. To test this, we determined the effects of deleting checkpoint genes on cell cycle progression. Cell cycle profiles of cac1Δ hir1Δ mec1Δ cells were identical to those of cac1Δ hir1Δ cells, indicating that the DNA damage checkpoint was not responsible for the cell cycle delay; identical results were obtained upon deletion of the DNA damage checkpoint genes RAD53 or RAD9 (data not shown). In contrast, the cell cycle profile of cac1Δ hir1Δ mad2Δ cells revealed a partial relief of the cac1Δ hir1Δ mitotic delay (Fig. 1B): a significantly greater percentage of cac1Δ hir1Δ mad2Δ cells completed mitosis 105 min (marked in dark gray) after α-factor release than did cac1Δ hir1Δ cells at the same time point. Therefore, cac1Δ hir1Δ cells activated the spindle assembly checkpoint.
To confirm that removal of the spindle checkpoint in cac1Δ hir1Δ cells relieved the block to anaphase onset, the kinetics of spindle elongation after release from an S-phase arrest were measured (Fig. 1D). Consistent with the flow cytometry data, cac1Δ hir1Δ cells experienced a 45–60-min delay in the onset of anaphase as determined by the percentage of cells in the population with preanaphase spindles. Furthermore, cac1Δ hir1Δ mad2Δ cells entered anaphase at a rate intermediate between that of wild-type and cac1Δ hir1Δ cells, indicating that MAD2 acts to regulate the rate of anaphase entry in cac1Δ hir1Δ cells. Together, the flow cytometry and spindle elongation data indicated that spindle assembly checkpoint activation contributed to the G2/M delay in cacΔ hirΔ cells. These observations suggested that attachment of chromosomes to the mitotic spindle was defective in cacΔ hirΔ cells.
Increased chromosome missegregation in cacΔ hirΔ cells
We hypothesized that cacΔ hirΔ double mutant cells activate the spindle assembly checkpoint because of malformation of centromeric chromatin, resulting in improper centromere/kinetochore function. To test whether kinetochore function was impaired, the fidelity of chromosome segregation was measured in cac1Δ hir1Δ cells. Yeast strains disomic for a fragment of Chromosome III were monitored for chromosome loss (1:0) and nondisjunction (2:0) events using a colony color assay to measure the chromosome missegregation rate per cell division (Shero et al. 1991). Chromosome missegregation frequencies for yeast strains grown at 30°C are shown in Table 1. In our wild-type strain, the reporter minichromosome was missegregated at a rate of <1 per 2 × 105 cell divisions. Whereas cac1Δ and hir1Δ cells showed no dramatic increases in chromosome missegregation relative to wild-type cells, cac1Δ hir1Δ cells experienced a dramatic increase in both chromosome loss and nondisjunction rates (97-fold and 78-fold, respectively) relative to the wild-type strain. These missegregation rates are comparable to those reported for mutations (ctf13-30 and ctf19Δ) in other kinetochore component genes (Doheny et al. 1993; Hyland et al. 1999).
Table 1.
Chromosome fragment missegregation
| Strain
|
Relevant genotype
|
Chromosome loss (1:0 segregation; CHL)
|
Nondisunction (2:0 segregation; NDJ)
|
Total missegregation (CHL + NDJ)
|
|---|---|---|---|---|
| PKY847 | Wild type | 1.9 × 10−5(1.0) | <1.9 × 10−5(1.0) | 1.9 × 10−5(1.0) |
| PKY2216 | cac1Δ | 4.3 × 10−5(2.2) | <2.1 × 10−5(<1.1) | 4.3 × 10−5(2.2) |
| PKY865 | hir1Δ | 1.9 × 10−5(1.0) | <1.9 × 10−5(<1.0) | 1.9 × 10−5(1.0) |
| PKY2217 | cac1Δhir1Δ | 1.9 × 10−3(97) | 1.5 × 10−3(78) | 3.4 × 10−3(180) |
| PKY1531 | mad2Δ | 1.9 × 10−4(9.6) | <3.7 × 10−5(<1.9) | 1.9 × 10−4(9.6) |
| PKY2218 | cac1Δhir1Δmad2Δ | 8.5 × 10−3(440) | 5.6 × 10−3(290) | 1.4 × 10−2(730) |
Chromosome loss and nondisjunction events at 30°C were scored in haploid strains bearing a nonessential chromosome fragment. The chromosome fragment CFIII(D8B.d) consists of CEN6, ∼125 kb of the left arm of Chromosome III, and the LEU2 and ade3-2p genes. Chromosome missegregation events were determined by the half-sector colony color assay (Materials and Methods). Numbers in bold indicate fold differences relative to the wild-type strain.
Consistent with previous reports, mad2Δ cells displayed a modest increase in chromosome loss rates (Li and Murray 1991). However, deletion of MAD2 in cac1Δ hir1Δ cells resulted in synergistic increases in chromosome missegregation rates: loss rates increased to 440-fold and nondisjunction rates increased to 290-fold above wild-type rates (Table 1). These data are reminiscent of the synergistic increase in chromosome missegregation observed upon combination of a mad2-1 mutation and a crippled CDEII element (Pangilinan and Spencer 1996), and strongly suggest the presence of compromised kinetochore structures in cac1Δ hir1Δ cells.
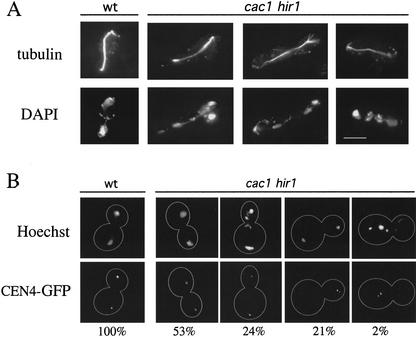
Microscopic examination showed that a subset of late anaphase cac1Δ hir1Δ cells grown at 16°C displayed severe chromosome missegregation events (Fig. 2A). Cells with aberrant DNA morphology often had fragmented nuclear bodies, resulting in unequal DNA staining between mother and bud and the presence of lagging chromosomes at the bud neck. These aberrant events were observed in 32% of cac1Δ hir1Δ anaphase cells at 16°C, compared with a frequency of <1% for wild-type, cac1Δ, and hir1Δ cells. These defects were prevalent in cacΔ hirΔ cells grown at 16°C but not at 30°C, consistent with the more severe growth rate and cell cycle phenotypes at this temperature (Fig. 1A,C). To directly examine the fate of individual chromosomes under these conditions, we visualized the centromere of Chromosome IV containing an array of lacO sites adjacent to CEN4 in cells that express a GFP–LacI fusion protein (Straight et al. 1996; Biggins et al. 1999). These cells allowed for direct observation of chromosome missegregation events, which were detected in cacΔ hirΔ but not wild-type or single mutant cells (Fig. 2B). After 24 h at 16°C, 24% of cac1Δ hir1Δ cells segregated CEN4 normally but displayed gross missegregation of other chromosomes as observed by Hoechst dye staining, 21% missegregated CEN4 but had a normal pattern of total DNA segregation, and 2% displayed defects in both CEN4 and total DNA segregation. We conclude that cells require the action of CAF-I and Hir proteins to maintain high-fidelity chromosome segregation during mitosis, and that kinetochore function becomes cold-sensitive in the absence of these proteins.
Figure 2.
Chromosome missegregation in cac1Δ hir1Δ cells. (A) Aberrant DNA segregation in cac1Δ hir1Δ cells at 16°C. Cells treated as in Figure 1C were stained for tubulin (YOL134) and DNA (DAPI). Yeast strains are PKY346 (wild type) and PKY1168 (cac1Δ hir1Δ). Bar, 5 μm. (B) Localization of centromere DNA. After growth at 16°C for 24 h, fixed cells were stained with Hoechst dye to visualize DNA, and CEN4 DNA was visualized using a lacO array/lacI-GFP system. For each genotype, the percentages of mid-to-late anaphase cells with the morphologies shown are indicated below the photographs. Yeast strains used were SBY214, wt; PKY1473, cac1Δ; PKY1475, hir1Δ; PKY1477, cac1Δ hir1Δ.
