Abstract
Previously, identification of promoters regulated by mammalian transcription factors has relied upon overexpression studies. Here we present the identification of a large set of promoters that are bound by E2F in physiological conditions. Probing a human CpG microarray with chromatin immunoprecipitated using an antibody to E2F4, we have identified 68 unique target loci; 15% are bidirectional promoters and 25% recruit E2F via a mechanism distinct from the defined consensus site. Interestingly, although E2F has been shown previously to regulate genes involved in cell cycle progression, many of the new E2F target genes encode proteins involved in DNA repair or recombination. We suggest that human CpG microarrays, in combination with chromatin immunoprecipitation, will allow rapid identification of target promoters for many mammalian transcription factors.
Keywords: E2F, chromatin immunoprecipitation, CpG island microarray, transcription
The precise transcriptional regulation of numerous mammalian genes is required to maintain normal cellular proliferation and differentiation. In fact, many neoplasias and developmental disorders have been linked directly to the aberrant expression of a single transcription factor, which ultimately leads to the deregulation of numerous target genes (Boyd and Farnham 1999). Recent studies have coupled transcription factor overexpression with cDNA microarray analysis in an attempt to gain insight into the network of genes that are regulated by a given factor. However, such studies suffer from two main disadvantages. First, the genes identified in a cDNA microarray analysis are not necessarily direct targets of the overexpressed transcription factor. Rather, it is possible that the deregulated gene expression is due solely to indirect effects resulting from alterations in signal transduction cascades. Second, it is unclear whether the same genes influenced by an overexpressed transcription factor represent the true target genes of that factor at the physiologically relevant concentrations present within the normal cellular environment. Therefore, to address these issues, it is critical to develop a technique in which direct targets of site-specific transcription factors can be identified under biologically relevant conditions.
The E2F family of transcription factors is composed of six members that heterodimerize with DP proteins to form a DNA-binding transcriptional activator. The E2F/DP heterodimer is thought to play a critical role in cell cycle progression through its ability to regulate the expression of target genes that include cyclins, Cdks, and components of the DNA synthesis machinery. Many of the previously characterized E2F target genes are highly expressed at the G1/S phase boundary and are transcriptionally regulated during the cell cycle by a mechanism that is thought to involve the inactivation of the E2F complex through its association with the retinoblastoma (Rb) protein family. Despite the great strides made in an effort to understand the role E2F plays in cellular proliferation and differentiation, there is still much to be uncovered. This is highlighted by recent studies that suggest that E2F may be involved in regulating transcription at the G2 transition of the cell cycle and that E2F perhaps regulates hundreds of genes that were not recognized previously as target genes (Ishida et al. 2001; Muller et al. 2001). However, these recent studies have relied upon E2F overexpression coupled to cDNA microarray analysis, therefore making it unclear whether E2F is involved directly in the regulation of this new subset of potential target genes. Thus, our goal was to examine the composition of E2F target genes by utilizing an unbiased method to isolate genomic sites directly bound by physiological levels of E2F in living cells. We found that by probing a CpG island microarray with chromatin immunoprecipitated using an E2F4 antibody, we were able to quickly and efficiently isolate a large number of E2F target genes to aid in the analysis of E2F function in living cells.
Results
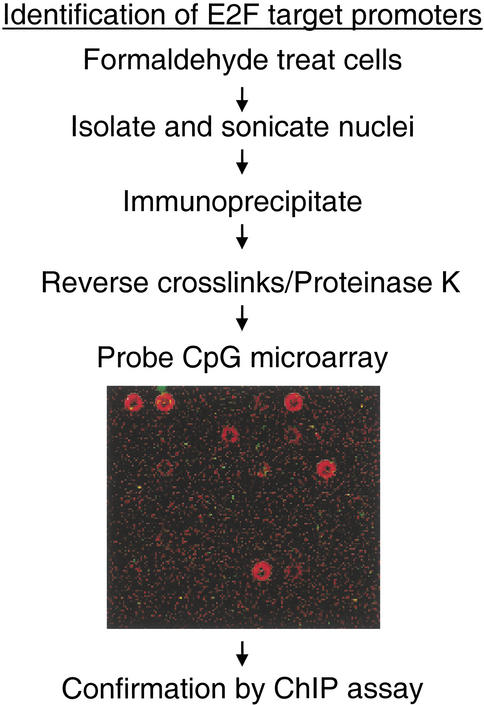
Our goal was to use a technique that would allow for the analysis of both known promoters and promoters for previously uncharacterized mRNAs. It was also important that the technique would allow for the rapid identification of a large number of target promoters. Previously, we utilized a modified chromatin immunoprecipitation protocol to identify several promoters directly bound by the E2F family of transcription factors in vivo (Weinmann et al. 2001). Unfortunately, this method was very laborious and did not allow for a more global analysis of E2F-regulated genes. The ability to combine chromatin immunoprecipitation with microarray analysis is the most promising means to increase both the speed of analysis and the number of targets isolated. In fact, such studies have been performed successfully in the yeast system using yeast genomic microarrays (Ren et al. 2000; Iyer et al. 2001; Lieb et al. 2001). However, a similar analysis in mammalian cells could not be performed because the development of a microarray that represents a major portion of intergenic regions has been hindered due to the vastly greater size of mammalian genomes. To date, the only commercially available human microarrays contain cDNA sequences and would be of little use for the analysis of promoter regions. To circumvent this problem, we have utilized a DNA microarray containing human genomic fragments that were isolated due to their high CpG content (Cross et al. 1994). CpG islands often correspond to promoter regions (Antequera and Bird 1993) and in fact, provide the most reliable measure for promoter prediction (Ioshikhes and Zhang 2000; Hannenhalli and Levy 2001). Therefore, we reasoned that probing a CpG island microarray with chromatin immunoprecipitated with an antibody to a human transcription factor would provide a high throughput method for the identification of in vivo target promoters (Fig. 1).
