Abstract
The Artemis nuclease is defective in radiosensitive severe combined immunodeficiency patients and is required for the repair of a subset of ionising radiation induced DNA double-strand breaks (DSBs) in an ATM and DNA-PK dependent process. Here, we show that Artemis phosphorylation by ATM and DNA-PK in vitro is primarily attributable to S503, S516 and S645 and demonstrate ATM dependent phosphorylation at serine 645 in vivo. However, analysis of multisite phosphorylation mutants of Artemis demonstrates that Artemis phosphorylation is dispensable for endonuclease activity in vitro and for DSB repair and V(D)J recombination in vivo. Importantly, DNA-dependent protein kinase catalytic subunit (DNA-PKcs) autophosphorylation at the T2609–T2647 cluster, in the presence of Ku and target DNA, is required for Artemis-mediated endonuclease activity. Moreover, autophosphorylated DNA-PKcs stably associates with Ku-bound DNA with large single-stranded overhangs until overhang cleavage by Artemis. We propose that autophosphorylation triggers conformational changes in DNA-PK that enhance Artemis cleavage at single-strand to double-strand DNA junctions. These findings demonstrate that DNA-PK autophosphorylation regulates Artemis access to DNA ends, providing insight into the mechanism of Artemis mediated DNA end processing.
Keywords: Artemis, autophosphorylation, DNA-PK, nonhomologous end-joining
Introduction
DNA double-strand breaks (DSBs) can lead to cell death or mutagenic genomic rearrangements if left unrepaired or misrepaired. Nonhomologous DNA end joining (NHEJ), a major DSB repair mechanism in mammalian cells, requires six ‘core' proteins: the Ku70 and Ku80 (Ku) heterodimer, the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) and the complex of Xrcc4, DNA ligase IV and XLF (Meek et al, 2004; Hefferin and Tomkinson, 2005; Ahnesorg et al, 2006; Buck et al, 2006). Cells defective in any of these components are radiosensitive, DSB repair deficient and impaired in V(D)J recombination, a process that requires NHEJ. The Artemis nuclease has been described as an additional NHEJ component and is mutated in individuals with radiosensitive severe combined immunodeficiency (RS-SCID) (Moshous et al, 2001). Artemis cleaves DNA hairpin intermediates during V(D)J recombination in an ATM-independent manner (Ma et al, 2002); however, it mediates the repair of a fraction (∼10%) of DSBs incurred after ionising radiation (IR) in an ATM-dependent manner (Riballo et al, 2004). Current models suggest that Artemis functions to process the ends of otherwise nonligatable DSBs prior to ligation by core NHEJ factors (Lobrich and Jeggo, 2005).
The nuclease activity of Artemis is conferred by β-Lactamase (aa1–135) and β-CASP (aa155–385) domains within its N-terminus. In vitro, Artemis has intrinsic 5′–3′ single-stranded DNA exonuclease activity and, in the presence of ATP and DNA-PKcs, gains DNA endonuclease activity that specifically targets single-stranded to double-stranded DNA (ssDNA–dsDNA) junctions (including 5′ or 3′ overhangs, hairpins, flaps, bubbles, loops and gaps) (Ma et al, 2002, 2005a). The mechanism of Artemis activation in vivo is unclear, although Artemis is rapidly hyperphosphorylated in an ATM-dependent manner after exposure to DSB-inducing agents (Poinsignon et al, 2004; Riballo et al, 2004; Zhang et al, 2004; Chen et al, 2005; Ma et al, 2005b; Wang et al, 2005). ATM and other phosphatidylinositol 3-kinase like kinases (PIKKs), including DNA-PKcs, preferentially phosphorylate serine or threonine followed by glutamine (S/T-Q) motifs. Artemis contains 10 such sites, of which eight are located in the C-terminal 200 amino acids. Artemis cDNA mutated in seven of these sites was able to complement the radiosensitivity of Artemis-deficient cells (Poinsignon et al, 2004). Despite this, other studies have suggested that phosphorylation of Artemis by DNA-PKcs leads to endonuclease activation (Ma et al, 2002, 2004, 2005b).
DNA-PKcs undergoes autophosphorylation within two distinct regions: the ABCDE (T2609, S2612, T2620, S2624, T2638 and T2647) and PQR cluster (S2023, S2029, S2041, S2051, S2053 and S2056) (Chan et al, 2002; Douglas et al, 2002; Ding et al, 2003; Cui et al, 2005). Phosphorylation site mutants for the ABCDE cluster of DNA-PKcs fail to rescue the radiosensitivity, DSB repair defect or V(D)J recombination deficiency of DNA-PKcs mutant cells, implicating DNA-PKcs autophosphorylation as a critical step within NHEJ in vivo. We have suggested that DNA-PKcs autophosphorylation is required for ‘remodelling' of the DNA-PK holoenzyme (comprised of DNA, DNA-PKcs and Ku), to enable ligation of bound DNA ends by Xrcc4-DNA Ligase IV (Block et al, 2004; Meek et al, 2004; Reddy et al, 2004). Moreover, regulation of DSB end accessibility by DNA-PKcs autophosphorylation at ABCDE and PQR may influence the ‘choice' between NHEJ and HR (Cui et al, 2005). Notwithstanding these models, the precise mechanistic role of DNA-PKcs and its autophosphorylation in NHEJ remains to be substantiated.
Ku has been shown to be dispensable for DNA-PKcs stimulated Artemis endonuclease activity in vitro (Ma et al, 2002). Since Ku is essential for NHEJ in vivo (Taccioli et al, 1994; Zhu et al, 1996; Gu et al, 1997; Nussenzweig et al, 1997), stimulates DNA-PKcs protein kinase activity in vitro (Gottlieb and Jackson, 1993; Hartley et al, 1995) and is required for higher order DNA-PK holoenzyme formation (Merkle et al, 2002; Calsou et al, 2003), it is unclear how to reconcile its lack of function with respect to Artemis activation.
Here, we examine the impact of DNA-PKcs, Ku and ATM on Artemis activity in vitro and DSB repair in vivo. We demonstrate that Ku is required for DNA-PKcs to support Artemis endonuclease activity at physiological salt concentrations and that ATM is incapable of mediating Artemis endonuclease activity in vitro. We identify the major ATM/DNA-PK phosphorylation sites within Artemis and demonstrate ATM-dependent phosphorylation of S645 in vivo. However, we show that DNA-PKcs autophosphorylation at the ABCDE cluster rather than Artemis phosphorylation is required for Artemis endonuclease activity. Further, we show that autophosphorylated DNA-PKcs remains stably associated with duplex DNA bearing large single-stranded DNA overhangs until cleavage by Artemis. We present a model for the cooperative role of Artemis and DNA-PK in DNA end processing.