Genetic interactions with kinetochore alleles
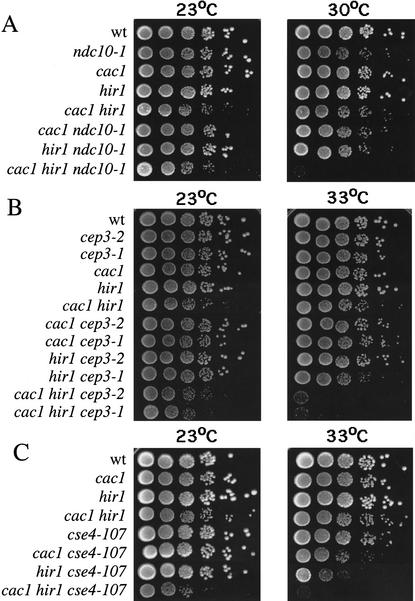
Because of the increased chromosome missegregation rates in cac1Δ hir1Δ cells, we predicted that deleting CAC and HIR genes in cells with conditional alleles of kinetochore components would result in synthetic nonviability phenotypes. Assembly of the CBF3 protein complex onto centromeres is absolutely required for high-fidelity chromosome segregation in budding yeast (Hyman and Sorger 1995). Figure 3, A and B, shows plating efficiencies of cells containing all possible combinations of cac1Δ and hir1Δ deletions and the CBF3 alleles ndc10-1, cep3-1, and cep3-2 at both permissive and semipermissive temperatures. Indeed, a striking degree of lethality was observed at semipermissive temperatures when CBF3 alleles were combined with both cac1Δ and hir1Δ mutations. As observed for the cell cycle and chromosome missegregation phenotypes, these genetic synergies depended on the simultaneous absence of both CAC and HIR genes. These studies also revealed that only a subset of CBF3 mutations require CAC and HIR gene function to remain viable: no effects were observed upon combining cac1Δ hir1Δ mutations with the ctf13-30, ndc10-2, or skp1-4 alleles (data not shown).
Figure 3.
cac1Δ hir1Δ cells display genetic interactions with kinetochore mutations. Yeast strains of the indicated genotypes were grown to log phase at 23°C, a temperature permissive for all kinetochore alleles used. Cells were then serially diluted, plated onto YPAD agar, and incubated at the indicated temperatures (23°C for 3 d; 30°C for 2 d; and 33°C for 2 d). At least two independent segregants for each genotype were tested in this manner. Shown are growth phenotypes of strains containing cac1Δ and hir1Δ gene deletions combined with (A) ndc10-1; (B) cep3-1 and cep3-2; and (C) cse4-107 alleles.
Figure 3 shows interactions between cac1Δ and hir1Δ mutations and an allele of the CEN-specific histone H3 variant CSE4 (Fig. 3C), the yeast homolog of mammalian centromere protein A (CENP-A; Stoler et al. 1995; Meluh et al. 1998). The cse4-107 allele contains the point mutation Q219D (Chen et al. 2000) in helix 3 of the histone fold domain. Helix 3 forms the four-helix bundle comprising the Cse4–Cse4 (or Cse4–H3) contact site within centromeric nucleosomes (Luger et al. 1997; Glowczewski et al. 2000; Yoda et al. 2000). The Q219D mutation is therefore predicted to destabilize centromeric nucleosomes. Similar to the phenotypes observed with the ndc10-1, cep3-1, and cep3-2 alleles, we observed a complete loss of cell viability in cac1Δ hir1Δ cse4-107 triple mutant cells at semipermissive temperatures. Therefore, cac hir cells are sensitive to defects in multiple kinetochore proteins, and these interactions are not restricted to CBF3 subunits. We note that allele-specific genetic interactions with kinetochore components have previously been observed for other kinetochore proteins such as Ctf19 (Hyland et al. 1999), the Aurora-like kinase Ipl1 (Biggins et al. 1999), and the Duo1/Dam1/Dad1 complex that links spindle integrity and kinetochore function (Cheeseman et al. 2001; Enquist-Newman et al. 2001).
Synthetic dosage lethality has been previously observed upon overexpression of wild-type kinetochore proteins in cells with weakened kinetochore function (Kroll et al. 1996; Biggins et al. 1999; Hyland et al. 1999). To further test genetic interactions between CAF-1 and kinetochore proteins, we assessed sensitivity of kinetochore mutants to the overexpression of CAC1. Galactose-induced overexpression of CAC1 led to synthetic dosage lethality in ndc10-1, ndc10-2, and cep3-1 cells, but not in wild-type cells (data not shown). Again, there was some specificity in this phenotype because galactose-induced overexpression of CAC1 did not cause lethality in cells bearing a temperature-sensitive mutation in IPL1, the Aurora-like kinase required for kinetochore regulation (Biggins et al. 1999). Taken together, our synthetic phenotype and overexpression data strongly indicate that CAF-1 and Hir proteins play an important role in kinetochore function.
Localization of CAF-1 and Hir1 to centromeric chromatin
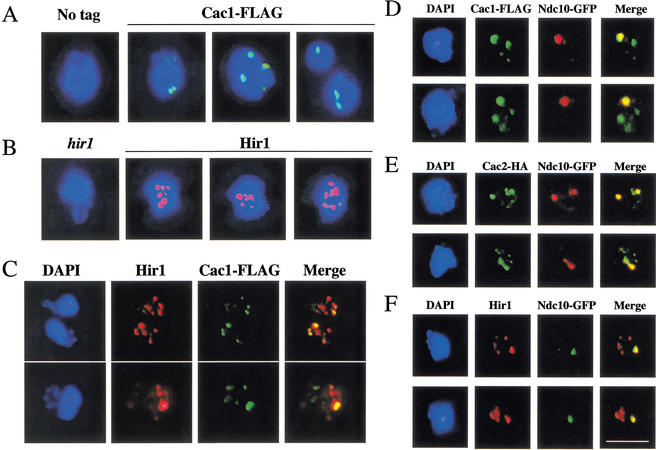
To determine whether CAF-1 and Hir proteins act directly to affect chromatin structures at centromeric loci, we tested for localization of these proteins to centromeres. Indirect immunofluorescence experiments with chromosome spread preparations (Loidl et al. 1998) detected endogenous levels of both Cac1 and Hir1 proteins. Cac1 was organized into discrete subnuclear foci: each nucleus typically displayed two–four brightly staining foci; other more weakly staining sites were also observed (Fig. 4A). Hir1 was also organized into foci, but in this case, a higher number of total foci (6–10) was observed (Fig. 4B). We tested whether CAF-1 subunits or Hir1 colocalized with centromeric nuclear domains, which are organized into discrete clusters near the spindle pole body region (Guacci et al. 1997; Goshima and Yanagida 2000; He et al. 2000; Jin et al. 2000). Indeed, both the Cac1 and Cac2 subunits of CAF-1 colocalized with the kinetochore protein Ndc10 (Goh and Kilmartin 1993; Meluh and Koshland 1997) in chromosome spread preparations from asynchronous cells (Fig. 4D,E). Additionally, we have detected this colocalization in cells arrested in G1 phase with α-factor or in M phase with nocodazole (data not shown). Typically, the strongest staining Cac1 and Cac2 foci showed the highest degree of overlap with centromeric regions. Identical results were obtained using Cse4-HA as a centromere marker (data not shown). Colocalization of both Cac1 and Cac2 with kinetochore proteins suggests that these proteins are directed to centromeric chromatin as members of the CAF-I complex. Similar results were obtained when nuclei were stained for Hir1 (Fig. 4F); again, Ndc10 was seen to colocalize with a subset of Hir1 staining regions. We also tested whether the multiple foci of Cac1 and Hir1 staining were coincident, and we observed that a subset was not (Fig. 4C). Together, these data indicate that CAF-I and Hir proteins can localize independently to multiple genomic loci, but both are recruited to the centromeres.