Figure 1.
Identification of E2F4 targets using a CpG island microarray. Presented is a flow chart describing the methodology used to examine E2F targets. A portion of the CpG island microarray probed with E2F4 (red:Cy5) and the control no antibody (green:Cy3) chromatin is also shown.
A microarray containing 7776 CpG islands was probed with chromatin immunoprecipitated using an antibody to human E2F4 or a no-antibody control. We detected ∼100 spots at which the signal obtained with the E2F4 chromatin was at least twofold higher than the signal obtained probing with the control chromatin. We felt it critical to confirm that the identified clones were bound by E2F in vivo and did not represent DNAs that were false positives due to repeated elements and/or nonspecific precipitation. Therefore, we randomly chose 16 clones for analysis in independent, standard chromatin immunoprecipitation (ChIP) experiments using antibodies to E2F1, E2F4, and RNA polymerase II, as well as a no-antibody control.
The clones contained on the microarray were isolated on the basis of their high CpG content and were not further characterized prior to spotting on the microarray. Therefore, we needed to determine the sequence of the E2F-positive clones to determine their identity. It is important to note that the CpG clone sequence spotted on the microarray provided us with the genomic localization of the ChIP DNA fragments. Because the size of the chromatin generated in the ChIP portion of the procedure was ∼1–2 kb, a positive CpG island clone indicates that a bound E2F site is located somewhere within 1–2 kb of the actual CpG island clone on the microarray. Therefore, to identify the location of the E2F-binding element, the genomic sequences surrounding the CpG island clone needed to be examined as well. Throughout the text, reference to a clone indicates the CpG island and surrounding region.
We first sequenced 16 E2F4-positive CpG clones and examined the surrounding sequence for evidence of a consensus E2F site (TTTSSCGC). Interestingly, of these 16 clones, 5 contained a consensus E2F-binding site in the surrounding region (Table 1; clones 1–5), 5 contained a site with a single base pair mismatch located in the T stretch of the consensus (clones 6–10), and 6 did not contain a sequence closely resembling the consensus site (clones 11–16). For all 16 clones, we prepared primer sets spanning the closest match to a consensus E2F site or alternatively, we chose primers that would span ∼300 bp upstream of the transcription start site of its corresponding gene, because E2F sites are most often found in close proximity to the start site of transcription (Kel et al. 2001). Due to the size of the chromatin used in these experiments, we believe that E2F binding to a site within 1 kb of the primer pair could be easily detected. We found that 14 of the 16 clones (88%) identified by CpG island microarray hybridization were in fact true positives, as shown by a higher specific signal in the E2F4-immunoprecipitated chromatin relative to the no-antibody control (Fig. 2A). Of the two false positives, one (THOX2) appeared to be located adjacent to a clone containing a high-affinity E2F-binding site (CpG18G8), suggesting a potential problem with cross contamination. Of the 14 confirmed positives, 11 showed high-affinity E2F binding in vivo (high-affinity binding is defined as an E2F4 signal intensity greater than or equal to a standarized aliquot of the total signal). All five clones containing a consensus E2F site were confirmed to bind E2F with high affinity in independent ChIP experiments. Of the five clones that contained a single base pair mismatch to the consensus, three showed high-affinity E2F binding in living cells. Interestingly, of the six clones that did not contain any recognizable E2F site, five were bound by E2F in vivo, with three of these bound with high affinity.
Table 1.
E2F targets identified by ChIP-CpG microarray analysis
| Clone
|
Gene
|
Accession No.
