Abstract
Short-lived cytokine mRNAs contain an AU-rich destabilizing element (ARE). AUF1 proteins bind the ARE, undergo shuttling, and promote cytoplasmic ARE-mRNA decay through a poorly understood mechanism. We therefore identified AUF1-interacting proteins that may play a role in ARE-mRNA decay. We used mass-spectrometry to identify 14-3-3σ protein as an AUF1-interacting protein. 14-3-3σ binds selectively and strongly to p37 AUF1 and to a lesser extent to the p40 isoform, the two isoforms most strongly associated with ARE-mRNA decay, but not to the two larger isoforms, p42 and p45. The 14-3-3σ interaction site on p37 was mapped to a region found only in the two smaller AUF1 isoforms and which overlaps a putative nuclear localization signal (NLS). Stable overexpression of 14-3-3σ significantly increased cytoplasmic accumulation of p37 AUF1 and reduced the steady-state level and half-life of a reporter ARE-mRNA. siRNA silencing of AUF1 eliminated the effect of 14-3-3σ overexpression. 14-3-3σ therefore binds to p37 AUF1, retains it in the cytoplasm probably by masking its NLS, and enhances rapid turnover of ARE-mRNAs.
Keywords: 14-3-3, AU rich element, AUF1, hnRNPD, RNA stability
Introduction
Many short-lived mRNAs, particularly those encoding proto-oncogenes and cytokines, contain an AU-rich element (ARE) in the 3′ untranslated region (UTR) of the mRNA. A bioinformatic analysis found that AREs are present in approximately 8% of human mRNAs (Bakheet et al, 2001). The ARE functions as a cytoplasmic destabilizing element (Guhaniyogi and Brewer, 2001), but there is little mechanistic or regulatory understanding of its function. Insertion of a prototypical ARE from the GM-CSF mRNA 3′UTR confers a short half-life to otherwise stable reporter mRNAs and removal of the ARE confers stability (Shaw and Kamen, 1986).
Although a number of ARE-binding proteins have been identified, most were identified based on UV-crosslinking studies using cell extracts and radio-labeled ARE-RNA probes. Only a few of the many identified proteins have been shown to regulate rapid decay of ARE-mRNAs in vivo. These include HuR (Myer et al, 1997), tristetraproline (TTP) (Carballo et al, 1998, 2000), KSRP (Min et al, 1997), and AUF1 (hnRNP D) (Brewer, 1991). Overexpression of HuR stabilizes reporter mRNAs containing AREs of various types (Fan and Steitz, 1998), such as the c-fos ARE (Peng et al, 1998). TTP has been shown to be important for the destabilization of reporter mRNAs containing AREs from TNF-α and GM-CSF among others (Carballo et al, 1998, 2000; Lai et al, 1999). Similarly, KSRP has a reported destabilizing effect on ARE-mRNAs. Silencing of KSRP expression results in the stabilization of reporter mRNAs containing AREs from IL2, c-fos, and TNF-α 3′UTRs (Gherzi et al, 2004).
AUF1 was first identified by its ability to accelerate c-myc mRNA decay in a crude in vitro decay system (Brewer, 1991). Subsequent cloning of AUF1 yielded four isoforms based on molecular weight: p37, p40, p42, and p45. All four AUF1 isoforms are generated from a single mRNA by alternative splicing (Wagner et al, 1998). These isoforms differ in the presence of an N-terminal insertion of a 19 amino-acid exon 2 (p40), a C-terminal insertion of a 49 amino-acid exon 7 (p42), or both insertions (p45). The p37 isoform is the core AUF1 protein and contains neither exon 2 nor exon 7. The exon insertions are thought to alter RNA binding affinity of the AUF1 proteins, as p37 AUF1 demonstrates the highest affinity for the c-fos ARE, whereas p40 AUF1 has the lowest (Wagner et al, 1998). The highest affinity binding for ARE sequences is possessed by p37 AUF1 among the four isoforms, which also correlates to the highest destabilizing activity for reporter ARE-mRNAs (Loflin et al, 1999; Sarkar et al, 2003a, 2003b). Steady-state distribution of AUF1 proteins shows that they are predominantly nuclear, although we have shown that all four isoforms interact and display nuclear–cytoplasmic shuttling activity. An intact C-terminal domain (CTD) in p37 and p40 AUF1 proteins contains a putative nuclear localization signal (NLS), which is required for nuclear import. The CTD is interrupted by exon 7 in the two larger AUF1 isoforms, which abolishes nuclear import activity (Sarkar et al, 2003a).