Figure 4.
Cac1, Cac2, and Hir1 colocalize with centromere protein Ndc10. All strains were grown to log phase at 30°C and prepared for chromosome spreads as described (Loidl et al. 1998). (A) Spread nuclei of yeast strains PKY028 (CAC1) and PKY1288 (CAC1-FLAG) were stained with anti-FLAG antibodies. Staining of Cac1-FLAG (green) is superimposed on DNA staining (DAPI, blue). (B) Spread nuclei of yeast strains PKY087 (hir1Δ) and PKY028 (HIR1) were stained with anti-Hir1 antibody. Staining of Hir1 (red) is superimposed on DNA staining (DAPI, blue). (C) Spread nuclei of strain PKY1288 (CAC1-FLAG) were processed for double-label immunofluorescence using anti-FLAG and anti-Hir1 antibodies. (D) Spread nuclei of yeast strain PKY2067 (CAC1-FLAG, NDC10-GFP) were stained with anti-FLAG (green) and anti-GFP antibodies (red). (E) Spread nuclei of yeast strain PKY2046 (CAC2-HA, NDC10-GFP) were stained using anti-HA (green) and anti-GFP antibodies (red). (F) Spread nuclei of yeast strain PKY2069 (HIR1, NDC10-GFP) were stained using anti-Hir1 (red) and anti-GFP antibodies (green). Bar, 5μm.
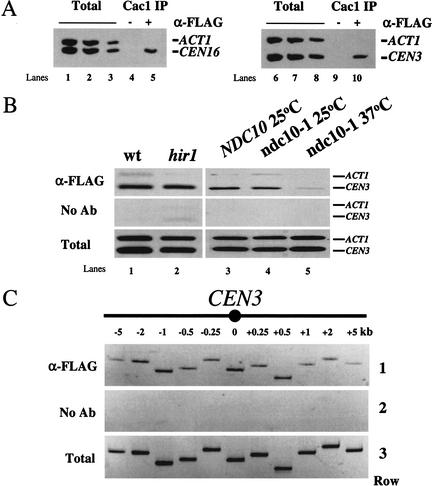
In a second approach, chromatin immunoprecipitation (ChIP) was used to confirm the localization of Cac1 to centromeric loci (Fig. 5A). Consistent with the immunolocalization data, immunoprecipitation of Cac1 resulted in the preferential recovery of the core centromeric regions for CEN3 and CEN16 relative to the control locus ACT1. We hypothesized that recruitment of Cac1 to centromeres was dependent on other kinetochore proteins and tested this by further ChIP experiments. Enrichment of Cac1 at CEN3 was independent of the presence of the HIR1 gene (Fig. 5B). In contrast, a conditional ndc10 allele reduced enrichment of Cac1 at CEN3 DNA (Fig. 5B), as previously observed for many kinetochore proteins (Goshima and Yanagida 2000; Enquist-Newman et al. 2001; He et al. 2001). Together, these localization data indicate that CAF-I and Hir1 are independently recruited constituents of centromeric chromatin. The presence of Cac1 and Hir1 at centromeres suggests that they act directly to contribute to kinetochore function.
Figure 5.
Cac1 is associated with centromeric chromatin. Formaldehyde cross-linked chromatin (1 h fixation) was prepared from cells grown to log phase at 30°C. (A) Chromatin prepared from yeast strain PKY1288 (CAC1-FLAG) was immunoprecipitated in the presence of anti-FLAG antibody or mock treated. PCR was performed on both total input and immunoprecipitated chromatin to visualize recovery of the core centromeric region of CEN3 (249 bp) and CEN16 (310 bp) relative to the ACT1 locus (367 bp). Dilution of total input chromatin: (lanes 1,6) 1:32; (lanes 2,7) 1:64; (lanes 3,8) 1:128. Approximately one-fifth of the eluate from immunoprecipitation was used for PCR analysis: (lanes 4,9) mock immunoprecipitation, (lanes 5,10) the experimental immunoprecipitation. (B) Cac1 localization at CEN3 requires NDC10 but not HIR1. Chromatin was prepared from yeast strains (lanes 1,3) PKY2199 (CAC1-FLAG); (lane 2) PKY2385 (CAC1-FLAG hir1Δ); and (lanes 4,5) PKY2200 (CAC1-FLAG ndc10-1); and analyzed as in A. Total input chromatin was diluted 1:64 prior to PCR analysis. (C) Distribution of CAC1 across the CEN3 region. Chromatin was prepared from yeast strain PKY1288 (CAC1-FLAG), and PCR was performed as in A. As in all experiments, CEN DNA in these FLAG antibody immunoprecipitations was enriched relative to the ACT1 negative control locus (data not shown). (Row 1) Anti-FLAG precipitated chromatin, (row 2) mock-precipitated chromatin, and (row 3) total input chromatin (1:64 dilution) was analyzed for recovery of fragments at or flanking the core centromeric region of CEN3 as indicated on the diagram (not to scale).
Kinetochore proteins are distributed across centromeric chromatin in characteristic patterns. For example, the yeast CENP-A and CENP-C homologs, Cse4 and Mif2, are localized specifically over the core centromere DNA with little if any distribution on the flanking regions containing phased nucleosomes (Meluh and Koshland 1997; Meluh et al. 1998). In contrast, the cohesin protein Mcd1/Scc1 covers a region of several kilobases around centromeres (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999). To determine whether Cac1 distribution was similar to either of these patterns, we performed ChIP and analyzed recovery of chromatin across a 10-kb region flanking CEN3 (Fig. 5C). We observed that Cac1 was enriched across the entire CEN-proximal region analyzed with subtle fluctuations, suggesting that CAF-I contributes to the full length of the specialized centromeric chromatin structure.
Defects in centromeric chromatin in cacΔ hirΔ cells
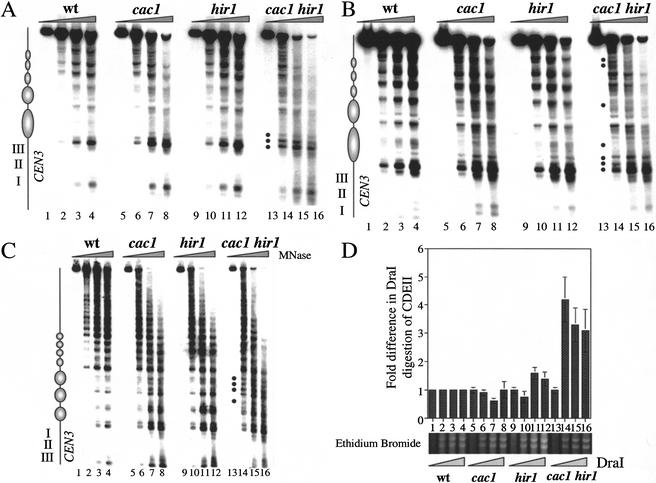
Our data predicted that simultaneous absence of CAF-I and Hir proteins would alter centromeric chromatin structure. To test for changes in the regions flanking the core centromere sequences, we performed indirect end-label analysis of micrococcal nuclease (MNase)-digested chromatin (Fig. 6A–C). As observed previously, a ladder of phased nucleosomes flanked both sides of the core centromeric sequences in wild-type cells (Bloom and Carbon 1982). At 30°C on the CDEIII-proximal side of CEN3, these digestion patterns were largely unchanged in cac1Δ or hir1Δ single mutant cells (Fig. 6A). In contrast, in cac1Δ hir1Δ double mutant cells, alterations in this chromatin structure occurred at multiple sites (see Fig. 6A, bullets). The most prominent changes included the ratio of three hypersensitive sites immediately adjacent to the CDEIII element. cac1Δ hir1Δ cells also displayed a general reduction in the periodicity in the digestion pattern. At 16°C, the periodicity in this digestion pattern was also slightly reduced in cac1Δ cells, but not nearly to the degree observed in the double mutant cells (Fig. 6B; data not shown). On the CDEI-proximal side of CEN3, significant changes were observed in cac1Δ hir1Δ cells only at 16°C, with the appearance of several new digestion sites (Fig. 6C). However, we note that the centromere-distal phased nucleosomes were unperturbed in the cac1Δ hir1Δ cells, showing specificity in the structural alterations. Consistent with our genetic data, we conclude that the structure of both core and flanking centromeric chromatin is most severely altered in cells that lack both CAF-I and Hir proteins, in a temperature-dependent manner that correlates with kinetochore function.