|
Consensus E2F site
|
Bidirectional
|
|---|---|---|---|---|
| 1 | TTK (4) | NM 003318 | yes: TTTCCCGC *R | |
| 2 | H2AFP (3) | NM 021064 | yes: TTTCGCGC | yes |
| H2BFR | NM 021058 | |||
| 3 | TYMS (3) | NM 001071 | yes: TTTCCCGC *R | |
| 4 | FLJ10287 (2) | NM 019083 | yes: TTTGGCGC | |
| 5 | Gar22 | NM 006478 | yes: TTTCCCGC | |
| 6 | FLJ11029 (2) | NM 018304 | 1: TCTCCCGC | yes |
| FLJ12758 | AK022820 | |||
| 7 | RECQL | NM 002907 | 1: CTTCCCGC *R | yes |
| CGI-141 | NM 016072 | |||
| 8 | similar to FLJ10891 | BC000807 | 1: CTTCGCGC | |
| 9 | CpG12B10 | Z56578 | 1: TCTCCCGC | |
| 10 | CpG66E11 | Z65907 | 1: TATCCCGC | |
| 11 | CpG32C1 | Z55309 | no | |
| 12 | CpG18G8 | Z57695 | no | |
| 13 | DBPA | L29064 | no | |
| 14 | UXT | NM 004182 | no | |
| 15 | CYP27B1 | NM 000785 | no | |
| 16 | THOX2 | AF230496 | no | |
| 17 | ESTs | AW503861 | yes: TTTCCCGC *R | |
| 18 | RAD51 (5) | NM 002875 | 1: ATTCCCGC *R | |
| 19 | ESTs (5) | AA303712 | no | |
| 20 | DKFZp586C1942 | AL136941 | 1: TTACGCGC *R | |
| 21 | DHPS | NM 013407 | no | |
| 22 | N-Ras related gene | NM 007158 | 1: CTTCCCGC | |
| 23 | SAAS | NM 013271 | no | |
| 24 | EPAS1 | NM 001430 | 1: TGTGGCGC | |
| 25 | clone sc75-F2 | 1: ATTCCCGC | ||
| 26 | predicted | 1: TTGCGCGC *R | ||
| 27 | MTHFD1 | NM 005956 | yes: TTTCCCGC | |
| 28 | DLEU1 | NM 005887 | 1: TCTCCCGC | yes |
| DLEU2 | NM 006021 | |||
| 29 | H4F2 (3) | NM 003548 | 1: CTTCCCGC | |
| 30 | MFAP1 (2) | NM 005926 | yes: TTTCCCGC | yes |
| FLJ12973 | NM 024908 | |||
| 31 | HPR6.6 | NM 006667 | 1: TCTGGCGC | |
| 32 | GTF2H4 | NM 001517 | 1: TTCGCCGC | |
| 33 | DDX11 | NM 030655 | 1: GTTCGCGC | |
| 34 | RBBP5 | NM 005057 | 1: TGTCGCGC | |
| 35 | RPA3 | NM 002947 | yes: TTTCCCGC | |
| 36 | clone PY1-H5 | 1: CTTGGCGC | ||
| 37 | UBCH10 | NM 007019 | 1: TCTGCCGC | |
| 38 | HP1α (5) | NM 012117 | 1: TTAGGCGC *R | yes |
| HNRPA1 | NM 002136 | |||
| 39 | FOXD3 | NM 012183 | 1: TTCCCCGC *R | |
| 40 | fls353 | AB024704 | 1: GTTCGCGC | |
| 41 | FLJ12190 | NM 025071 | 1: GTTCCCGC | |
| 42 | ESTs | AL529783 | no | |
| 43 | H3FL | NM 003537 | 1: TCTCGCGC *R | yes |
| H2AFM | NM 003513 | |||
| 44 | clone DL3-C10 | 1: TCTGCCGC | ||
| 45 | CDC25C | NM 022809 | no | |
| 46 | ESTs | BG418908 | no | |
| 47 | SNRPC | NM 003093 | 1: CTTCCCGC | |
| 48 | SMARCA5 | NM 003601 | 1: ATTCCCGC | |
| 49 | FLJ10466 | NM 018100 | 1: TTCCCCGC | |
| 50 | MGC11266 | NM 024322 | 1: ATTCCCGC *R | |
| 51 | MCM10 homolog | AB042719 | 1: TCTGGCGC *R | |
| 52 | ESTs | AL039875 | no | |
| 53 | NASP | AF035191 | no | |
| 54 | ESTs | AL515034 | 1: CTTGGCGC | |
| 55 | RPS16 | NM 001020 | 1: TTAGCCGC | |
| 56 | ESTs | AI672018 | 1: TTCCGCGC | |
| 57 | DKFZP564M082 | NM 014042 | 1: TGTGCCGC *R | |
| 58 | HT007 | NM 018380 | yes: TTTCCCGC | |
| 59 | DKFZp564C0482 | AL050353 | 1: CTTCCCGC *R | yes |
| ALG5 | NM 013338 | |||
| 60 | clone sc11-E2 | no | ||
| 61 | FANCD2 | AF340183 | yes: TTTCCCGC *R | |
| 62 | ESTs | BC006444 | no | |
| 63 | BM037 | NM 018454 | yes: TTTGGCGC | yes |
| OIP5 | AF025441 | |||
| 64 | clone sc43-H2 | no | ||
| 65 | clone sc88-B4 | 1: TCTCCCGC | ||
| 66 | clone PY2-F8 | 1: GTTCGCGC *R | ||
| 67 | FLJ11193 | NM 018356 | 1: ATTGGCGC | yes |
| RN3 | AF189011 | |||
| 68 | clone DL2-C3 | yes: TTTCCCGC |
A table listing the 68 unique loci identified as E2F targets in the CpG island microarray analysis is shown. Listed are the corresponding gene name and accession number for the most likely gene regulated by the CpG island clone identified in this study, as determined by genomic database searches. The number of times a clone was isolated (if >1) is indicated in parenthesis next to the name of the corresponding gene. The sequences of all identified consensus E2F sites are indicated, as are the sequences of all sites in which one of the three T's is altered (indicated by 1 followed by the sequence). The *R indicates the sequence is present in the reverse orientation, relative to the start site, in the genome. The presence of a bidirectional promoter (a promoter driving the expression of two mRNAs in opposing directions) is also indicated in the table, with both genes being listed for that clone.
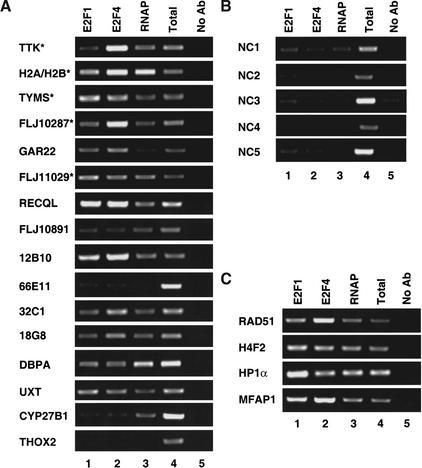
Figure 2.