Despite the pivotal role of AUF1 in the regulation of ARE-mRNA decay, the absence of enzymatic activity in these proteins suggests that AUF1 functions by binding to other protein factors that interact with AUF1 to facilitate ARE-mRNA decay. We have previously shown that AUF1 forms a complex with poly(A) binding protein (PABP), initiation factor eIF4G and heat shock proteins Hsp/Hsc70 (Laroia et al, 1999). During heat shock, HSP70 can sequester AUF1 in the nucleus and stabilize ARE-mRNAs (Laroia et al, 1999). Recently, lactate dehydrogenase (LDH) has been found to directly bind to AUF1 in vitro (Pioli et al, 2002), although its effect on ARE-mRNA stability is still unknown. In this study, we report the identification of a new AUF1 binding protein, 14-3-3σ, a member of the 14-3-3 family of proteins. 14-3-3 proteins function in the regulation of subcellular localization of their binding partners (reviewed in Muslin and Xing, 2000). We show that 14-3-3σ binds directly and largely selectively to p37 AUF1, with some interaction with p40 and little interaction with the two larger AUF1 isoforms. The specificity for p37 AUF1 binding was mapped to a unique region in the C-terminus of the p37 isoform, near a region that contains an NLS. 14-3-3σ binding to p37 AUF1 masks its NLS, promotes the cytoplasmic accumulation of nuclear p37 AUF1, and promotes more rapid cytoplasmic degradation of ARE-mRNAs.
Results
14-3-3σ is an AUF1-binding protein that interacts primarily with the p37 isoform
To identify novel p37 AUF1-interacting proteins, we utilized a recovery assay in which HeLa cell lysate was precleared with Ni2+-NTA resin, treated exhaustively with RNase then mixed with recombinant His-p37 previously coupled to NTA resin. The resin–His-p37 complex was recovered and associated proteins were washed, resolved by SDS–PAGE and visualized by silver stain. A single prominent band migrating at approximately 30 kDa was observed in multiple independent experiments (Figure 1A, arrow). The band was subsequently excised for proteolysis and peptide sequencing analysis by mass spectrometry. Mass spectrometric analysis revealed two potential p37 AUF1-interacting proteins in the 30 kDa band. One protein was identified as LDH (data not shown), which was found previously by biochemical purification to bind directly to p37 AUF1 (Pioli et al, 2002). As LDH binding to AUF1 was previously reported, it was not further studied here. The other protein was identified as 14-3-3σ, a member of the 14-3-3 family of acidic proteins. To date, seven 14-3-3 isoforms have been identified in human cells, all of which share a high degree of amino-acid sequence conservation (Yaffe, 2002). Unlike the other 14-3-3 family members which are ubiquitously expressed, 14-3-3σ expression is more restricted but includes epithelial cells (Leffers et al, 1993) (HeLa cells are epithelial in origin). The 14-3-3 protein was identified as the sigma isoform based on peptide regions unique to 14-3-3σ (Figure 1B). To further verify the identification of 14-3-3σ, and to exclude contamination by the sigma isoform during isolation, proteolysis or mass spectrometry, the resin His-p37 proteins were resolved by SDS–PAGE and subjected to immunoblot analysis with various 14-3-3 antibodies (Figure 1C). 14-3-3σ was present in the His-p37 isolate, but not in the control lane lacking His-p37 (Figure 1C, upper panel). In contrast, although the γ and β/ζ isoforms of 14-3-3 were also present in the HeLa lysate, these isoforms did not interact with His-p37 AUF1 (Figure 1C, middle panels). As a positive control for these studies, His-p37 was recovered and shown to interact with PABP (Figure 1C, lower panel), a known p37 AUF1-binding protein (Laroia et al, 1999). Thus, 14-3-3σ appears to be an authentic binding partner of p37 AUF1.