Figure 6.
Centromeric chromatin phenotypes in cac1Δ hir1Δ cells. Nuclei were prepared from yeast strains PKY346 (wt), PKY1100 (cac1Δ), PKY1154 (hir1Δ), and PKY1168 (cac1Δ hir1Δ) as indicated and digested with nucleases as follows. (A–C) Indirect end-label analysis of CEN3 chromatin. Nuclei were incubated with 0.6 U MNase (Sigma) at 32°C for 0 min (lanes 1,5,9,13), 5 min (lanes 2,6,10,14), 10 min (lanes 3,7,11,15), or 15 min (lanes 4,8,12,16) prior to isolation of genomic DNA, restriction enzyme digestion, and DNA blot hybridization with probes as described in the Materials and Methods. (A) CDEIII-proximal side of CEN3 in cells grown at 30°C. Samples were digested with ClaI. (B) CDEIII-proximal side of CEN3 in cells shifted to 16°C for 36 h prior to isolation of nuclei. Samples were digested with ClaI. (C) CDEI-proximal side of CEN3 in nuclei prepared from cells grown at 16°C. Samples were digested with BamHI. (D) DraI accessibility to CDEII within CEN3. Nuclei were incubated with DraI (0, 50, 100, or 150 U/mL) at 37°C for 30 min. DNA was purified, digested with EcoRI, and subjected to Southern blot hybridization. The fold differences in digestion relative to wild-type cells represent the average of three independent experiments with standard deviations indicated by the error bars. (Lower panel) A region of the same gel used for the Southern blot visualized by ethidium bromide staining, indicating similar extents of DraI digestion of total chromatin in all four strains.
As an independent assay for chromatin structure at CEN3, we measured restriction enzyme accessibility at the core of the CEN3 locus (Saunders et al. 1988). A cluster of three closely spaced DraI restriction endonuclease sites at the CDEII region of CEN3 is protected from digestion in the presence of the centromere/kinetochore complex. Conditional mutations in CSE4 and HHF1 (histone H4), or depletion of histones H2B or H4 causes an increase in the accessibility of the DraI cluster (Saunders et al. 1990; Meluh et al. 1998), thus indicating that the integrity of centromeric nucleosomes is required for CDEII protection. Nuclei prepared from wild-type, cac1Δ, hir1Δ, and cac1Δ hir1Δ cells grown at 30°C were incubated with increasing amounts of DraI enzyme. DNA purified from these nuclear preparations was analyzed by Southern blot hybridization to quantitate CDEII accessibility. Figure 6D shows the DraI accessibility at CEN3 CDEII relative to that observed in wild-type cells. We observed that cac1Δ and hir1Δ cells showed levels of CDEII accessibility that in all cases were less than twofold different from that observed in wild-type cells. In contrast, a fourfold increase in CDEII accessibility was observed in cac1Δ hir1Δ cells relative to that of wild type. DraI digestion of total chromatin occurred to the same degree in all samples and was independent of strain genotype (Fig. 6D, lower panel; data not shown). These data and the MNase experiments (Fig. 6A) lead us to conclude that CAF-1 and Hir proteins are required to maintain the integrity of core and flanking centromeric chromatin.
Alteration of Cse4 localization in cac1Δ hir1Δ cells
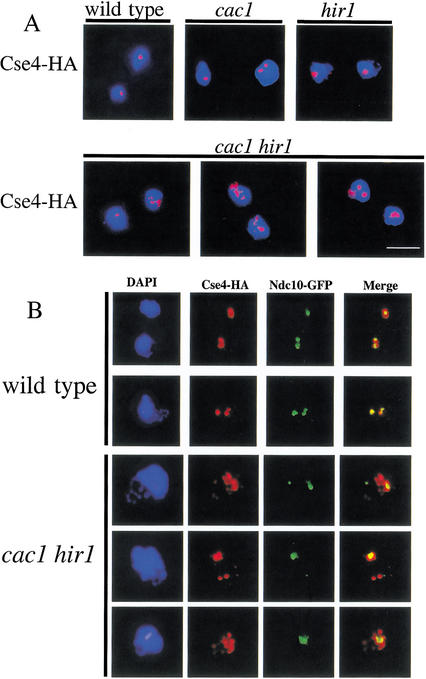
The observed alteration in centromeric chromatin structure in cac1Δ hir1Δ cells (Fig. 6) together with the genetic interactions with cse4-107 (Fig. 3C) suggested that the cellular pattern of Cse4 localization might be altered in cacΔ hirΔ cells. Chromosome spreads prepared from wild-type, cac1Δ, hir1Δ, and cac1Δ hir1Δ cells grown at 30°C were immunostained to detect the presence of Cse4-HA (Fig. 7A). The vast majority of wild-type, cac1Δ, and hir1Δ nuclei showed that Cse4 localization was largely restricted to 1–2 foci per nucleus, consistent with previous observations of centromeric clustering (Guacci et al. 1997; Jin et al. 1998; Meluh et al. 1998). In contrast, a strikingly different pattern of Cse4 localization was observed in cac1Δ hir1Δ double mutants. An average of 32% of cac1Δ hir1Δ nuclei showed a less discrete localization (>2 foci) of Cse4, compared with a frequency of <4% observed for wild-type, cac1Δ, and hir1Δ nuclei.
Figure 7.
Extracentromeric localization of Cse4 in cac1Δ hir1Δ cells. (A) Dispersion of Cse4. Yeast strains PKY2084 (CSE4-HA), PKY2296 (cac1Δ CSE4-HA), PKY2298 (hir1Δ CSE4-HA), and PKY2179 (cac1Δ hir1Δ CSE4-HA) were grown to log phase at 30°C and prepared for chromosome spreads. Nuclei were stained with anti-HA antibody. Staining of Cse4-HA (red) is superimposed on DNA staining (DAPI, blue). Bar, 5 μm. (B) Extracentromeric localization of Cse4. Yeast strains PKY2444 (CSE4-HA NDC10-GFP) and PKY2453 (cac1Δ hir1Δ CSE4-HA NDC10-GFP) were treated as in A and were stained to detect Cse4-HA (red) and Ndc10-GFP (green).
The observed declustering of Cse4 could be explained by loss of sister chromatid cohesion, the loss of centromere clustering, or altered deposition of Cse4. We tested each of these possibilities. Using chromosomes marked with CEN-linked LacI-GFP arrays (Straight et al. 1996; Biggins et al. 1999), we observed no defects in sister chromatid cohesion in cac1Δ hir1Δ cells arrested in G2/M (Fig. 2B; data not shown). Furthermore, we observed that the Mcd1/Scc1 cohesin subunit was localized to centromeres in cac1Δ hir1Δ cells (data not shown). Together, these data indicated that changes in Cse4 localization did not reflect a loss of chromosome cohesion.
To test for declustering of centromeres, we examined the relative nuclear localization of Cse4 and the CBF3 subunit Ndc10. We observed that Cse4 colocalized with Ndc10 in all strains tested except for cac1Δ hir1Δ cells (Fig. 7B; data not shown). In cac1Δ hir1Δ cells, significant amounts of Cse4 were not coincident with the punctate localization of Ndc10. Therefore, in cac1Δ hir1Δ cells centromeres are still clustered, but some Cse4 exists in extracentromeric locations. Chromatin immunoprecipitation experiments showed that Cse4 was localized indistinguishably to the core centromere region of CEN3 in wild-type (Meluh et al. 1998; Glowczewski et al. 2000) and cac1Δ hir1Δ lysates, and in neither case was Cse4 associated with 5-kb regions flanking CEN3 (data not shown). Therefore, we conclude that the extracentromeric deposition of Cse4 occurred at loci distal from the centromeres in cac1Δ hir1Δ cells. These data reinforce our conclusion that CAF-1 and Hir proteins are required for proper formation and function of kinetochores.