Confirmation of E2F targets. A representative chromatin immunoprecipitation experiment using HeLa cells is shown (A–C). Antibodies against E2F1 (lane 1), E2F4 (lane 2), RNA polymerase II (lane 3), or no antibody (lane 5) were used. Following DNA purification, samples were subjected to PCR with primers designed to the individual loci. A fixed portion of the total input (0.2%) was also examined by PCR (lane 4). (A) Examination of 16 randomly chosen positive clones identified as E2F targets in the CpG island microarray by ChIP. The first five clones contain a consensus E2F site, the next five clones contain a 7/8-bp match to the consensus, and the last six clones contain no recognizable E2F site. (*) Loci that were multiple hits in the microarray analysis. (B) Examination of five randomly chosen negative clones by ChIP. (C) Loci identified by multiple hits in the CpG island microarray are examined in a ChIP experiment.
As a negative control to ensure that not all clones spotted on the microarray would bind nonspecifically to E2F in the ChIP assay, we randomly chose five clones that did not test positive in the initial hybridization with the E2F4-precipitated chromatin. Sequence analysis of these five clones revealed that one contained a consensus E2F site in the surrounding region. Again, primers were designed spanning the closest match to an E2F consensus site. The analysis of these clones using the ChIP assay revealed that they did not constitute high-affinity E2F-binding sites (Fig. 2B). Of particular interest is the fact that the negative clone that contained a consensus site was not bound by E2F with high affinity in vivo, rather only a low-level signal was detected (Fig. 2B; NC1). Database analysis suggests that the consensus E2F site found in the corresponding gene to this clone is located downstream of the transcription start site in the first intron. We have shown previously that E2F sites in the exons of the Myc and Hox3D genes are not bound with high affinity in vivo (Weinmann et al. 2001). Thus, both our past and present studies support the hypothesis that E2F sites found in promoter regions are bound with a higher occupancy rate than E2F sites in other regions of the genome. All of the 16 positive and 5 negative clones were also examined in an additional chromatin immunoprecipitation experiment; results similar to those shown in Figure 2 were obtained (data not shown).
Having confirmed that our method for detecting E2F target promoters was reliable, we sequenced all of the positive clones and identified their chromosomal location using the University of California, Santa Cruz human genomic database (http://genome.cse.ucsc.edu/). Several conclusions concerning these clones can be drawn. First, a majority of the clones are promoters as indicated by their proximity to the start of an mRNA. This data strongly suggests that our hypothesis that a CpG island microarray can serve as a promoter-enriched microarray is valid. Because the clones were not characterized prior to spotting onto the microarray, it is likely that some CpG islands are represented multiple times on the array. However, it is unlikely that any one CpG island represents a significant proportion of the almost 8000 clones. Therefore, we reasoned that multiple hits found in the positive clone population most likely correspond to true E2F targets. Sequence analysis indicated that CpG islands corresponding to the promoter region for nine known genes and one EST cluster were identified multiple times. Five of the nine known genes that were identified multiple times were present in the first sixteen randomly chosen clones and were confirmed to be E2F targets in the experiments shown in Figure 2A. We tested the additional four promoters that were identified multiple times and found that each one showed high-affinity E2F binding in vivo (Fig. 2C). Therefore, we conclude that multiple, independent positive signals corresponding to a given promoter provides high confidence that the CpG clone is a true positive.
Due to the over-representation of 10 loci and the lack of genomic sequence verification on several clones, only 68 different loci are represented in the positive clones; a complete list of the 68 identified loci can be found in Table 1. A total of 19% of these clones contain a perfect match to the consensus E2F site and 56% of the promoter regions contain a 7 out of 8-bp match to the consensus (with the mismatch being located in the T stretch of the consensus). E2F has been shown to regulate promoters such as B-myb via a site in which one of the T's in the consensus is replaced by a C (Lam and Watson 1993). Therefore, it is likely that many of the positive clones that correspond to promoters with at least a 7 out of 8-bp match to the consensus are in fact regulated by E2F. Of interest are the clones that do not have a close match to a consensus E2F site. Although some may be false positives, we have shown that a high percentage of these clones are bound by E2F in vivo. For example, we have analyzed 12 different clones that do not have a close match to an E2F site within 1 kb of the start site of transcription. In independent chromatin immunoprecipitation assays, we found that nine of these clones showed robust binding to E2Fs (clones 11–14, 19, 42, 46, 53, and 62 from Table 1), one showed weak binding (clone 15), and two did not show E2F binding within 1 kb of the start site (clones 16 and 23) (Fig. 2A; data not shown). This indicates that the vast majority of the identified clones that lack a recognizable E2F site are bona fide in vivo E2F target promoters. These data suggest that there are at least two classes of E2F-regulated promoters, those containing a close match to the well-characterized consensus sequence and those to which E2F is recruited via an alternative mechanism.