Figure 1.
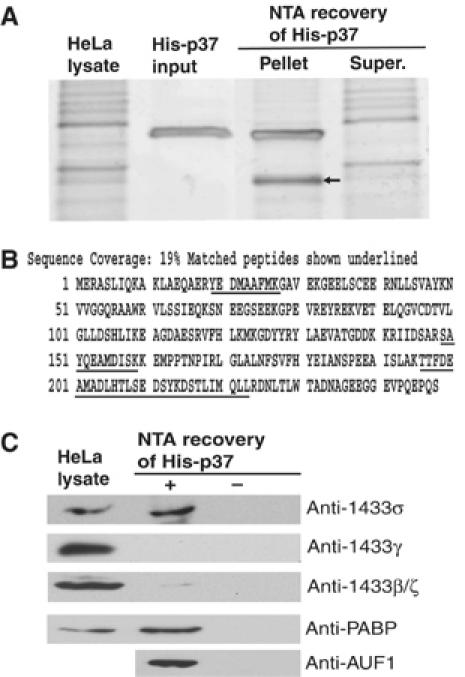
14-3-3σ is a p37 AUF1-interacting protein. (A) Recombinant His-tagged p37 protein was purified from E. coli by NTA resin and mixed with whole-cell HeLa lysate. His-p37 AUF1 was recovered by NTA resin chromatography, proteins resolved by SDS–PAGE, and visualized by silver stain. The 30 kDa band (arrow) was excised and sequenced by mass spectrometry. (B) Peptides present in the 30 kDa band (underlined) were mapped to regions of 14-3-3σ protein and identified as corresponding to the sigma (σ) isoform. (C) To verify interaction of 14-3-3σ with His-p37 AUF1, proteins associated with His-p37 were recovered by NTA resin chromatography, resolved by SDS–PAGE, and detected by immunoblot using anti-14-3-3 antibodies, anti-AUF1, and anti-PABP, a known p37 AUF1-interacting protein, and visualized using the enhanced chemiluminescence system (ECL).
We next investigated whether the interaction between 14-3-3σ and p37 AUF1 is direct, or achieved via other protein partners. Exhaustive degradation with RNaseA excluded the possibility that p37 AUF1 and 14-3-3σ interact via an RNA moiety. In an in vitro binding assay, purified recombinant GST-14-3-3σ was mixed with increasing amounts of purified, recombinant His-p37 AUF1. GST-14-3-3σ was recovered by glutathione Sepharose-GSH resin chromatography, and the association of His-p37 AUF1 was examined by immunoblot analysis using anti-AUF1 antibody. In vitro binding analysis revealed that p37 AUF1 interacts with 14-3-3σ directly, and does so in a dose-dependent manner (Figure 2A). There was no interaction between AUF1 and GST alone (Figure 2A). Because the His-p37 recovery assay suggested that only the sigma isoform, but not the γ or β/ζ isoforms, of 14-3-3 binds to p37 AUF1 (Figure 1C), studies were also carried out to further determine whether the interaction between p37 AUF1 and 14-3-3σ is specific for this isoform or extends to other 14-3-3 proteins. There is good evidence that 14-3-3σ is somewhat unique among 14-3-3 proteins, both in its mechanism of protein interaction and its low level of binding to target proteins that interact with the more conventional members of the 14-3-3 family (Bridges and Moorhead, 2005). In this regard, 14-3-3ζ (zeta) is representative of the conventional 14-3-3 proteins. It is ubiquitously expressed and largely nuclear whereas 14-3-3σ is predominantly cytoplasmic and associated with specific interactions related to p53 function, cell cycle control and possibly ionizing radiation responses (Leffers et al, 1993; Hermeking et al, 1997). Nevertheless, both are nuclear-cytoplasmic shuttling proteins (Yaffe, 2002). 14-3-3η (eta) is a widely expressed isoform that interacts with cyclin-dependent kinases, but has been shown to bind to the ARE-binding protein TTP (Johnson et al, 2002). TTP also interacts with several other 14-3-3 proteins including the sigma-isoform (Johnson et al, 2002). Under identical binding conditions, recombinant purified p37 AUF1 was found to interact strongly with 14-3-3σ, to a lesser extent with 14-3-3η, but not with 14-3-3ζ (Figure 2B). Thus, p37 AUF1 shows strong specificity for 14-3-3σ, with some binding to the eta isoform that binds to TTP, another key ARE-binding protein.