Results and discussion
Artemis endonuclease activity is supported by DNA-PKcs, Ku and ATP but not by ATM
For these studies, we utilised insect cell expressed human Artemis. Artemis endonuclease activity was assayed using 25 base pair (bp) duplex DNA with 15 nucleotides (nt) of 5′ single-stranded overhang as a substrate (Figure 1A). The radiolabel (32P-α-dCTP) was incorporated at the 3′ end of the longer strand to preclude the impact of Artemis 5′–3′ exonuclease activity (Figure 1A). Consistent with previous findings (Ma et al, 2002), Artemis alone had no detectable endonuclease activity but efficiently cleaved the ssDNA–dsDNA junction in the presence of DNA-PKcs but the absence of Ku (Figure 1B, lanes 1–3). The lack of requirement for Ku was surprising given that the DNA-PK holoenzyme is necessary for NHEJ in vivo. However, when the salt concentration was increased from 10 to 50 mM to a more physiological concentration (100 mM), the ability of DNA-PKcs alone to stimulate Artemis endonuclease activity was abolished and was restored following addition of Ku (Figure 1B, lanes 4–8). DNA-PKcs protein kinase activity reflected these results: DNA-PKcs without Ku was highly active towards Artemis at 10 mM KCl while being essentially inactive at 75 mM KCl unless Ku was present (Supplementary Figure 1A). The lack of Ku dependency is most likely explained by the ability of DNA-PKcs to bind DNA in low, nonphysiological salt conditions (Hammarsten and Chu, 1998). Although previous studies provided insightful evidence for a role of DNA-PKcs in Artemis endonuclease activation (Ma et al, 2002), we now demonstrate the importance of Ku to this process, consistent with in vivo findings.
Figure 1.
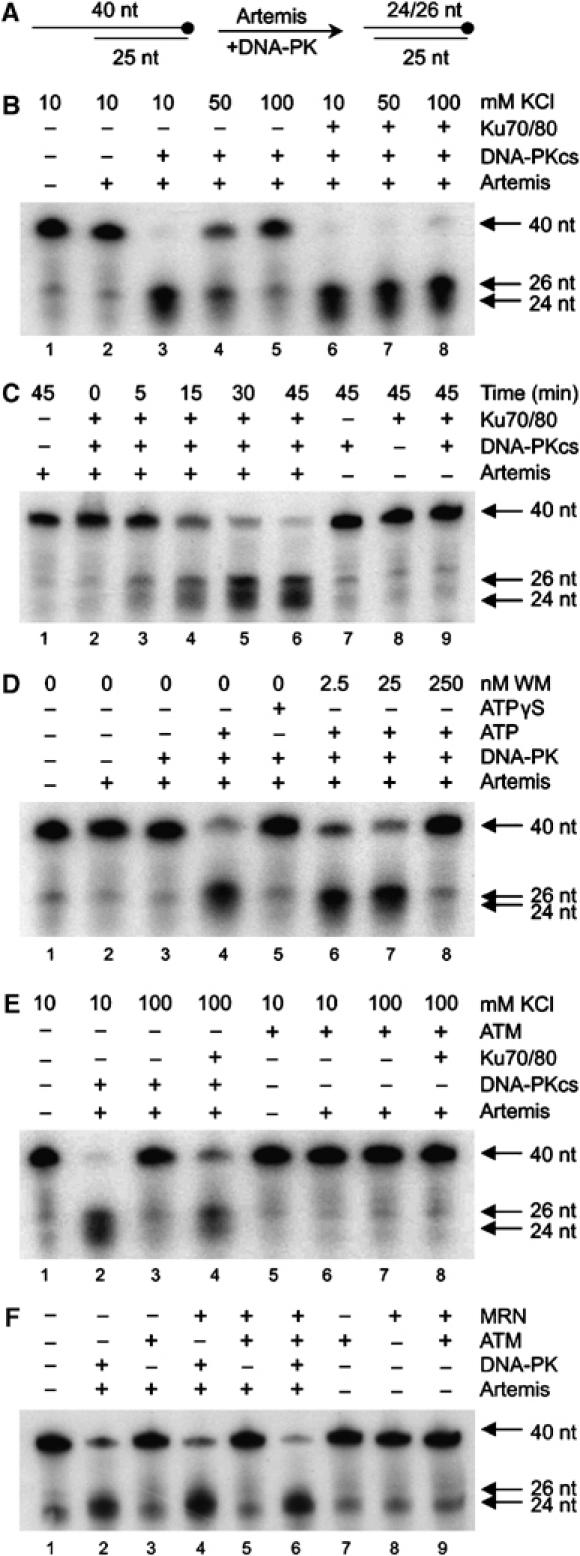
Artemis endonuclease activity requires DNA-PKcs, Ku, ATP and is not supported by ATM. (A) Substrate utilised. (B) Artemis (3.9 pmol) was assayed with DNA-PKcs (0.525 pmol) or the DNA-PK holoenzyme (0.525 pmol) with 10, 50 or 100 mM KCl. All reactions contained 0.25 mM ATP. (C) Purified Artemis (3.9 pmol) was assayed alone or with DNA-PKcs (0.525 pmol) and/or the Ku70/80 heterodimer (0.525 pmol) for the indicated times. Assays contained 75 mM KCl and 0.25 mM ATP. (D) Artemis (3.9 pmol) was assayed with the DNA-PK holoenzyme (0.525 pmol), 75 mM KCl and either no ATP, 0.25 mM ATP or 0.25 mM ATPγS. Indicated concentrations of wortmannin (WM) were incubated with DNA-PK for 5 min on ice before addition. (E) Artemis (3.9 pmol) was assayed with DNA-PKcs (0.262 pmol) or ATM (0.262 pmol) in the presence or absence of the Ku70/80 (0.262 pmol) heterodimer, as indicated. Reactions contained 0.25 mM ATP and either 10 or 100 mM KCl, as indicated. (F) Artemis (3.9 pmol) was assayed with either DNA-PK (0.262 pmol) or ATM (0.262 pmol) in the presence or absence of 0.2 pmol of the MRN complex. Reactions contained 0.25 mM ATP and 100 mM KCl. All assays are representative of data from multiple experiments.
We next characterised Artemis activity under Ku-dependent conditions. Artemis, Ku, DNA-PKcs or DNA-PK (DNA-PKcs+Ku) alone had no detectable endonuclease activity (Figure 1C, lanes 1, 7–9). However, Artemis in the presence of DNA-PK efficiently cleaved the substrate into 24 and 26 nt fragments. Thus, in the presence of DNA-PK Artemis targets the ssDNA–dsDNA junction at the n+1 and n–1 positions, where n equals the first dsDNA nt (Figure 1C, lanes 2–6). As previously shown for DNA-PKcs, DNA-PK stimulation of Artemis endonuclease activity requires its protein kinase activity since assays performed without ATP, with nonhydrolysable ATPγS or with inhibitory concentrations of the PIKK inhibitor wortmannin (WM) were unable to support Artemis activity (Figure 1D). Given that Artemis-dependent DSB repair in vivo is ATM dependent, we examined the ability of purified, active ATM to support Artemis endonuclease activity. Under low ionic strength (10 mM KCl) or physiological salt conditions (100 mM KCl), in the presence or absence of Ku, ATM was unable to promote Artemis endonuclease activity (Figure 1E). Since the Mre11/Rad50/Nbs1 (MRN) complex enhances ATM protein kinase activity in vitro (Lee and Paull, 2004) and is postulated to recruit ATM to DSB ends in vivo (Uziel et al, 2003), we also examined whether ATM together with MRN could support Artemis endonuclease activity. Although the MRN complex stimulated ATM protein kinase activity towards Artemis (Supplementary Figure 2), it failed to enable ATM to support Artemis endonuclease activity (Figure 1F). We conclude that, despite their overlapping substrate specificities, DNA-PK but not ATM can modify Artemis activity. Thus, DNA-PK has a unique property promoting Artemis endonuclease activity.