Discussion
We show here a novel role for chromatin assembly factor I and the Hir proteins: they contribute to the formation of functional kinetochores. This is the first demonstration that histone deposition proteins contribute to the specialized function of centromeric chromatin. Our data are consistent with previous experiments showing that mutations in core histone proteins (Smith et al. 1996; Pinto and Winston 2000) or nucleosome remodeling enzymes (Tsuchiya et al. 1998) also interfere with formation of centromeric chromatin. Together, these data support the idea that despite the simple primary sequence requirements for the budding yeast centromere, a more elaborate chromatin structure is the underlying functional unit required for kinetochore activity.
Cellular localization of CAF-I and Hir proteins
In vertebrate cells, CAF-I colocalizes with sites of DNA synthesis during S phase (Krude 1995). After completion of DNA replication, CAF-I is then relocalized to centric heterochromatin in a manner dependent on binding heterochromatin protein-1 (HP-1; Murzina et al. 1999; Taddei et al. 1999). However, HP-1 homologs are not present in budding yeast, and it had previously not been determined whether CAF-I in S. cerevisiae is localized to any specific loci. Here, we show yeast CAF-I is localized to centromeric chromatin in asynchronous cell populations, and we have observed this localization in cells arrested outside of S phase. Together, these data indicate that there may be an evolutionarily conserved role for CAF-I at centromeres outside of S phase.
Both CAF-I subunits and Hir proteins are organized into discrete nuclear foci, and only a subset of these colocalized with centromeres. Because of the data linking CAF-I to heterochromatic gene silencing, a reasonable hypothesis would have been that these extracentromeric foci represented the clustered telomeres or HM loci. However, our preliminary data show that only a minor subset of CAF-I foci colocalized with the Sir3 protein, a marker for silent regions in immunofluorescence experiments (Gotta et al. 1996; data not shown). These data indicate that cells contain several unidentified loci enriched in CAF-I. Likewise, Hir1 is organized into multiple foci, many of which are not coincident with Cac1 staining. In chromatin immunoprecipitation experiments, Hir1 localizes to the HTA1–HTB1 intergenic region, a locus that requires Hir proteins for transcriptional repression (Osley and Lycan 1987; Z. Nackerdien and M.A. Osley, unpubl.). This locus, together with other chromosomal regions that require the activity of Hir proteins to build repressive chromatin structures, are likely to represent some of the extracentromeric foci. Complete elucidation of the biological roles of CAF-I and Hir proteins will require identification of all their sites of localization.
Complexity of centromere requirement for chromatin assembly factors
Several lines of genetic evidence strongly predict that Hir proteins are histone-deposition factors, and the orthologous mammalian HIRA protein binds histones (Lorain et al. 1998). First, HIR1 is highly homologous to the CAC2-encoded subunit of CAF-I, indicating conserved gene function (Kaufman et al. 1998). Second, the synergistic loss of heterochromatic gene silencing and slow-growth phenotypes of cacΔ hirΔ double mutant cells indicate overlapping functions of Hir proteins with those of CAF-I (Kaufman et al. 1998; Qian et al. 1998). Epistasis studies at HML have shown that HIR genes contribute to silencing at this locus via the same genetic pathway as the known histone-binding and -deposition factor ASF1 (Tyler et al. 1999; Sharp et al. 2001). Furthermore, Hir1 and Hir2 proteins interact physically with Asf1 in vitro and in cell extracts (Sharp et al. 2001) and in two-hybrid experiments (Sutton et al. 2001). The simple genetic prediction from these data was that cacΔ asf1Δ double mutant cells would show centromere phenotypes similar to those of cacΔ hirΔ cells. Instead, cacΔ asf1Δ cells showed a G2/M delay independent of spindle assembly checkpoint activation and segregated the CFIII(D8B.d) reporter minichromosome at frequencies similar to wild-type cells (data not shown). Therefore, centromeres differ from the known silent regions by requiring HIR genes, but not ASF1 in the absence of CAF-I function. Further study will be needed to better understand this bifurcation of the HIR/ASF1 chromatin assembly pathway.
Role of CAF-I and Hir proteins at the centromere
Centromeres may serve as repositories for CAF-I when it is not active during S-phase DNA replication or recruited to sites of DNA repair (Martini et al. 1998). However, we show here that CAF-I and the Hir proteins are components of centromeric chromatin and are required for proper centromere structure and function. Therefore, we propose active roles for CAF-I and Hir proteins in building functional kinetochores.
Given the known biochemical activities of CAF-I, a logical idea would be that CAF-I acts as a histone-deposition factor at CEN DNA. Such a role would not be necessarily restricted to S phase: recent data have suggested that budding yeast centromeric chromatin undergoes reversible oscillations caused by tension imposed by the spindle apparatus (He et al. 2000; Pearson et al. 2001). Based on the length of centromeric chromatin that undergoes such deformations and measurement of the maximal deformation of the centromeric DNA, it has been calculated that centromeric DNA sequences can be stretched to a length corresponding to B-form DNA, which implies that existing nucleosomes would be lost. Evidence for loss of nucleosomes upon physical stretching of chromatin fibers has also been obtained in single-molecule experiments (Cui and Bustamante 2000). We therefore hypothesize that there is a continual need for replenishment of histones at yeast centromeres, and that this may be provided in part by CAF-I and Hir proteins. Also, we note that CAF-I contributes to maintaining the stability of the silent state at telomeres and HM loci (Monson et al. 1997; Enomoto and Berman 1998). Because this silencing is maintained throughout the cell cycle (Loo and Rine 1995), these data are consistent with the idea that CAF-I can replace nucleosomes in dynamic chromatin structures outside of S phase.
The presence of a proper chromatin configuration at the centromere may be important for the establishment or maintenance of tension, which is required for anaphase entry in higher organisms (Rieder et al. 1994; Li and Nicklas 1995). Tension requires both cohesion between sister chromatids and bipolar attachment to the mitotic spindle. The core centromere DNA sequence is required to establish and maintain sister chromatid cohesion, and cohesion proteins have been localized to centromeric chromatin (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999). However, we observed no defects in sister chromatid cohesion or loading of the cohesin protein Mcd1/Scc1 onto centromeric DNA in cacΔ hirΔ cells (data not shown). Furthermore, localization of CAF-I to centromeres during G1 phase when sister chromatids do not yet exist, suggest that CAF-I has roles other than assisting cohesion. It is therefore likely that reformation of nucleosomes at CEN DNA impacts kinetochore function independently of cohesion.
Instead, the ordered array of nucleosomes flanking the core centromere could be crucial for the proper positioning of core kinetochore components, without which attachment to the mitotic spindle could be perturbed. An analogous situation exists at origins of DNA replication, where movement of nucleosomes neighboring the binding site for the Origin Recognition Complex reduces the efficiency of replication initiation (Simpson 1990; Lipford and Bell 2001). An alternative hypothesis is that the structure acts to enforce the extraordinarily specific localization of Cse4 to the CDEII region as opposed to other AT-rich sites in the genome. In cacΔ hirΔ cells, the centromere-specific histone Cse4 is deposited to regions outside of centromeric chromatin, as defined by colocalization experiments with the CBF3 subunit Ndc10 (Fig. 7). Therefore, although CAF-I and Hir proteins are not required for Cse4 deposition, in their absence the Cse4-deposition mechanism is significantly perturbed. The specialized chromatin structure existing at yeast centromeres may therefore act to ensure the specificity of Cse4 deposition at centromeres.