Figure 2.
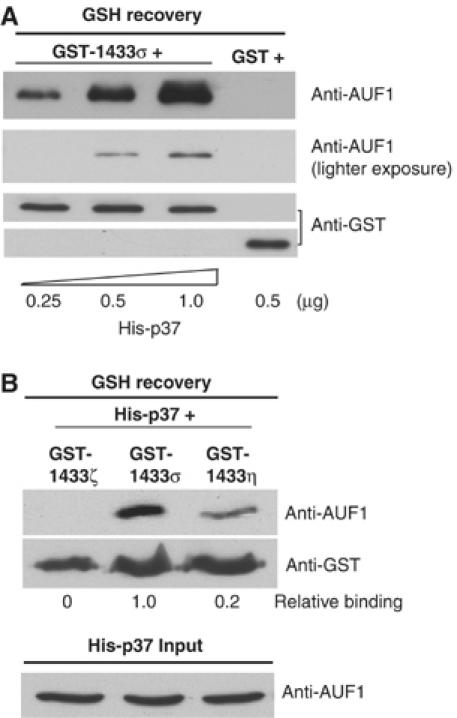
Selective, direct, and dose-dependent interaction between 14-3-3σ and p37 AUF1. (A) Purified, recombinant GST tagged 14-3-3σ protein was mixed with increasing amounts of purified, recombinant His-tagged p37 AUF1, both expressed in E. coli. GST alone, in place of GST-14-3-3σ, was used as a negative control. GST proteins were recovered by GSH-Sepharose chromatography, washed and association of p37 AUF1 examined by immunoblotting with anti-AUF1 or anti-GST antibody and visualized by ECL. Two different exposures for AUF1 interaction are shown. GST-14-3-3σ and GST polypeptide were resolved in two different gels as shown in the two lower panels. (B) Equal amounts of purified recombinant GST-14-3-3ζ, GST-14-3-3σ, or GST-14-3-3η proteins were mixed with purified, recombinant His-p37 AUF1. GST proteins were recovered by GSH-Sepharose chromatography and the association of p37 AUF1 was examined as in (A). Typical results of three independent studies are shown. Densitomtery was used for quantiation of immunoblots.
14-3-3σ interacts with the p37 isoform of AUF1
Next, the interaction between endogenous AUF1 and 14-3-3σ protein was investigated. HeLa cells express abundant amounts of 14-3-3σ as well as AUF1 proteins. We therefore immunoprecipitated 14-3-3σ protein from HeLa whole-cell lysates, which was resolved by SDS–PAGE and immunoblotted for interacting AUF1 isoforms. Association of p37 with 14-3-3σ was confirmed, but interestingly, there was little detectable interaction with the other three isoforms of AUF1, p40, p42 or p45 (Figure 3A). To further examine whether the three larger isoforms of AUF1 interact with 14-3-3σ, an in vitro binding assay was performed. GST-14-3-3σ or GST alone were mixed with equal molar amounts of His-tagged p37, p40, p42, or p45 AUF1 proteins, and then recovered with GSH resin. The predominant isoform of AUF1 which bound to 14-3-3σ was the p37 protein, and to a much lesser extent p40 AUF1. There was no detectable interaction between 14-3-3σ and the two larger AUF1 isoforms, p42 and p45 (Figure 3B).
Figure 3.