Mapping the DNA-PK and ATM phosphorylation sites in Artemis
An examination of the impact of phosphorylation on Artemis activity requires identification of the phosphorylation sites. Of the 14 DNA-PKcs phosphorylation sites in Artemis previously reported, none were S/T-Q sites (Ma et al, 2005b). Edman degradation and mass spectrometry demonstrated that the phosphorylation of Artemis (by DNA-PK) under physiological salt conditions primarily occurs at S503, S516 and S645 (Supplementary Figure 3).
Single S>A mutants were generated at these three sites and all remaining SQ sites in Artemis. While DNA-PK efficiently phosphorylated all Artemis S>A mutants, ATM was unable to efficiently phosphorylate Artemis containing either S503A, S516A or S645A mutations, demonstrating ATM specificity for these sites and an apparent interdependency for phosphorylation at these sites (Figure 2B). DNA-PK, in contrast, independently targets multiple sites within Artemis. Of the 10 S/T-Q sites in Artemis, eight (S362, 516, 534, 538, 548, 553, 562 and 645) are located in the C-terminal half of the protein. Examination of N (aa1–502) and C- (aa386–692) terminal fragments and a full-length protein mutated for all the potential C-terminal phosphorylation sites (9A mutant, S362, 503, 516, 534, 538, 548, 553, 562 and 645 mutated to alanines) as substrates for either DNA-PK and ATM demonstrated that Artemis phosphorylation occurred exclusively within the C-terminus and was attributable to the identified sites (Figure 2C). Identical data were found using insect cell expressed wild type (WT) and 9A Artemis (not shown). Of note, while this manuscript was under review, Soubeyrand et al (2006) identified six DNA-PK phosphorylation sites within Artemis in agreement with our findings but in contrast to the non-SQ sites previously identified by Ma et al (2005b). Given that Soubeyrand et al (2006) also utilised physiologically relevant ionic conditions (100 mM KCl) to prepare phosphorylated Artemis, controversy over the identity of Artemis phosphorylation sites is most probably explained by technical differences in salt concentration.
Figure 2.
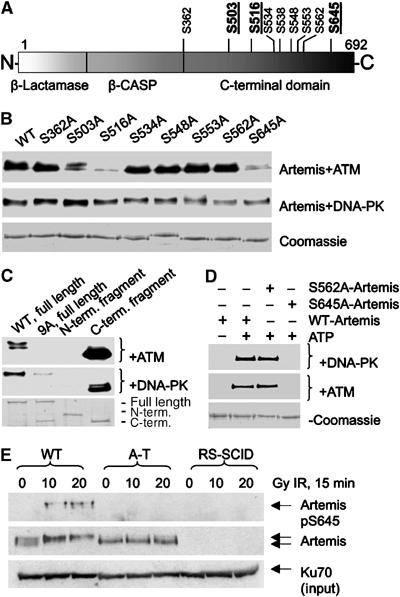
Mapping the DNA-PK and ATM phosphorylation sites in Artemis. (A) A schematic of Artemis indicating the DNA-PK phosphorylation sites (underlined sites were identified by MS). (B) Purified DNA-PK or ATM was incubated with WT or S>A mutants of GST-Artemis under standard assay conditions. Reactions were visualised by autoradiography. The lower panel represents the Coomassie stained GST-Artemis. (C) WT GST-Artemis, GST-Artemis 9A (serines 362, 503, 516, 534, 538, 548, 553, 562 and 645 to alanine), amino acids 1–502 (N terminal fragment) or amino acids 386–692 (C-terminal fragment) were phosphorylated by purified DNA-PK or ATM as described above. (D) DNA-PK (upper panel) or ATM (bottom panel) was incubated with WT GST-Artemis, S562A or S645A GST-Artemis as described above. Reactions were immunoblotted with αArtemis phosphoserine 645 (αArtemis pS645). (E) 48BR (WT), AT1BR (A-T) or FO2-385 (RS-SCID) cells were irradiated and harvested 15 min later. Whole-cell extract (350 μg) was immuno-precipitated with α-artemis (mouse) and immunoblotted with αArtemis pS645 with dephosphopeptide. Sixty micrograms of input were immunoblotted for Artemis and Ku70 as loading controls.
To examine phosphorylation in vivo, we generated a phosphospecific antibody to Artemis S645 (αArtemis pS645). αArtemis pS645 was immunoblotted against GST-Artemis containing either S645A or S562A mutations (Figure 2D) and specifically detected WT and S562A Artemis but not S645A Artemis. To examine Artemis S645 phosphorylation in vivo, WT (48BR), ATM-deficient (A-T) (AT1BR) and Artemis-deficient (RS-SCID) (FO2-385) primary fibroblasts were irradiated with 0, 10 or 20 Gy IR and, 15 min post irradiation, cell extracts were immunoprecipitated for Artemis and immunoblotted with αArtemis pS645 and αArtemis (Figure 2E). IR induces an ATM-dependent mobility shift in Artemis. αArtemis pS645 selectively detects a signal from irradiated WT cells, and not from unirradiated cells or irradiated A-T or RS-SCID cells. Further specificity was confirmed by phosphatase treatment and addition of competing phospho-peptide (see Supplementary Figure 4A). Notably, WT cells treated with the DNA-PK specific kinase inhibitor NU7441 showed normal induction of αArtemis pS645 after IR, suggesting that Artemis phosphorylation in vivo is not dependent on DNA-PK activity (Supplementary Figure 4B). αArtemis pS645 was nonspecific by immunofluorescence or immunoblotting without prior Artemis immunoprecipitation (data not shown). We conclude that Artemis S645 is an in vivo ATM phosphorylation site.
Mutation of the DNA-PK/ATM phosphorylation sites in Artemis does not impact upon Artemis activity in vitro or in vivo
Having examined Artemis phosphorylation in vitro and in vivo, we next monitored its functional impact. To verify that 9A Artemis encompasses the major in vivo phosphorylation sites, we examined its IR-induced hyperphosphorylation in vivo. Following transient transfection of 9A Artemis cDNA (cloned into pCI-neo-c-Myc), the mobility of 9A-Artemis remained unperturbed by irradiation in contrast to WT Artemis (Figure 3A). Thus, IR-induced Artemis hyperphosphorylation occurs at one or more of the identified sites.
Figure 3.