Materials and methods
Strains, plasmids, and growth conditions
Yeast strains used in this study are listed in Table 2. Previously described deletion alleles in the W303 background include cac2Δ::TRP1 (Kaufman et al. 1997), cac1Δ::hisG-URA3-kanR-hisG (Game and Kaufman 1999), hir1Δ::HIS3 (Recht et al. 1996), and hmlΔ::LEU2 (Kaufman et al. 1998). The trp1::CAC1-FLAG::TRP1 allele in which the native CAC1 promoter drives Cac1-FLAG expression was constructed by inserting the BamHI/EcoRI fragment from pPK184 (Sharp et al. 2001) into similarly digested pRS404 (Sikorski and Hieter 1989) to generate pPK264. pPK264 was digested with MfeI to direct integration into the TRP1 locus. The cac1::CAC1-FLAG::URA3 allele in which the native CAC1 promoter drives Cac1-FLAG expression at the native CAC1 locus was constructed by inserting the BamHI/EcoRI fragment from pPK184 (Sharp et al. 2001) into pRS406 (Sikorski and Hieter 1989) to generate pPK229. pPK229 was digested with BamHI + EspI, filled in with Klenow + dNTPs and religated to form pPK318. pPK318 was digested with EcoNI to direct integration to the CAC1 locus. The CAC2 gene with a triple HA tag (Krawitz et al. 2002) was inserted into a URA3 integration vector with the GPD promoter (Schena et al. 1991), generating pPK106, which was integrated at the URA3 locus. All tagged CAC alleles were shown to complement deletion mutations for telomeric silencing. ndc10::NDC10-GFP::HIS3 (Zeng et al. 1999) was generated in the W303 strain background. The cse4Δ::kanMX deletion in W303 was a gift of Sue Biggins (Fred Hutchinson Cancer Research Center, Seattle, WA). The CFIII(D8B.d) fragment was generated in W303-based strains by digesting pDK352 (Shero et al. 1991) with NotI prior to transformation. Leu+ transformants were checked by pulse-field gel electrophoresis for the presence of a 125-kb chromosomal fragment not present in untransformed cells.
Table 2.
Yeast strains used in this study
| Name
|
Genotype
|
Source
|
|---|---|---|
| PKY346 | MATa hmlΔ::LEU2 | Kaufman et al. 1998 |
| PKY1100 | MATa hmlΔ::LEU2 cac1Δ::hisG | This study |
| PKY1154 | MATa hmlΔ::LEU2 hir1Δ::HIS3 | This study |
| PKY1168 | MATa hmlΔ::LEU2 cac1Δ::hisG hir1Δ::HIS3 | This study |
| PKY1340 | MATa hmlΔ::LEU2 cac2Δ::TRP1 hir1Δ::HIS3 | This study |
| PKY1257 | MATa hmlΔ::LEU2 cac1Δ::hisG hir1Δ::HIS3 mad2Δ::URA3 | This study |
| PKY1265 | MATa hmlΔ::LEU2 mad2Δ::URA3 | This study |
| RHC15 | MATa mad2Δ::URA3 | Andrew Murray |
| SBY164 | MATa ndc10-1 | Sue Biggins |
| SBY165 | MATa ndc10-2 | Sue Biggins |
| SBY163 | MATα ctf13-30 | Sue Biggins |
| SBY168 | MATa cep3-1 | Sue Biggins |
| SBY162 | MATa cep3-2 | Sue Biggins |
| SBY597 | MATa/MATα cse4Δ::kanMX/CSE4 | Sue Biggins |
| PKY2160 | MATa cse4Δ::kanMX + pcse4-107HA-URA3 | This study; Molly Fitzgerald-Hayes |
| PKY028 | MATa | Kaufman et al. 1997 |
| PKY1288 | MATa cac1Δ::hisG trp1::CAC1-FLAG::TRP1 | This study |
| PKY087 | MATa hir1Δ::HIS3 | Kaufman et al. 1998 |
| PKY2084 | MATα cse4Δ::kanMX + pCSE4-HA-TRP1 | This study; Pamela Meluh |
| PKY2296 | MATa cac1Δ::hisG-URA3-hisG cse4Δ::kanMX + pCSE4-HA-TRP1 | This study |
| PKY2298 | MATα hir1Δ::HIS3 cse4Δ::kanMX + pCSE4-HA-TRP1 | This study |
| PKY2179 | MATα cac1Δ::hisG-URA3-hisG hir1Δ::HIS3 cse4Δ::kanMX + pCSE4-HA-TRP1 | This study |
| PKY2069 | MATa ndc10::NDC10-GFP::HIS3 | This study |
| PKY2067 | MATa cac1Δ::hisG trp1::CAC1-FLAG::TRP1 ndc10::NDC10-GFP::HIS3 | This study |
| PKY2046 | MATα cac2Δ::TRP1 ura3::GPD-CAC2-HA::URA3 ndc10::NDC10-GFP::HIS3 | This study |
| PKY847 | MATα CFIII(D8B.d) ade3Δ | This study |
| PKY2216 | MATα cac1Δ::hisG CFIII(D8B.d) ade3Δ | This study |
| PKY865 | MATα his1Δ::HIS3 CFIII(D8B.d) ade3Δ | This study |
| PKY2217 | MATa cac1Δ::hisG hir1Δ::HIS3 CFIII(D8B.d) ade3Δ | This study |
| PKY1531 | MATα mad2Δ::URA3 CFIII(D8B.d) ade3Δ | This study |
| PKY2218 | MATa cac1Δ::hisG hir1Δ::HIS3 mad2Δ::URA3 CFIII(D8B.d) ade3Δ | This study |
| SBY214 | MATa his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::lacO(256 repeats)::TRP1 bar1-1 lys2Δ | Sue Biggins |
| PKY1473 | MATa cac1Δ::hisG his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::lacO(256 repeats)::TRP1 | This study |
| PKY1475 | MATa hir1Δ::HIS3 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::lacO(256 repeats)::TRP1 | This study |
| PKY1477 | MATa cac1Δ::hisG hir1Δ::HIS3 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::lacO(256 repeats)::TRP1 | This study |
| PKY2199 | MATa HMR::ADE2 cac1::CAC1-FLAG::URA3 | This study |
| PKY2200 | MATa HMR::ADE2 cac1::CAC1-FLAG::URA3 ndc10-1 | This study |
| PKY2385 | MATa cac1::CAC1-FLAG::URA3 hir1Δ::HIS3 | This study |
| PKY2444 | MATa ndc10::NDC10-GFP::HIS3 cse4Δ::kanMX (pCSE4-HA-TRP1) | This study |
| PKY2453 | MATa cac1Δ::hisG-URA3-hisG hir1Δ::HIS3 ndc10::NDC10-GFP::HIS3 cse4Δ::kanMX (pCSE4-HA-TRP1) | This study |
| PKY2390 | MATa hmlΔ::LEU2 MCD1-HA | This study; Vincent Guacci |
| PKY2427 | MATa hmlΔ::LEU2 MCD1-HA cac1Δ::hisG hir1Δ::HIS3 | This study |
All strains are in the W303 background and contain the leu2-3, 112; ura3-1; his3-11, 15; trp1-1; ade2-1; and can1-100 mutations.
The plasmids pCSE4-HA-TRP1 and pcse4-107HA-URA3 have been described (Meluh et al. 1998; Chen et al. 2000). Genetic interactions between cac1Δ and hir1Δ gene combinations with the cse4-107HA allele were determined to be independent of the HA tag, because cac1Δ and hir1Δ gene combinations with CSE4-HA displayed no temperature-sensitivity. A CAC1-FLAG allele driven by the GAL promoter was constructed in two steps. First, an N-terminal fragment (amino acids 1–331) from CAC1 flanked by 5′ BamHI and 3′ NheI sites was generated by PCR and cloned into BamHI + NheI-digested pVF1 (pRS416 [Sikorski and Hieter 1989] containing the GAL1-10 promoter inserted into the EcoRI–BamHI sites), generating pPK293. Second, the FLAG-tagged C terminus of CAC1 from pPK184 (Sharp et al. 2001) was excised with BlpI + Bsp120I and inserted into BlpI + NotI-digested pPK293, generating pPK299.