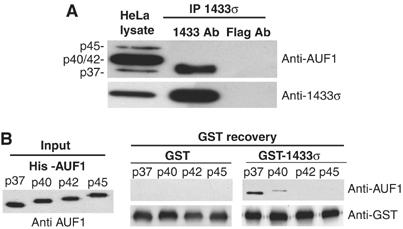
14-3-3σ interacts primarily with the p37 isoform of AUF1. (A) Using whole-cell HeLa lysate, 14-3-3σ was immunoprecipitated (IP) with a monoclonal antibody, resolved by SDS–PAGE and detected by immunoblot with AUF1 antibody. As a control, an equal amount of lysate was subjected to IP with anti-Flag antibody. (B) Equal amounts of purified recombinant GST-14-3-3σ or GST as a control were mixed with equal amounts of purified recombinant His-tagged p37, p40, p42 or p45 AUF1 proteins, followed by recovery using GSH-Sepharose chromatography. The association of AUF1 protein isoforms with 14-3-3σ was detected by immunoblot analysis using AUF1 antibody and ECL. Typical results from at least three independent experiments are shown.
A region in the CTD of p37 AUF1 is crucial for 14-3-3σ interaction
The p37 AUF1 region required for interaction with 14-3-3σ was determined by utilizing deletion mutants of the N-terminal 78 amino acids (His-p37 ΔN78) or the C-terminal 45 amino acids (His-p37 ΔC45). Similar amounts of p37 proteins were used in each reaction (Figure 4A, input). Wild type or mutant recombinant His-p37 AUF1 proteins were mixed with equal amounts of GST-14-3-3σ protein. Interaction of GST-14-3-3σ and His-p37 AUF1 was detected by recovery, SDS–PAGE and direct protein staining. Both wild type and His-p37 ΔN78 AUF1 proteins bound 14-3-3σ, whereas no interaction was detected with His-p37 ΔC45 (Figure 4A, NTA recovery). Similar results were observed when GSH resin was used to recover GST-14-3-3σ, followed by detection of full length or deletion mutants of His-p37 AUF1 (data not shown). These data demonstrate that p37 AUF1 interacts with 14-3-3σ through its CTD.
Figure 4.
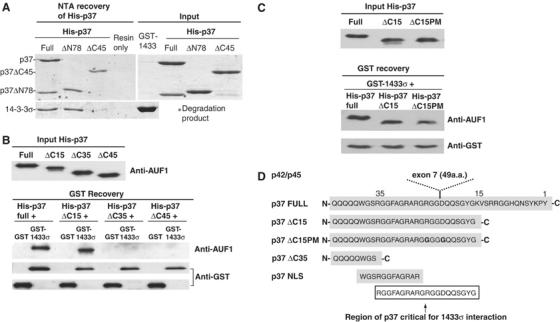
A 20 amino-acid sequence in the CTD of p37 AUF1 is critical for interaction with 14-3-3σ. (A) Equal amounts of purified recombinant full-length proteins His-p37, His-p37 containing a deletion in the N-terminal 78 amino acids (ΔN78), or His-p37 containing a deletion in the C-terminal 45 amino acids (ΔC45), were mixed with purified recombinant GST-14-3-3σ. GST-14-3-3σ alone was used as a negative control. Following NTA resin recovery of His-tagged AUF1 proteins (left panel), the association of 14-3-3σ to full-length or deletion mutants of His-p37 AUF1 was visualized by SDS–PAGE/Coomassie blue staining of the gel (bottom panel). The level of input proteins is shown (right panel). (B) Full-length, ΔC45, ΔC35 or ΔC15 His-p37 AUF1 were mixed with either GST-14-3-3σ or GST alone as a control. Following GSH-Sepharose recovery of GST proteins, the association of 14-3-3σ to full length or deletion mutants of His-p37 AUF1 was visualized by immunoblot analysis using AUF1 antibody and ECL. Typical results of at least three independent experiments are shown. (C) Same as in (B), except His-p37 ΔC15PM was also included. (D) A schematic representation of the C-terminus of p37 AUF1, the location of exon 7 insertion, the putative NLS, and location of the 20 amino acid (a.a.) region in the CTD of p37 AUF1 found to be critical for p37 AUF1-14-3-3σ interaction. Amino-acid numbers are oriented from the C-terminus as position 1.