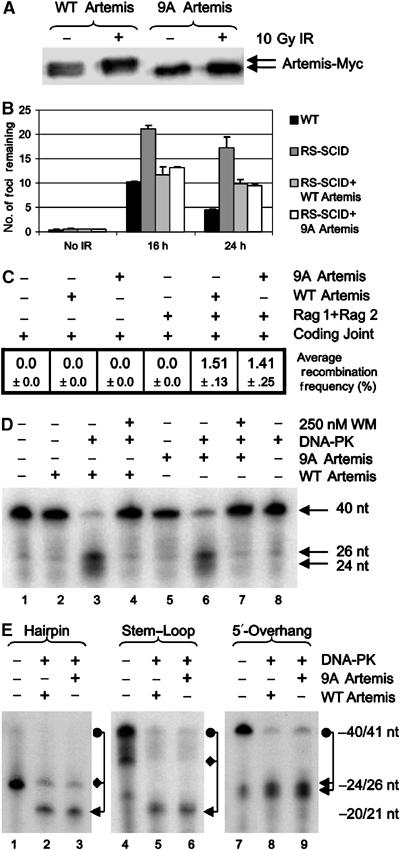
Artemis phosphorylation mutants are proficient for endonuclease activity and complement RS-SCID cells for DSB repair. (A) MRC5Vi cells were transfected with c-Myc-tagged WT or 9A Artemis, irradiated with 0 or 10 Gy IR, harvested after 2 h and cell extracts immunoblotted for c-Myc (see Materials and methods). (B): WT (48BR) or Artemis deficient (CJ179) primary cells were transfected with vector alone, c-Myc-tagged WT Artemis or 9A-Artemis (see Materials and methods). Cells were untreated or irradiated with 10 Gy IR and harvested after 16 or 24 h. Cells were immunostained with αMyc and α53BP1 antibodies. Transfected cells (Myc positive) were counted for 53BP1 foci. (C) Artemis-deficient MEFs were transfected with coding joint substrate, Rag1 and Rag2 and WT or 9A Artemis as indicated. After 72 h, coding joints were recovered and transformed into Escherichia coli and plated onto Bluo-Gal containing plates. Blue colonies, representing Artemis-mediated recombination events, were scored relative to white colonies to calculate recombination frequencies (%). The mean of three independent experiments is shown. (D) WT or 9A Artemis (3.9 pmol) was incubated with or without DNA-PK holoenzyme (0.525 pmol) or WM. All reactions contained 75 mM KCl and 0.25 mM ATP. (E) WT or 9A Artemis (3.9 pmol) were incubated with DNA-PK (0.525 pmol) and either a 40 nt hairpin, a 41-nt stem-loop or the 5′-overhang substrate. Due to the faster mobility of hair-pinned DNA (even under denaturing conditions), the uncleaved hairpin and stem-loop substrates migrate at two distinct sizes indicated by the circle (expected size) and diamond (nonlinear mobility).
We then examined whether WT and 9A Artemis could complement the previously described Artemis-dependent DSB repair defect (Riballo et al, 2004). RS-SCID cells (CJ179-hTERT) were transfected with WT or 9A Artemis cDNA and assayed for the disappearance of 53BP1 foci, a monitor of DSB repair, after exposure to 10 Gy IR (Figure 3B). As expected, Artemis-defective CJ179-hTERT cells transfected with empty vector had elevated numbers of 53BP1 foci (an additional 10–15 foci/cell, 10% of the estimated induced DSBs) 24 h after IR relative to WT cells, demonstrating their characterised repair defect (Figure 3B). Expression of either WT or 9A Artemis restored CJ179hTERT cells to a WT phenotype (Figure 3B), suggesting that Artemis remains active despite its lack of phosphorylation. Further examination of Artemis containing S>A mutations in all 10 SQ sites as well as S503 (11A Artemis) showed identical results to 9A Artemis (data not shown). Thus, loss of every SQ/TQ site within the protein does not compromise function in vivo. To examine whether phosphorylation site mutated Artemis could support V(D)J recombination, Artemis-deficient MEFs were transiently transfected with a V(D)J coding joint substrate plasmid (pHRec-CJ), Rag1 and Rag2 cDNAs and either WT or 9A Artemis cDNA (Figure 3C and Supplementary Figure 5). Both WT and 9A Artemis supported equivalent levels of V(D)J recombination, demonstrating that both proteins are proficient at hairpin cleavage. These findings are consistent with and extend previous reports that Artemis S>A protein mutated in seven of the 10 SQ sites complements radiosensitivity conferred by defective Artemis (Poinsignon et al, 2004).
Finally, insect cell expressed 9A and WT Artemis displayed comparable overhang endonuclease activity (in the presence of DNA-PK) (Figure 3D), and both 9A and WT Artemis opened hairpin or stem-loop substrates with equal proficiency (Figure 3E). Together these findings provide strong evidence that Artemis phosphorylation is dispensable for endonuclease activity.
DNA-PK protein kinase activity is prerequisite for, but dispensable during, the Artemis endonuclease reaction
Since WM inhibits Artemis endonuclease activity (Ma et al, 2002; Figures 1D and 3D), our findings raised the possibility that if Artemis phosphorylation is dispensable for its endonuclease function, then the observed effects could be due to phosphorylation of DNA-PKcs and/or Ku. To examine this, we initially asked whether pre-autophosphorylated DNA-PK could support the endonuclease activity of subsequently added Artemis. We separated the reaction into distinct phases, first preincubating DNA-PK, ATP and the DNA substrate (phase 1) before adding Artemis (phase 2). Remarkably, DNA-PK that was autophosphorylated prior to the addition of Artemis still supported Artemis activity (Figure 4A). This was surprising since we had previously shown that DNA-PKcs autophosphorylation leads to loss of protein kinase activity and dissolution of the DNA-PK holoenzyme (Chan and Lees-Miller, 1996; Douglas et al, 2001; Merkle et al, 2002). This suggested that the putative dissociation of the holoenzyme either did not occur or did not affect subsequent Artemis activity. In contrast, antagonising autophosphorylation by addition of a phosphatase perturbed the ability of DNA-PK to confer endonuclease activity on Artemis (Figure 4B), providing the first evidence that the completed process of DNA-PK autophosphorylation is a prerequisite for Artemis to act as an endonuclease.
Figure 4.