Doubling times of strains in rich media at 30°C were determined by counting cells from sonicated samples with a hemocytometer. This method revealed that a cac1Δ hir1Δ strain had a significantly longer doubling time compared to single mutants or wild-type cells. Thus, the previous observation of a wild-type doubling time of some cacΔ hirΔ strains (Kaufman et al. 1998) was an underestimate because optical density measurements had been used.
Cell cycle analysis involving α-factor arrest required that all strains analyzed be made isogenic for the hmlΔ::LEU2 mutation. cacΔ hirΔ double mutants are defective in silencing at the mating loci (Kaufman et al. 1998; Sharp et al. 2001). However, cacΔ hirΔ hmlΔ strains capable of expressing only a gene mating information arrested more efficiently in α-factor. Samples were prepared for flow cytometry as described (http://www.fhcrc.org/∼gottschling/facs.html). Flow cytometry profiles were analyzed and compiled using WinMDI 2.8 software.
Chromosome missegregation assays
Microscopic analysis of chromosome missegregation was performed using the lacO/lacI-GFP system (Straight et al. 1996; Biggins et al. 1999). CUP1-driven lacI-GFP expression was induced with 250 μM CuSO4 for 30 min prior to cell harvest. Cells were fixed in 3.7% formaldehyde for 10 min at 4°C, washed twice in cold 0.1 M potassium phosphate (pH 7.5), resuspended in the same buffer, and sonicated briefly. Chromosome missegregation was scored by comparing CEN4 position (GFP fluorescence) with nuclear staining (Hoechst).
Quantitative analysis of chromosome missegregation was performed as described (Shero et al. 1991). In brief, cells were grown to log phase at 30°C and plated onto YPD media containing 4% glucose. Between 50,000 and 80,000 colonies were plated for each strain at a colony density of 200–300 colonies per plate. Cells were incubated at 30°C for 2–3 d and then placed at 4°C to assist color development. Chromosome missegregation events were then monitored using half-sector analysis to quantitate chromosome loss or nondisjunction events in the first cell division of colony formation. Pink and white half-sectored colonies were scored as chromosome loss events; red and white half-sectored colonies were scored as chromosome nondisjunction events. The number of white colonies was subtracted from the total number of cells plated to determine chromosome missegregation frequencies.
Immunofluorescence microscopy
Chromosome spreads were performed as described previously (Loidl et al. 1998). The primary antibodies used were mouse anti-FLAG (Sigma, 1:2000 dilution), rabbit anti-GFP (Seedorf et al. 1999, 1:3000 dilution), mouse anti-GFP (Roche, 1:1000 dilution), rabbit anti-Hir1 (1:2000 dilution), 12CA5 mouse anti-HA (1:10,000 dilution), and rabbit anti-HA (Clontech; 1:40 dilution). The affinity-purified secondary antibodies used were FITC-conjugated goat anti-mouse antibody (Cappel/Organon Technika, 1:500 dilution), rhodamine-conjugated goat anti-rabbit antibody (Cappel/Organon Technika, 1:500 dilution), and Cy3-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories, 1:750 dilution). Indirect immunofluorescence was performed on spheroplasted yeast cells as described previously (Pringle et al. 1991). The YOL1/34 rat anti-tubulin antibody (Accurate Chemical and Scientific Corp.) was used at a dilution of 1:200; FITC-conjugated anti-rat secondary antibody (Cappel/Organon Technika) was used at a dilution of 1:500. Microscopy was performed on a Nikon Eclipse E600 equipped with a Hamamatsu CCD camera controlled by ImageQuant software.
Several control experiments were performed for the antibody colocalization experiments. First, immunofluorescent signals were all tag-dependent. Second, no bleedthrough of any fluorophore was observed in secondary filters. Likewise, control experiments were performed with the anti-Hir1 polyclonal antibody on hir1Δ and HIR1 strains to show that staining was dependent on Hir1. Secondary antibodies used in double-labeling experiments were also tested to determine that there was no cross-reactivity to the reciprocal primary antibody.
Accessibility of centromeric chromatin
Yeast nuclei were prepared essentially as described previously (Hirschhorn et al. 1992), except that cell pellet resuspension steps after Zymolyase treatment were eliminated to preserve delicate spheroplasts. Instead, wash buffers were laid gently on top of the spheroplast pellet. Pellets were placed facing inward during spins so that centrifugal force mediated buffer exchange in the pellet. Spheroplasts were lysed in a 20-fold excess of 9% Ficoll-400, 20 mM PIPES (pH 6.3), 0.5 mM CaCl2. After the final wash, nuclei were resuspended in MNase digestion buffer using 3–4 strokes of a Dounce homogenizer (loose pestle). A detailed protocol of these procedures is available upon request. For indirect end-labeling experiments, nuclei were digested with 0.6 U/mL MNase (Sigma) at 32°C for 0, 5, 10, and 15 min. Digestions were terminated by the addition of one-quarter volume 5% SDS, 20 mM EDTA (pH 8.0), 20 mM EGTA (pH 8.0). After organic extraction and precipitation, genomic DNA was digested with ClaI to visualize nucleosome positioning over a 1.5-kb region adjacent to the CDEIII element. Blots were hybridized using a PCR-generated probe corresponding to SGD coordinates 113757–114293 on Chromosome III. To analyze nucleosome positioning proximal to an 8.5-kb region adjacent to CDEI, genomic DNA from MNase-treated samples was purified and digested with BamHI. Blots were hybridized with a PCR-generated probe corresponding to the DraI–BamHI region of CEN3 (Chromosome III, SGD coordinates 114318–114934).
The DraI assay to assess CDEII accessibility of CEN3 was performed as described (Saunders et al. 1990; Meluh et al. 1998). The 0.9-kb probe used was generated by PCR and corresponds to the BamHI–HindIII region of CEN3 (Chromosome III, SGD coordinates 114943–115835). DraI cutting at CDEII results in the appearance of a 2.9-kb fragment in addition to the 5.1-kb genomic fragment after EcoRI digestion. The signal strength of the bands was quantitated from a phosphorimage exposure using Bio-Rad Quantity One software. The degree of DraI digestion was calculated as % CDEII cut (signal for 2.9-kb fragment) relative to the total signal (2.9-kb fragment + 5.1-kb fragment) for each strain tested, normalized to the values obtained for the wild-type strain.
Chromatin immunoprecipitation
In vivo cross-linking and chromatin immunoprecipitation were performed as described (Meluh and Broach 1999). Immunoprecipitation was performed on the diluted chromatin solution with anti-FLAG antibody or anti-HA antibody at a concentration of 5 μg/mL. PCR analysis of total and immunoprecipitated chromatin was performed using primers to amplify the ACT1 promoter (Chromosome VI, SGD coordinates 54660–55027), CEN3 (Chromosome III, SGD coordinates 114316–114564), and CEN16 (Chromosome XVI, SGD coordinates 555852–556162). Sequences of oligonucleotides used to amplify CEN3-proximal regions are available upon request.
Acknowledgments
We thank Jasper Rine, Pamela Silver, David King, Sue Biggins, Iain Cheeseman, Doug Koshland, Andrew Murray, Pamela Meluh, Vincent Guacci, and Molly Fitzgerald-Hayes for sharing reagents; Zeena Nackerdien and Dessislava Dimova for assistance in raising and characterizing the anti-Hir1 antibody; Fred Winston, Jasper Rine, Iain Cheeseman, Priya Budde, and Peter Garber for thoughtful discussions; and Stephanie Cheung, Jay Chuang, and Gizem Rizki for excellent technical assistance. This work was supported by Department of Energy funds awarded to P.D.K. and administered through the Lawrence Berkeley National Laboratory, National Science Foundation Grant MCB-9982909 to P.D.K., and by National Institutes of Health grants 1 R01 GM55712 to P.D.K. and GM40118 to M.A.O. A.A.F. was supported by a predoctoral fellowship from the National Science Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pdkaufman@lbl.gov; FAX (510) 486-6488.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.925302.