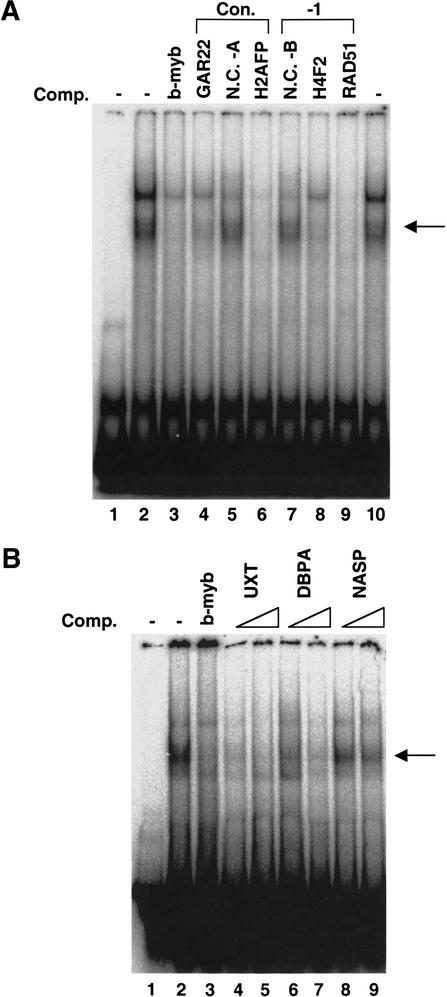
It is possible that cooperative binding between E2F and another factor allows binding in vivo that would not be predicted based solely upon sequence inspection. Alternatively, E2F may be able to directly and independently bind to a sequence distinct from the previously derived consensus site. To begin to characterize the E2F binding to the newly identified target promoters, we performed electromobility shift assays (Fig. 3). We chose seven different promoters identified by our ChIP–CpG microarray analysis for further investigation; two of these contain a consensus E2F site, two contain a 7 out of 8-bp match to the consensus, and three do not contain a recognizable E2F-binding site. We first tested whether the identified consensus site or the 7 out of 8-bp match to the consensus site were, in fact, responsible for recruiting E2F to the promoters. We prepared PCR fragments from the GAR22, H2AFP, H4F2, and RAD51 promoters that spanned the putative E2F sites. As negative controls, we also prepared two fragments that did not contain any matches to an E2F-binding site. The well-characterized E2F site from the B-myb promoter was used as a probe. Incubation of this probe with HeLa nuclear extract resulted in a upward shift of the probe, creating a band that we have shown previously by antibody supershift analysis to be composed of E2F/DP complexes (Weinmann et al. 2001). Binding of E2F to the target promoters was assayed by the ability of the unlabeled PCR fragments to compete for E2F binding to the radiolabeled probe. As shown in Figure 3A, the fragments containing the consensus or 1-bp mismatch E2F sites competed for binding (lanes 4, 6, 8, and 9), whereas the fragments that lacked an E2F site did not compete (lanes 5 and 7).
Figure 3.
In vitro confirmation of E2F binding to newly identified target promoters. (A) An electromobility shift competition experiment was performed using the E2F site from the B-myb promoter as a probe. Lane 1 contains the probe alone. In lanes 2–10, the probe was incubated with HeLa nuclear extract and either no competitor DNA (lanes 2,10), the unlabeled B-myb E2F site oligonucleotide (lane 3), fragments containing E2F sites from the identified promoters indicated above the gel image (lanes 4,6,8,9), or fragments which do not contain E2F sites (lanes 5,7). NC-A (negative control A) and NC-B (negative control B) are PCR fragments generated using the same method as the specific competitors. These fragments were derived from promoters that contain a consensus E2F site (NC-A) or a 7 out of 8-bp match to the consensus E2F-binding site promoter (NC-B), but the fragments lack the region containing the E2F site. Arrows to the right of the gel image in A and B indicate the specific E2F/DP complex. (B) An EMSA competition experiment was performed using the E2F site from the B-myb promoter as a probe. Lane 1 contains the probe alone. In lanes 2–9, the probe was incubated with HeLa nuclear extract and either no competitor DNA (lane 2), the unlabeled B-myb E2F site oligonucleotide (lane 3), or two different concentrations of fragments from the identified promoters UXT (lanes 4,5), DBPA (lanes 6,7), and NASP (lanes 8,9). Increasing competitor concentrations are indicated above the gel image.
Of greater interest are the clones that bind E2F in vivo but have no recognizable E2F site (Fig. 3B). We prepared 300-bp fragments near the transcription start site of the UXT, DBPA, and NASP promoters to be used as competitors in an EMSA competition experiment. The UXT promoter fragment effectively competed E2F binding at both low and high DNA competitor concentrations, the DBPA promoter fragment competed strongly only at higher DNA concentrations (this promoter also showed weaker binding in vivo), and the NASP promoter fragment did not compete well for binding of E2F to the B-myb probe (Fig. 3B). We note that the NASP promoter fragment that did not compete in vitro did display reproducible high-affinity in vivo E2F binding. Although the UXT, DBPA, and NASP promoters all lack a recognizable E2F site, they may recruit E2F via distinct mechanisms. Perhaps E2F binds directly and independently to the UXT and DBPA promoters, but the binding of E2F to the NASP promoter requires protein–protein interactions that are not reproduced in vitro. In summary, using an independent method, we have confirmed that E2F does, in fact, bind to the identified promoters, even those that do not contain a consensus E2F site.