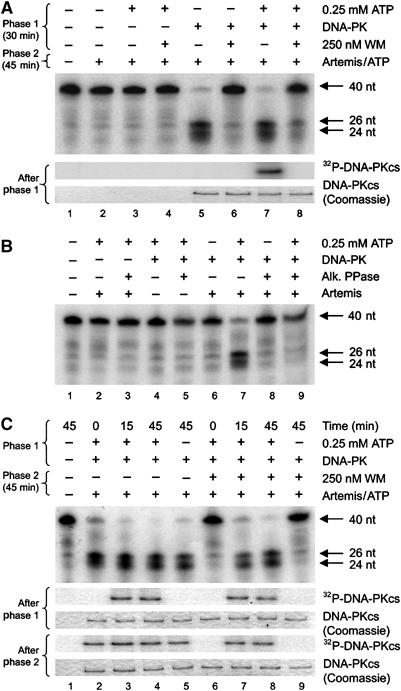
The kinase activity of DNA-PK is prerequisite for Artemis endonuclease activity but is dispensable during the nuclease reaction. (A) To initiate phase 1, the nuclease substrate was incubated with 0.25 mM ATP, 0.525 pmol DNA-PK and/or 250 nM WM. WM was incubated with DNA-PK for 5 min on ice before starting the reaction. Artemis (3.9 pmol) was added to initiate phase 2, and 0.25 mM ATP was added to any reactions where it was absent (to control for this variable). Identical phase 1 reactions were prepared using 2 μCi of 32P-γ-ATP and visualised by autoradiography (lower panels, A). (B) The nuclease substrate was incubated with 0.525 pmol DNA-PK, 0.25 mM ATP, Artemis (3.9 pmol) and/or 0.5 U alkaline phosphatase for 45 min, as indicated. (C) The nuclease substrate was preincubated (phase 1) for 15 or 45 min with 0.525 pmol DNA-PK and 0.25 mM ATP as indicated. After preincubation, reactions were returned to ice and 250 nM WM was added. Once phase 1 was complete, Artemis (3.9 pmol) and ATP (to 0.25 mM final) were added to initiate phase 2. Identical reactions were prepared as in (A) and visualised by autoradiography (lower panels, C).
We next examined whether DNA-PK activity was required during the endonuclease reaction. We preincubated DNA-PK, ATP and the DNA substrate for varying times (phase 1) before adding WM to inhibit DNA-PK protein kinase activity and finally Artemis to initiate the nuclease reaction (phase 2) (Figure 4C). Strikingly, addition of WM to reactions containing autophosphorylated DNA-PK did not affect Artemis endonuclease activity (Figure 4C, lanes 7 and 8). In contrast, reactions that did not undergo DNA-PK autophosphorylation during phase 1 were unable to support Artemis activity (Figure 4C, lanes 6 and 9). These data consolidate our findings that Artemis phosphorylation is dispensable for endonuclease function. Instead, they strongly suggest that DNA-PK autophosphorylation is required to remodel the DNA (or the orientation of protein domains around the DNA) to enable intra-strand cleavage by Artemis.
DNA-PKcs autophosphorylation modulates its orientation and association with DNA, regulating Artemis activity
We then assessed whether the maintenance of DNA-PK in an autophosphorylated state is required to support Artemis endonuclease activity. DNA-PK and the DNA substrate were preincubated with ATP (phase 1) prior to addition of WM and incubation with or without alkaline phosphatase (phase 2). Artemis was then added to initiate the standard nuclease reaction (phase 3). As in Figure 4A, Artemis retained activity when WM was added subsequent to DNA-PK autophosphorylation (Figure 5A, upper panel, lane 6). Little endonuclease activity was observed where DNA-PK autophosphorylation was antagonised by alkaline phosphatase (Figure 5A, upper panel, lane 5). Importantly, loss of phosphate after the completion of autophosphorylation (confirmed by autoradiography and immunoblot, Figure 5A, lower panels) did NOT perturb Artemis endonuclease activity, conferring only a size alteration of one cleavage product (25 nt instead of 24 nt) (Figure 5A, upper panel, lanes 4 and 8). This suggested that autophosphorylation was required for conformation changes within DNA-PK but that the subsequent phosphate removal does not completely ‘reset' these changes. Further, the partial alteration in substrate cleavage position (caused by DNA-PKcs dephosphorylation) suggests that autophosphorylation may impact upon the orientation of DNA-PKcs with the DNA, allowing Artemis access. Identical results were found using a stem-loop DNA substrate (Supplementary Figure 6), indicating that this phenomenon is not restricted to single-stranded overhangs.
Figure 5.
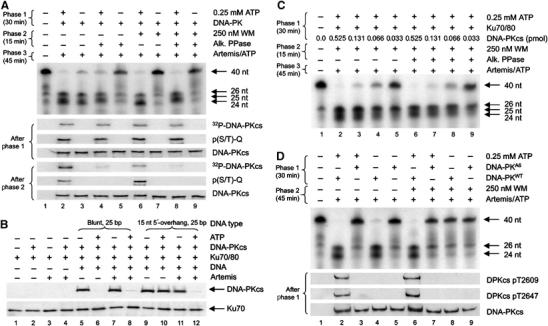
The process of DNA-PK autophosphorylation enables Artemis to target DNA-PK-associated DNA ends. (A): To initiate phase 1, 0.25 pmol of substrate was incubated with 0.525 pmol DNA-PK and 0.25 mM ATP. The addition of 250 nM WM and 0.5 U of alkaline phosphatase initiated phase 2. Phase 3 was initiated by addition of 3.9 pmol of Artemis and ATP (to 0.25 mM final, all reactions). Duplicate reactions were carried out with cold ATP or 32P-γ-ATP, stopping the reactions after either phase 1 or phase 2 with SDS sample buffer. Cold ATP reactions were immunoblotted for phospho-S/T-Q and total DNA-PKcs, while 32P-γ-ATP reactions were processed for autoradiography (lower six panels). (B) 0.25 mM ATP, 0.25 pmol DNA, 0.525 pmol of DNA-PK and/or Ku were incubated for 20 min at 30°C. Artemis (3.9 pmol) was added for 30 min at 37°C. Reactions were then immunoprecipitated for Ku70 and immunoblotted for DNA-PKcs and Ku70. (C) To initiate phase 1, precisely 0.232 pmol of the nuclease substrate was incubated with the indicated amount of DNA-PKcs, 0.525 pmol of Ku70/80 and 0.25 mM ATP. The addition of 250 nM WM and 0.5 U of alkaline phosphatase initiated phase 2. Phase 3 was initiated by the addition of 3.9 pmol of Artemis and ATP (to 0.25 mM final, all reactions) to indicated reactions. (D) To initiate phase 1, approximately 0.25 pmol of the nuclease substrate was incubated with 0.525 pmol of either WT or the six-autophosphorylation (A6) site mutated (threonines 2609, 2620, 2638, 2647 and serines 2612, 2624 to alanine) DNA-PKcs, 0.525 pmol Ku70/80 and 0.25 mM ATP. Phase 2 was initiated by the addition of 250 nM WM, 3.9 pmol of Artemis and ATP (to 0.25 mM final, all reactions). Duplicate reactions were carried out, stopping the reactions after phase 1 and immunoblotting for DNA-PKcs (DPKcs) pT2609, DNA-PKcs pT2647 and total DNA-PKcs (lower three panels).