References
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes & Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Composite motifs and repeat symmetry in S. pombe centromeres: Direct analysis by integration of NotI restriction sites. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- Cui Y, Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc Natl Acad Sci. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Cranston G, Allshire RC. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics. 1999;153:1153–1169. doi: 10.1093/genetics/153.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the reestablishment, of silencing at the yeast silent mating loci. Genes & Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders M, Berman J. RLF2, a subunit of yeast chromatin assembly factor I, is required for telomeric chromatin function in vivo. Genes & Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- Enquist-Newman M, Cheeseman IM, Van Goor D, Drubin DG, Meluh PB, Barnes G. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol Biol Cell. 2001;12:2601–2613. doi: 10.1091/mbc.12.9.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J, Kaufman PD. Role of Saccharomyces cerevisiae Chromatin Assembly Factor-I in repair of ultraviolet radiation damage in vivo. Genetics. 1999;151:485–497. doi: 10.1093/genetics/151.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowczewski L, Yang P, Kalashnikova T, Santisteban MS, Smith MM. Histone–histone interactions and centromere function. Mol Cell Biol. 2000;20:5700–5711. doi: 10.1128/mcb.20.15.5700-5711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh PY, Kilmartin JV. NDC10: A gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI. Transcriptional silencing in fission yeast. J Cell Physiol. 2000;184:311–318. doi: 10.1002/1097-4652(200009)184:3<311::AID-JCP4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: Evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnenberger KM, Carbon J, Clarke L. Identification of DNA regions required for mitotic and meiotic functions within the centromere of Schizosaccharomyces pombe chromosome I. Mol Cell Biol. 1991;11:2206–2215. doi: 10.1128/mcb.11.4.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore–microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes & Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- Hoyt MA. Exit from mitosis: Spindle pole power. Cell. 2000;102:267–270. doi: 10.1016/s0092-8674(00)00031-3. [DOI] [PubMed] [Google Scholar]
- Hyland KM, Kingsbury J, Koshland D, Hieter P. Ctf19p: A novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Sorger PK. Structure and function of kinetochores in budding yeast. Annu Rev Cell Dev Biol. 1995;11:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- Jin Q, Trelles-Sticken E, Scherthan H, Loidl J. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J Cell Biol. 1998;141:21–29. doi: 10.1083/jcb.141.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin QW, Fuchs J, Loidl J. Centromere clustering is a major determinant of yeast interphase nuclear organization. J Cell Sci. 2000;113:1903–1912. doi: 10.1242/jcs.113.11.1903. [DOI] [PubMed] [Google Scholar]
- Kamakaka RT, Bulger M, Kaufman PD, Stillman B, Kadonaga JT. Post-replicative chromatin assembly by Drosophila and human Chromatin Assembly Factor-I. Mol Cell Biol. 1996;16:810–817. doi: 10.1128/mcb.16.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor 1: A molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cell lacking chromatin assembly factor-I. Genes & Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Cohen JL, Osley MA. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of Chromatin Assembly Factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/s0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- Kayne PS, Kim UJ, Han M, Mullen JR, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Rose AM. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat Cell Biol. 1999;1:514–521. doi: 10.1038/70309. [DOI] [PubMed] [Google Scholar]
- Krawitz, D.C., Kama, T., and Kaufman, P.D. 2002. Chromatin Assembly Factor-I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- Loidl J, Klein F, Engebrecht J. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. Methods Cell Biol. 1998;53:257–285. doi: 10.1016/s0091-679x(08)60882-1. [DOI] [PubMed] [Google Scholar]
- Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- Lorain S, Quivy JP, Monier-Gavelle F, Scamps C, Lecluse Y, Almouzni G, Lipinski M. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol Cell Biol. 1998;18:5546–5556. doi: 10.1128/mcb.18.9.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maggert KA, Karpen GH. Acquisition and metastability of centromere identity and function: Sequence analysis of a human neocentromere. Genome Res. 2000;10:725–728. doi: 10.1101/gr.10.6.725. [DOI] [PubMed] [Google Scholar]
- Martini E, Roche DM, Marheineke K, Verreault A, Almouzni G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J Cell Biol. 1998;143:563–575. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: Essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Megee PC, Mistrot C, Guacci V, Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes & Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Broach JR. Immunological analysis of yeast chromatin. Methods Enzymol. 1999;304:414–430. doi: 10.1016/s0076-6879(99)04025-2. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Monson EK, de Bruin D, Zakian VA. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: Interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- Osley MA, Lycan D. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol Cell Biol. 1987;7:4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Biol Cell. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto I, Winston F. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 2000;19:1598–1612. doi: 10.1093/emboj/19.7.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Qian Z, Huang H, Hong JY, Burck CL, Johnston SD, Berman J, Carol A, Liebman SW. Yeast Ty1 retrotransposition is stimulated by a synergistic interaction between mutations in Chromatin Assembly Factor-I and histone regulatory (Hir) proteins. Mol Cell Biol. 1998;18:4783–4792. doi: 10.1128/mcb.18.8.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy JP, Grandi P, Almouzni G. Dimerization of the largest subunit of chromatin assembly factor 1: Importance in vitro and during Xenopus early development. EMBO J. 2001;20:2015–2027. doi: 10.1093/emboj/20.8.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J, Dunn B, Raff A, Osley MA. Functional analysis of histones H2A and H2B in transcriptional repression in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2545–2553. doi: 10.1128/mcb.16.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Spierer P. Position effect variegation and chromatin proteins. BioEssays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histonedeacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M, Fitzgerald-Hayes M, Bloom K. Chromatin structure of altered yeast centromeres. Proc Natl Acad Sci. 1988;85:175–179. doi: 10.1073/pnas.85.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders MJ, Yeh E, Grunstein M, Bloom K. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol Cell Biol. 1990;10:5721–5727. doi: 10.1128/mcb.10.11.5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Picard D, Yamamoto KR. Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol. 1991;194:389–398. doi: 10.1016/0076-6879(91)94029-c. [DOI] [PubMed] [Google Scholar]
- Seedorf M, Damelin M, Kahana J, Taura T, Silver PA. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JS, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- Shero JH, Koval M, Spencer F, Palmer RE, Hieter P, Koshland D. Analysis of chromosome segregation in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:749–773. doi: 10.1016/0076-6879(91)94057-j. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- Smith J, Caputo E, Boeke J. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM, Yang P, Santisteban MS, Boone PW, Goldstein AT, Megee PC. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol Cell Biol. 1996;16:1017–1026. doi: 10.1128/mcb.16.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes & Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Sullivan KF. A solid foundation: Functional specialization of centromeric chromatin. Curr Opin Genet Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- Sun X, Wahlstrom J, Karpen G. Molecular structure of a functional Drosophila centromere. Cell. 1997;91:1007–1019. doi: 10.1016/s0092-8674(00)80491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A, Bucaria J, Osley MA, Sternglanz R. Yeast ASF1 protein is required for cell-cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Roche D, Sibarita JB, Turner BM, Almouzni G. Duplication and maintenance of heterochromatin domains. J Cell Biol. 1999;147:1153–1166. doi: 10.1083/jcb.147.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tsuchiya E, Hosotani T, Miyakawa T. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 1998;26:3286–3292. doi: 10.1093/nar/26.13.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-I and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Wiens GR, Sorger PK. Centromeric chromatin and epigenetic effects in kinetochore assembly. Cell. 1998;93:313–316. doi: 10.1016/s0092-8674(00)81157-5. [DOI] [PubMed] [Google Scholar]
- Xu H, Kim UJ, Schuster T, Grunstein M. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5249–5259. doi: 10.1128/mcb.12.11.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Kahana JA, Silver PA, Morphew MK, McIntosh JR, Fitch IT, Carbon J, Saunders WS. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]