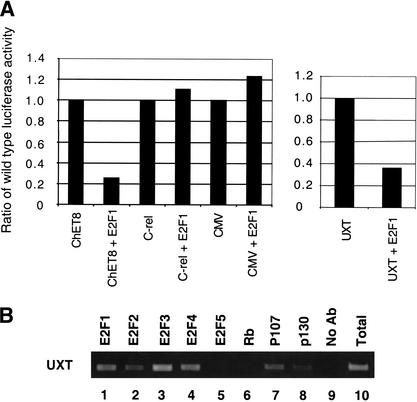
We also wished to further determine the functional consequence of E2F binding using an independent assay. We chose to examine the UXT promoter for this analysis because it binds E2F both in vitro (Fig. 3B) and in vivo (Fig. 2A), but lacks a recognizable E2F consensus binding site. Thus, an analysis of the UXT promoter may provide novel insight into the role E2F plays at this subset of promoters. Previously, we isolated the ChET8 promoter using a ChIP cloning strategy (Weinmann et al. 2001). This promoter also lacks a site closely resembling the E2F consensus site and surprisingly, E2F overexpression repressed ChET8 promoter activity in transient transfection analysis (Weinmann et al. 2001; Fig. 4A). Interestingly, E2F overexpression also repressed promoter activity of a UXT promoter–reporter construct (Fig. 4A). This repression is specific because two control promoter–reporter constructs were not affected by E2F overepression (Fig. 4A). Most E2F target promoters have been shown previously to be activated by E2F family members. In contrast, our findings suggest that the E2F target promoters lacking a site closely resembling the E2F consensus site may be regulated through a different mechanism. Therefore, the atypical set of E2F target promoters we have identified through our unbiased in vivo approach may help to define a new role for E2F in transcriptional regulation. It is possible that promoter context and/or nuclear environment play a larger role in the ultimate ability of E2F to act as an activator or repressor than was thought previously.
Figure 4.
Characterization of UXT promoter. (A) Fragments of the UXT, ChET8, c-Rel, or CMV promoters were cloned upstream of the luciferase reporter and used in transient transfection experiments. 3T3 cells were transfected with either a promoter luciferase reporter construct alone or with an E2F1 expression plasmid as indicated on the x-axis. The specific promoter used in each transfection is also indicated on the x-axis. The y-axis represents the relative activity of each construct transfected alone or with E2F1 overexpression. Ratios are relative to the individual construct activity without E2F overexpression. (B) A ChIP experiment utilizing antibodies to E2F1 (lane 1), E2F2 (lane 2), E2F3 (lane 3), E2F4 (lane 4), E2F5 (lane 5), Rb (lane 6), p107 (lane 7), p130 (lane 8), or a no-antibody control (lane 9) are shown. PCR was performed with a primer set specific to the UXT promoter. A standardized aliquot of the total input chromatin is also shown (lane 10).
We next wished to determine whether members of the Rb pocket protein family are recruited to the UXT promoter in vivo. As shown in Figure 4B, the pocket protein family members p107 and p130 are recruited to the UXT promoter as determined by ChIP analysis. It is also worth noting that similar to previously characterized E2F-regulated promoters, there does not seem to be a preferential recruitment of an individual E2F family member to the UXT promoter region.
The analysis of the UXT promoter by use of three independent assays provides validation that the promoters isolated using the ChIP–CpG cloning strategy are likely bona fide E2F targets. Standard ChIP experiments showed strong binding of E2F to the UXT promoter in living cells (Figs. 2A and 4B). In addition, EMSA competition experiments suggested that E2F can bind to the UXT promoter in vitro (Fig. 3B). Finally, E2F overexpression specifically repressed a UXT promoter–reporter construct, suggesting a functional consequence for the association of E2F with this promoter (Fig. 4A). In addition to providing validation that the target genes isolated using this approach are in fact true targets, these experiments also provide novel insight into our understanding of E2F-regulated transcription. It is interesting to note that ∼25% of the promoters isolated lack a site closely matching the E2F consensus site and that both promoters (UXT and ChET8) analyzed to date, which lack a consensus E2F site, are repressed by E2F overexpression. Determining the context-dependent requirements for the binding of E2F to these promoters and the mechanism of E2F repression will provide insight into this novel mechanism for E2F regulation.
Discussion
We have shown that chromatin immunoprecipitated with an antibody to E2F4 can be used as a hybridization probe for a CpG island microarray to allow for an unbiased and rapid identification of a large set of target promoters. The E2F family has been implicated previously in the regulation of genes involved in cell cycle control. However, as described below, most of the genes identified using our unbiased approach for the detection of E2F target promoters were not involved in cell cycle progression. Rather, our studies have pointed to new roles for E2F in the cell; in particular, we have linked the E2F family to DNA repair and recombination.
In addition to the analysis of sequence elements and context-dependent requirements for E2F binding to target genes in living cells, the large data set collected can also be used to examine potential commonalities of E2F-regulated genes. In fact, computer-based inspection revealed three distinct sequence elements (a 9-bp dyad, a 10-bp element containing a direct repeat, and an 18-bp element) that are each represented in a significant proportion of the target genes isolated in this analysis (M. Zhang and P. Farnham, unpubl.). Interestingly, 11 of the identified E2F target promoters contain all 3 novel sequence elements within 1 kb of the start site for transcription. The identity of these 11 promoters and the sequence from each promoter that matches the 3 elements can be found at http://mcardle.oncology.wisc.edu/farnham/. These findings illustrate the potential power of analyzing large data sets, which will make it possible to examine the nature of transcription factor target gene regulation in a more global perspective. It will be interesting to determine in future studies whether the common sequence elements are, in fact, involved in regulating a subset of E2F target genes that encode proteins involved in a common function.
It is also worth noting the unusually high frequency of bidirectional promoters that were found to be bound by E2F in this analysis (15%). Many gene clusters have been shown to regulate a common function and their coordinated expression is critical for maintaining a particular process. For instance, the histone genes are clustered and coordinated expression is beneficial to the assembly of chromatin structure. It will be interesting to determine in future studies whether E2F influences the expression of one or both of the genes in the various clusters and whether there is biological significance to the high frequency of bidirectional promoters regulated by E2F.