Previous studies have demonstrated that autophosphorylation causes dissociation of DNA-PKcs from Ku-bound DNA, loss of protein kinase activity and that phosphatase treatment can reverse these effects (Chan and Lees-Miller, 1996; Douglas et al, 2001; Merkle et al, 2002). Earlier studies examining the association of DNA-PKcs with DNA utilised DNA ends with no more than 5 nt of 5′ and/or 3′ ssDNA overhang (Hammarsten et al, 2000; DeFazio et al, 2002; Martensson and Hammarsten, 2002; Merkle et al, 2002), whereas the substrate used in our study contained 15 nt of 5′ ssDNA overhang. We therefore examined whether the 15 nt overhang stabilised the DNA-PK–DNA complex after autophosphorylation. We preincubated DNA-PKcs, Ku and/or ATP in the presence or absence of a blunt 25 bp DNA duplex or a 25 bp DNA duplex with a 15 nt 5′-overhang (i.e. the Artemis endonuclease substrate). Artemis was then added (or not) and complexes were immunoprecipitated with α-Ku70 antibodies prior to immunoblotting for DNA-PKcs and Ku70 (Figure 5B). Since Ku but not DNA-PKcs remains associated with DNA after autophosphorylation, the presence or absence of DNA-PKcs in the immunoprecipitates provides a monitor of dissociation. Consistent with previous findings (Merkle et al, 2002), DNA-PKcs but not Ku dissociates from blunt DNA after autophosphorylation (Figure 5B, lanes 5 and 6). In contrast, DNA-PK complexes assembled on DNA with 15 nt ssDNA overhangs remained stable after autophosphorylation (Figure 5B, lanes 9 and 10). Importantly, the subsequent addition of Artemis reversed the stability of the autophosphorylated DNA-PK complex (Figure 5B, lanes 11 and 12), providing a direct correlation between the removal of the ssDNA overhang and the dissociation of autophosphorylated DNA-PKcs from Ku-bound DNA. Of note, we were unable to observe the association of Artemis with DNA-PK complexes in vitro (data not shown), suggesting that any direct interactions between these proteins are transient.
These findings suggested that a 1:1 relationship between DNA-PK and the DNA would be required to support Artemis endonuclease activity. We examined the stoichiometric relationship between DNA-PK and the DNA substrate subject to Artemis cleavage. Using the conditions of lanes 6 and 8 of Figure 5A, the amount of DNA-PKcs added to the Artemis nuclease assay was incrementally reduced. Under conditions where DNA-PK was sub-stoichiometric to DNA (estimated to be 0.232 pmol), much reduced Artemis endonuclease activity was observed (Figure 5C). This supported the idea that to be targeted by Artemis, each DNA needs to be associated with a DNA-PK molecule whose autophosphorylation triggered conformational changes that were stable in the presence of protein phosphatase activity.
DNA-PK autophosphorylation at the ABCDE cluster is essential for Artemis endonuclease activity
Finally, to confirm our hypothesis, we examined whether DNA-PK with S>A mutations within the previously described ABCDE cluster (DNA-PKA6) (Ding et al, 2003; Block et al, 2004) could support Artemis endonuclease activity. While DNA-PKWT efficiently facilitated Artemis endonuclease activity under every autophosphorylation permissible condition (Figure 5D, lanes 2, 4, 6 and 8), the DNA-PKA6 mutant was unable to support Artemis activity under any circumstance (Figure 5D, lanes 3, 5, 7 and 9). Identical results were found using either stem-loop or hairpin DNA substrates (Supplementary Figures 6 and 7). As previously described (Ding et al, 2003; Block et al, 2004) and confirmed here (Supplementary Figure 8), DNA-PKA6 has normal protein kinase activity and can phosphorylate other sites within itself and other substrates, including Artemis. These data strongly suggest that autophosphorylation at the ABCDE cluster of DNA-PKcs is required for Artemis nuclease activity, possibly by altering holoenzyme conformation such that the ssDNA-dsDNA junction of the DNA is exposed and rendered susceptible to internal cleavage by Artemis. Moreover, where a single-stranded DNA overhang is present, this conformation is stable and essentially unaffected by subsequent loss of the ABCDE phosphates.
Conclusion
We have identified the major DNA-PK and ATM phosphorylation sites within Artemis (S503, S516 and S645) under physiologically relevant ionic conditions, and have shown that ATM-dependent Artemis phosphorylation at S645 occurs in vivo. ATM cannot substitute for DNA-PK to support Artemis activity in vitro, supporting the in vivo dependency upon DNA-PKcs. Under physiological ionic strength conditions, Ku was also required for Artemis endonuclease activity, also consistent with in vivo findings. Strikingly, we show that Artemis-dependent DSB repair and DNA-PK induced Artemis ‘activation' does not entail Artemis phosphorylation but rather involves DNA-PK autophosphorylation at the ABCDE cluster. We propose that DNA-PKcs autophosphorylation causes conformational changes that render the DNA amenable for Artemis intra-strand incision at the ssDNA-dsDNA junction.
A model for the coordinated activity of DNA-PKcs, Ku and Artemis during DNA end processing
We propose the following model for DNA-PK and Artemis mediated DNA end processing (Figure 6). Closely located IR-induced single-strand breaks (SSBs) can resolve into a DSB with lengthy ssDNA overhangs, which may contain additional damage refractory to single-strand annealing (or fill-in) and repair. The Ku70/80 heterodimer rapidly loads onto the DNA end, recruiting DNA-PKcs to form the DNA-PK holoenzyme, conferring end protection. DNA-PKcs autophosphorylation then ensues. In the absence of any overhang, autophosphorylated DNA-PK dissociates from Ku-bound DNA, perhaps actively in favour of subsequent NHEJ factors such as the XRCC4-DNA Ligase IV complex. Where an unannealed overhang is present, DNA-PK autophosphorylation has the added impact that it causes conformational changes that expose the junction between the single-stranded overhang and the DNA duplex, facilitating Artemis cleavage. DNA-PKcs dissociation can therefore occur on blunt DNA ends without any requirement for Artemis, but only after Artemis cleavage if the DNA has ssDNA overhangs of sufficient length and/or if they are not rapidly filled in or annealed. Once dissociated from DNA, the conformation of DNA-PKcs is reset by the action of protein phosphatases. Previous studies have provided evidence that DNA-PKcs autophosphorylation promotes its release from the DNA end. Our model provides an additional and unique component to autophosphorylation, namely facilitating Artemis end-processing.
Figure 6.
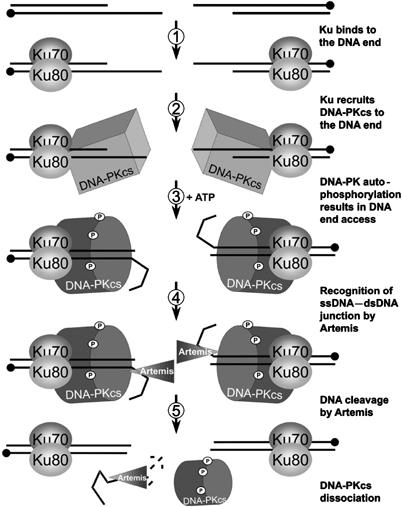
A model for DNA-PK and Artemis mediated DNA end processing. (1) Two SSBs resolve into a DSB with long overhangs. The Ku70/80 heterodimer binds the DNA end to confer protection and (2) recruit DNA-PKcs. (3) When bound to a DNA end, DNA-PK autophosphorylates and undergoes a conformational change that alters the orientation of the DNA such that (4) Artemis can now recognise the ssDNA–dsDNA junction of the overhang, make an intra-strand incision and (5) cleave the fragment via its exonuclease activity. With reduced affinity for the now blunt DNA, autophosphorylated DNA-PKcs eventually dissociates leaving Ku-bound DNA ends ready for further processing by NHEJ factors.
This model is consistent with other findings. Two SSBs on opposing strands within at least 30 nt of each other can resolve into a DSB (Vispe and Satoh, 2000). Since the majority of cellular DSBs arise via this mechanism, it is likely that DSBs with long overhangs occur frequently, a subset of which may preclude reannealing and rejoining due to the presence of additional damage. This subset may be those DSBs repaired in an Artemis-dependent manner. Consistent with this, Artemis is not needed for rejoining simple DSBs produced by etoposide (Riballo et al, 2004).
In vivo, ATM is dispensable for hairpin cleavage during V(D)J recombination but is required for Artemis-dependent DSB repair after IR, suggesting that ATM is not necessary for Artemis activation per se. Our finding that ATM cannot support Artemis endonuclease activity in vitro is consistent with this notion. In the context of DSB repair in vivo, it is possible that ATM indirectly (but essentially) regulates Artemis dependent repair via the activation of down stream effector proteins such as 53BP1 and/or the MRN complex. One possibility is that ATM is required for chromatin modifications that allow repair factors access to DSB sites. Another possibility is that while Artemis phosphorylation does not directly affect enzymatic ability, it could impact upon protein stability. Such a role may escape detection by our complementation assay that involves ectopic expression of Artemis.
Our model of DNA-PK function is consistent with other studies on DNA-PKcs. DNA-PKcs deficient cells complemented with the DNA-PKcsA6 remain severely impaired for V(D)J recombination, particularly with respect to the frequency of coding joint recombination events that require Artemis hairpin opening activity (Ding et al, 2003; Cui et al, 2005). DNA binding induces structural alterations in DNA-PKcs conformation (Boskovic et al, 2003) and the increased affinity of DNA-PKcs for duplex DNA with ssDNA tails versus blunt ends has been recently demonstrated using surface plasmon resonance (Jovanovic and Dynan, 2006). It has been postulated that DNA-PKcs contains an ssDNA-binding pocket adjacent to its DSB binding channel, and that ssDNA regions of sufficient length adhere to the adjacent pocket following duplex binding (Hammarsten et al, 2000; Pawelczak et al, 2005). DNA-PKcs may therefore have the ability to interact with dsDNA and ssDNA such that the junction between the two reorients upon autophosphorylation, exposing the dsDNA to ssDNA transition to the incising action of Artemis. Future studies examining the impact of autophosphorylation on DNA-PKcs structure (together with Ku) and its association with DNA ends containing lengthy single-stranded overhangs, stem-loops or hairpins will be required to consolidate these findings, and these are currently underway. In summary, our findings provide important insight into how Ku, DNA-PKcs and Artemis coordinate to mediate DNA end processing.
Materials and methods
Cells and tissue culture
48BR (WT), AT1BR (A-T) and FO2-385 (Artemis deficient, RS-SCID) primary fibroblasts, GM02188 (WT) or GM03189D (A-T) EBV-transformed lymphoblast and MRC5Vi (WT), Artemis-deficient MEFs and CJ179 (Artemis deficient, RS-SCID) transformed fibroblasts were as described (Riballo et al, 2004).
Reagents
See Supplementary data.
Cloning, mutagenesis and expression of human Artemis in baculoviral and bacterial systems
See Supplementary data.
Purification of human DNA-PKcs, Ku and ATM
See Supplementary data.
In vitro DNA-PK and ATM kinase assays
DNA-PK and ATM kinase assays were carried out as described (Goodarzi and Lees-Miller, 2004).
In vitro nuclease assay
To prepare the nuclease substrate, complementary oligonucleotides (5′-TTTTT-TTTTT-TTTTT-AAGCT-TGCAT-G CCTG-CAGGT-CGAC-3′ and 5′-GGTCG-ACCTG-CAGGC-ATGCA-AGCTT-3 ′) were annealed and labelled with 32P-α-dCTP using exonuclease-free Klenow polymerase. Radiolabelled oligonucleotides were purified and stored at 4°C in sterile water. Assays were carried out in a 5 μl volume containing 25 mM TRIS–HCl pH 8.0, 10 mM MgCl2, 1 mM DTT, 50 ng/μl BSA, approximately 0.15–0.25 pmol 32P-labelled DNA, 0.25 mM ATP and 75 mM KCl (unless otherwise indicated). Purified human Artemis, alkaline phosphatase (Promega), purified human DNA-PKcs, ATM and/or Ku70/80 were used at the specified amounts. Reactions were carried out at 37°C for 45 min (unless otherwise indicated), before being stopped with 5 μl of formamide loading buffer (96% (v/v) formamide, 10 mM EDTA pH 8.0, 0.2% Orange-G dye), a 1-min incubation at 100°C and rapid cooling on ice. Nuclease assays were resolved at 300 V on 0.5 mm thick, 15% PAGE mini gels containing 1 × TBE buffer and 7 M urea. Resolved gels were fixed (in 10% (v/v) methanol and 10% (v/v) acetic acid) before being dried and exposed to film.
Identification of in vitro DNA-PK phosphorylation sites on GST-Artemis
Purified, recombinant GST-Artemis (5.0 μg) was phosphorylated as described for XRCC4 (Yu et al, 2003). Tryptic peptides of phosphorylated Artemis were generated and analysed by Edman degradation, phosphoamino acid analysis and mass spectrometry as described previously for DNA-PKcs (Douglas et al, 2002).
Expression and detection of myc-tagged Artemis
WT or the 9A (serines 362, 503, 516, 534, 538, 548, 553, 562 and 645 to alanine) full-length Artemis cDNA were cloned into pCI-neo-c-Myc to generate c-Myc-Artemis. These clones were transfected into MRC5Vi cells and Artemis was immunoblotted as described (Riballo et al, 2004).
Complementation of Artemis deficient cells
48BR and CJ179 primary human cells were transfected with empty vector, WT or 9A Artemis c-Myc constructs (described above) using the AMAXA transfection system (according to the manufacturer's instructions, Gaithersburg, MD). At 24 h post-transfection, cells were either untreated (background) or exposed to 10 Gy IR and harvested at 16 or 24 h later. Cells were fixed and stained for both Myc and 53BP1. Cells positive for Myc (i.e. expressing the construct) were counted for 53BP1 foci as described (Kuhne et al, 2004).
VDJ recombination assay
See Supplementary data.
Antibodies
See Supplementary data.
Immunoprecipitating Artemis and Ku and immunoblotting
See Supplementary data.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure 8
Supplementary Data
Acknowledgments
We thank Dr Kathy Meek (Michigan State University) for helpful discussions and for providing cells expressing the DNA-PKcs ABCDE (A6) mutant, Dr Roland Kanaar and Dr Eddy van der Linden (Erasmus MC, Rotterdam) for generously providing purified MRN complex, Dr Graeme Smith (KuDos Pharmaceuticals) for DNA-PK inhibitors and D Boland of the SACRI Antibody Facility. AAG is supported by a Post-Doctoral Fellowship grant from the Alberta Heritage Foundation for Medical Research (AHFMR). SPLM is an AHFMR Scientist, an Investigator of the Canadian Institutes for Health Research (CIHR) and holds the Engineered Air Chair in Cancer Research. Work in the SPLM laboratory was funded by Grant #69–1467 from the CIHR. Work in the PAJ laboratory is funded by the MRC, the Leukaemia Research Fund, the Human Frontiers Science Foundation, the EU and the International Association for Cancer Research.