Of the 68 identified loci, 36 have been characterized previously; the others correspond to the promoter regions of expressed sequence tags (ESTs) and/or mRNAs that code for proteins of unknown function. Of the 36 genes that have been thoroughly characterized, 4 correspond to genes that have been implicated previously as E2F targets. These include the promoters for H2A/H2B (Oswald et al. 1996) and thymidylate synthase (DeGregori et al. 1995; Kasahara et al. 2000), which have been shown previously to be regulated by E2F. In addition, the mRNAs of GAR22 and RAD51 have been shown to be regulated upon overexpression of E2F family members (Ishida et al. 2001; Muller et al. 2001); our studies provide evidence that both of these genes are directly regulated by E2Fs. Furthermore, the isolation of known E2F target genes provides further validation that this technique can be used to successfully identify promoters bound and regulated by a specific transcription factor in living cells. Due to the limitations of designing a complete microarray containing sequences corresponding to the entire human genome, this technique cannot be used as an exhaustive search for every E2F-binding site within the genome, but rather, it allows for the isolation of a large subset of target genes. In addition, the data needs to be interpreted with caution concerning the exact role and extent a factor plays in the regulation of any individual target gene. It remains possible that a subset of promoters bound by E2F will not be regulated by E2F. However, because we identified these promoters as bound by E2F family members in a living cell under normal physiological conditions (i.e., without overexpression of an E2F), it is likely that many, if not most, will be regulated by E2F.
A summary of the characteristics of the identified genes can be found in Figure 5. We note that 6/36 (17%) of the characterized genes encode either histones (H2A/H2B, H3, H4F2) or chromatin remodeling factors (HP1α, SMARCA5, and NASP). In addition, 7/36 (19%) of the genes correspond to replication, recombination, or DNA repair proteins. The observation that almost one-half of the characterized genes regulate DNA structure or function may explain why overexpression of E2F can have severe biological consequences (Johnson et al. 1993; Qin et al. 1994; Shan and Lee 1994; Wu and Levine 1994; Kowalik et al. 1995; Lee and Farnham 2000). Our finding that E2F transcriptionally regulates DNA repair genes, in combination with the previous observation that E2F can physically interact with DNA repair proteins (Maser et al. 2001) suggests that there is an intricate involvement of the E2F family in the process of DNA repair. It is also worth noting that the identification of HP1α as an E2F target gene provides the interesting possibility of a negative feedback loop controlling E2F target gene expression. Recently, it was shown that HP1α is recruited to the cyclin E promoter through its ability to associate with the E2F/Rb complex (Nielsen et al. 2001). This interaction is thought to lead to the down-regulation of cyclin E transcription through alterations in chromatin structure. That both Rb (Shan et al. 1994) and HP1α (this study) are E2F targets suggests that E2F regulates the intricate timing of the expression of its target genes through negative feedback mechanisms.
Figure 5.
Characterization of E2F targets. A summary of the results obtained after sequence analysis of the positive clones. The results of the ChIP assay for the first 16 randomly chosen clones is presented, as is a list of loci represented by multiple hits, a list of known E2F targets, and a list of targets involved in chromatin structure and DNA replication/repair/recombination.
It has been postulated, on the basis of E2F overexpression and cDNA microarray analysis, that E2F may play a role in regulating genes during G2, however it was unknown whether E2F overexpression was directly or indirectly involved in the regulation of these genes (Ishida et al. 2001). We note that the promoter regions for the TTK and Cdc25C genes were identified in our study, and both of these genes have been shown to be expressed during the G2 phase of the cell cycle (Ishida et al. 2001). Our results provide direct evidence that E2F is bound to the promoters of at least two G2-regulated genes, making it possible that E2F is involved in gene regulation during G2. These findings again illustrate that E2F is most likely playing roles in cellular life that have been overlooked in previous analyses.
In summary, we suggest that the combination of chromatin immunoprecipitation and CpG microarrays will be useful for the identification of target genes for many mammalian transcription factors, and will be especially useful for the analysis of transcription factors for which only a few bona fide direct target promoters have been identified previously. Although transcription factors known to bind to and regulate CpG-rich promoters will be most easily studied using this method, it is worth noting that we have recently used chromatin immunoprecipitation and CpG microarray analysis to identify binding sites for another human transcription factor (A.S. Weinmann and P.J. Farnham, unpubl.). A subset of the identified binding sites are located several kilobases from the start site of the corresponding gene. Thus, this method can identify binding sites for factors that do not regulate CpG-rich promoters, as long as the regulatory regions are themselves CpG rich. This technique will also provide the means for a rapid collection of a large data set of high-affinity binding sites to be used in the development of an in vivo consensus site that perhaps may differ from a consensus developed using standard in vitro methods.
Materials and methods
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation assay was performed as described previously using HeLa cells (Weinmann et al. 2001) and a detailed protocol can be found at http://mcardle.oncology.wisc.edu/farnham/. Chromatin was sheared to an average length of 1–3 kb for this analysis.