References
- Ahnesorg P, Smith P, Jackson SP (2006) XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end joining. Cell 124: 301–313 [DOI] [PubMed] [Google Scholar]
- Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP (2004) Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res 32: 4351–4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovic J, Rivera-Calzada A, Maman JD, Chacon P, Willison KR, Pearl LH, Llorca O (2003) Visualization of DNA-induced conformational changes in the DNA repair kinase DNA-PKcs. EMBO J 22: 5875–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P (2006) Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299 [DOI] [PubMed] [Google Scholar]
- Calsou P, Delteil C, Frit P, Drouet J, Salles B (2003) Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment. J Mol Biol 326: 93–103 [DOI] [PubMed] [Google Scholar]
- Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ (2002) Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev 16: 2333–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Lees-Miller SP (1996) The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem 271: 8936–8941 [DOI] [PubMed] [Google Scholar]
- Chen L, Morio T, Minegishi Y, Nakada S, Nagasawa M, Komatsu K, Chessa L, Villa A, Lecis D, Delia D, Mizutani S (2005) Ataxia-telangiectasia-mutated dependent phosphorylation of Artemis in response to DNA damage. Cancer Sci 96: 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K (2005) Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol 25: 10842–10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFazio LG, Stansel RM, Griffith JD, Chu G (2002) Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J 21: 3192–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K (2003) Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol 23: 5836–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Moorhead GB, Ye R, Lees-Miller SP (2001) Protein phosphatases regulate DNA-dependent protein kinase activity. J Biol Chem 276: 18992–18998 [DOI] [PubMed] [Google Scholar]
- Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, Meek K, Alessi DR, Lees-Miller SP (2002) Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem J 368 (Part 1): 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Lees-Miller SP (2004) Biochemical characterization of the ataxia-telangiectasia mutated (ATM) protein from human cells. DNA Repair (Amst) 3: 753–767 [DOI] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP (1993) The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72: 131–142 [DOI] [PubMed] [Google Scholar]
- Gu Y, Jin S, Gao Y, Weaver DT, Alt FW (1997) Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci USA 94: 8076–8081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarsten O, Chu G (1998) DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci USA 95: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarsten O, DeFazio LG, Chu G (2000) Activation of DNA-dependent protein kinase by single-stranded DNA ends. J Biol Chem 275: 1541–1550 [DOI] [PubMed] [Google Scholar]
- Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP (1995) DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 82: 849–856 [DOI] [PubMed] [Google Scholar]
- Hefferin ML, Tomkinson AE (2005) Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 4: 639–648 [DOI] [PubMed] [Google Scholar]
- Jovanovic M, Dynan WS (2006) Terminal DNA structure and ATP influence binding parameters of the DNA-dependent protein kinase at an early step prior to DNA synapsis. Nucleic Acids Res 34: 1112–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M (2004) A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res 64: 500–508 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT (2004) Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304: 93–96 [DOI] [PubMed] [Google Scholar]
- Lobrich M, Jeggo PA (2005) Harmonising the response to DSBs: a new string in the ATM bow. DNA Repair (Amst) 4: 749–759 [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR (2004) A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell 16: 701–713 [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Lu H, Niewolik D, Schwarz K, Lieber MR (2005b) The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J Biol Chem 280: 33839–33846 [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108: 781–794 [DOI] [PubMed] [Google Scholar]
- Ma Y, Schwarz K, Lieber MR (2005a) The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 4: 845–851 [DOI] [PubMed] [Google Scholar]
- Martensson S, Hammarsten O (2002) DNA-dependent protein kinase catalytic subunit. Structural requirements for kinase activation by DNA ends. J Biol Chem 277: 3020–3029 [DOI] [PubMed] [Google Scholar]
- Meek K, Gupta S, Ramsden DA, Lees-Miller SP (2004) The DNA-dependent protein kinase: the director at the end. Immunol Rev 200: 132–141 [DOI] [PubMed] [Google Scholar]
- Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, Bazett-Jones DP, Lees-Miller SP (2002) The DNA-dependent protein kinase interacts with DNA to form a protein–DNA complex that is disrupted by phosphorylation. Biochemistry 41: 12706–12714 [DOI] [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP (2001) Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105: 177–186 [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Sokol K, Burgman P, Li L, Li GC (1997) Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc Natl Acad Sci USA 94: 13588–13593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelczak KS, Andrews BJ, Turchi JJ (2005) Differential activation of DNA-PK based on DNA strand orientation and sequence bias. Nucleic Acids Res 33: 152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinsignon C, de Chasseval R, Soubeyrand S, Moshous D, Fischer A, Hache RJ, de Villartay JP (2004) Phosphorylation of Artemis following irradiation-induced DNA damage. Eur J Immunol 34: 3146–3155 [DOI] [PubMed] [Google Scholar]
- Reddy YV, Ding Q, Lees-Miller SP, Meek K, Ramsden DA (2004) Non-homologous end joining requires that the DNA-PK complex undergo an autophosphorylation-dependent rearrangement at DNA ends. J Biol Chem 279: 39408–39413 [DOI] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell 16: 715–724 [DOI] [PubMed] [Google Scholar]
- Soubeyrand S, Pope L, De Chasseval R, Gosselin D, Dong F, de Villartay JP, Hache RJ (2006) Artemis phosphorylated by DNA-dependent protein kinase associates preferentially with discrete regions of chromatin. J Mol Biol 358: 1200–1211 [DOI] [PubMed] [Google Scholar]
- Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA (1994) Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 265: 1442–1445 [DOI] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y (2003) Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22: 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vispe S, Satoh MS (2000) DNA repair patch-mediated double strand DNA break formation in human cells. J Biol Chem 275: 27386–27392 [DOI] [PubMed] [Google Scholar]
- Wang J, Pluth JM, Cooper PK, Cowan MJ, Chen DJ, Yannone SM (2005) Artemis deficiency confers a DNA double-strand break repair defect and Artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair (Amst) 4: 556–570 [DOI] [PubMed] [Google Scholar]
- Yu Y, Wang W, Ding Q, Ye R, Chen D, Merkle D, Schriemer D, Meek K, Lees-Miller SP (2003) DNA-PK phosphorylation sites in XRCC4 are not required for survival after radiation or for V(D)J recombination. DNA Repair (Amst) 2: 1239–1252 [DOI] [PubMed] [Google Scholar]
- Zhang X, Succi J, Feng Z, Prithivirajsingh S, Story MD, Legerski RJ (2004) Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol Cell Biol 24: 9207–9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Bogue MA, Lim DS, Hasty P, Roth DB (1996) Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 86: 379–389 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure 8
Supplementary Data