CpG island microarray analysis
A total of 7776 CpG island clones derived from the CGI genomic library (UK Genome Mapping Project Centre) prescreened for human Cot-1 DNA were organized individually in 96-well culture chambers as master plates. CpG island inserts (0.2–2 kb) from these clones were amplified by PCR as described (Cross et al. 1994; Huang et al. 1999). The Affymetrix/GMS 417 microarrayer arrayed unpurified PCR products (∼0.02 μL per dot, 0.1 μg/μL), in the presence of 20% DMSO, as microdots (150 μm diameter spaced at 300 μm) on poly-L-lysine-coated microscope slides. Spotted DNA was post-processed and denatured before use. A total of 50 individual chromatin immunoprecipitations were performed, using 1 × 107 cells for each sample. Additionally, 50 individual ChIP reactions were performed in which the primary antibody was omitted to provide a no-antibody control. The combined E2F4-specific samples and the combined no-antibody samples were then labeled and used to probe the microarray as described previously (Yan et al. 2001). Incorporation of amino-allyl dUTP (aa-dUTP, Sigma) into 2 μg each of E2F4-precipitated DNA and control no-antibody DNA was conducted using the Bioprime DNA-labeling system protocol (Life Technologies). Cy5 and Cy3 fluorescent dyes were coupled to aa-dUTP-labeled E2F4-precipitated DNA and control DNA, respectively, and cohybridized to the microarray panel. Microarray protocols including the hybridization and post-hybridization washing procedures are according to protocols developed by DeRisi and colleagues and can be found at (http://www.microarrays.org). Hybridized slides were scanned with the GenePix 4000A scanner (Axon) and the acquired images were analyzed with the software GenePix Pro 3.0. CpG island tags having a Cy5/Cy3 ratio >2 were chosen as E2F4-specific signals (for review, see Yan et al. 2001).
Electromobility shift assays
In vitro E2F DNA-binding activity was assayed using EMSA competition experiments. Approximately 6 μg of HeLa nuclear extract was incubated with 2.5 μg of sonicated salmon sperm DNA and 2 μL of 5×-500 buffer (100 Hepes at pH 7.4, 500 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 35% glycerol, and 5 mM NaF) in a total volume of 18 μL for 10 min at room temperature. A 34-bp double-stranded oligonucleotide containing the E2F site from the B-myb promoter (end labeled using T4 polynucleotide kinase and [γ-32P]ATP) was then added in 2 μL of water and the incubation continued for 20 min. The competitor DNA was generated by PCR and included in the first incubation at a 50-fold molar excess to the labeled probe. The reactions were electrophoresed for ∼2 h on a 4% polyacrylamide gel that had been pre-electrophoresed for 30 min.
Transient transfection analysis
Transient transfection and luciferase analyses were performed as described previously (Weinmann et al. 2001).
Acknowledgments
This work was supported in part by Public Health Service grant CA45250 (to P.J.F.), CA07175 (a NCI Cancer Center Core grant), R33CA84701 (T.H-M.H.), training grant CA09681 (A.S.W.) from the National Institutes of Health, and the Molecular and Environmental Toxicology Center training grant NIEHS144KH84 (M.J.O.). We thank members of the Farnham laboratory for helpful discussions and technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL farnham@oncology.wisc.edu; FAX (608) 262-2824.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.943102.
References
- Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Farnham PJ. Identification of target genes of oncogenic transcription factors. Proc Soc Exp Biol Med. 1999;222:9–28. doi: 10.1111/j.1525-1373.1999.09992.x. [DOI] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S, Levy S. Promoter prediction in the human genome. Bioinformatics. 2001;17:S90–S96. doi: 10.1093/bioinformatics/17.suppl_1.s90. [DOI] [PubMed] [Google Scholar]
- Huang TH-M, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Human Mol Gen. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- Ioshikhes IP, Zhang MQ. Large-scale human promoter mapping using CpG islands. Nat Genet. 2000;26:61–63. doi: 10.1038/79189. [DOI] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factor SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Takahashi Y, Nagata T, Asai S, Eguchi T, Ishii Y, Fujii M, Ishikawa K. Thymidylate synthase expression correlates closely with E2F1 expression in colon cancer. Clin Cancer Res. 2000;6:2707–2711. [PubMed] [Google Scholar]
- Kel AE, Kel-Margoulis OV, Farnham PJ, Bartley SM, Wingender E, Zhang MQ. Computer-assisted identification of cell cycle-related genes - new targets for E2F transcription factors. J Mol Biol. 2001;309:99–120. doi: 10.1006/jmbi.2001.4650. [DOI] [PubMed] [Google Scholar]
- Kowalik TF, DeGregori J, Schwarz JK, Nevins JR. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam EW-F, Watson RJ. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TA, Farnham PJ. Exogenous E2F1 is growth inhibitory before, during, and after neoplastic transformation. Oncogene. 2000;19:2257–2268. doi: 10.1038/sj.onc.1203556. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein B, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Maser RS, Mirzoeva OK, Wells J, Olivares H, Williams BR, Zinkel R, Farnham PJ, Petrini JHJ. The MRE11 complex and DNA replication: Linkage to E2F and sites of DNA synthesis. Mol Cell Biol. 2001;21:6006–6016. doi: 10.1128/MCB.21.17.6006-6016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes & Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer U-M, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Oswald F, Dobner T, Lipp M. The E2F transcription ractor activates a replication-dependent Human H2A gene in early S phase of the cell cycle. Mol Cell Biol. 1996;16:1889–1895. doi: 10.1128/mcb.16.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X-Q, Livingston DM, Kaelin WG, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Shan B, Lee W-H. Deregulated expression of E2F1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B, Chang C-Y, Jones D, Lee W-H. The transcription factor E2F-1 mediates the autoregulation of RB gene expression. Mol Cell Biol. 1994;14:299–309. doi: 10.1128/mcb.14.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Bartley SM, Zhang MQ, Zhang T, Farnham PJ. The use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci. 1994;91:1–5. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan PS, Chen C-M, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, Huang TH-M. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]