Detailed mapping of the CTD region of AUF1 required for binding to 14-3-3σ was carried out. Additional truncation mutants lacking the C-terminal 15 amino acids (His-p37 ΔC15) or 35 amino acids (His-p37 ΔC35) were developed. Equal amounts of recombinant proteins were used for interaction analysis, GST-14-3-3σ was recovered by GSH-Sepharose chromatography, and interaction with His-p37 AUF1 proteins were detected by SDS–PAGE and immunoblot analysis (Figure 4B). Studies showed that the interaction between 14-3-3σ was preserved in His-p37 ΔC15 AUF1, but lost in His-p37 ΔC35 AUF1 (Figure 4B). There was no interaction between AUF1 proteins and GST alone. A 20 amino-acid sequence located between C-terminal position 15 to 35 of the p37 AUF1 CTD is therefore crucial for 14-3-3σ binding. Examination of the CTD sequence of p37 AUF1 revealed that an uninterrupted form of the 20 amino-acid sequence is absent in the two larger AUF1 isoforms p42 and p45, where it is disrupted by the insertion of exon 7. Although the CTD of the p40 AUF1 isoform also contains the uninterrupted 20 amino-acid fragment, p40 interacts with 14-3-3σ more weakly. Previous studies suggest that the p37 and p40 AUF1 isoforms have somewhat different structures, which may be conferred by the N-terminal insertion of exon 2 in p40 AUF1 (Laroia et al, 1999). For instance, the presence of exon 2 diminishes the ability of p40 to be ubiquitinated compared to p37 (Laroia et al, 1999), and the two proteins differ in their strength of interaction with canonical AUUUA motifs despite the presence of identical RNA binding regions (Wilson et al, 2001). Thus, either p37-specific structure or sequence could define the 14-3-3σ interaction site. We therefore further mutated residues R and D of the exon 7 insertion site (RGG/DQQ) of His-p37 ΔC15 to create His-p37 ΔC15PM (GGG/GQQ) (Figure 4C and D). Interaction analysis revealed that the two point mutations did not decrease interaction of His-p37 ΔC15 with 14-3-3σ (Figure 4C). Thus, it is most likely that the tertiary structure of the p37 CTD confers 14-3-3σ interaction. The 20 amino-acid sequence also overlaps a 12 amino-acid putative NLS found in p37 AUF1 (Figure 4D), which has been implicated in facilitating nuclear localization of p37 AUF1 (Sarkar et al, 2003a, 2003b). Interestingly, other 14-3-3 proteins have been shown to bind to the NLS of their respective binding partners and to alter their localization within the cell. For example, 14-3-3ɛ inhibits the nuclear import of Cdc25C by interfering with its NLS (Kumagai and Dunphy, 1999; Yang et al, 1999). Similarly, Cdc25B contains an NLS that is adjacent to the binding site for 14-3-3β, ɛ, η, and ζ (Davezac et al, 2000). We therefore determined whether binding of 14-3-3σ to p37 AUF1 masks its NLS and affects its localization and biological functions in the cell.
14-3-3σ promotes cytoplasmic accumulation of p37 AUF1
Since the 14-3-3σ binding site on p37 AUF1 overlaps the p37 NLS, and AUF1 proteins are predominantly localized to the nucleus but shuttle to the cytoplasm (Sarkar et al, 2003a, 2003b), we determined whether the binding of 14-3-3σ controls the intracellular distribution of AUF1. To study the distribution of AUF1, a CHO cell line was developed that stably expresses human 14-3-3σ protein (CHO-14-3-3σ) under the control of a tetracycline (Dox) inhibited promoter. The addition of Dox strongly silenced the expression of 14-3-3σ to almost undetectable levels at 3 days postaddition, determined by both immunoblot analysis and immunofluorescence staining of cells with anti-14-3-3σ antibody (Figure 5A). The level of 14-3-3σ mRNA after Dox treatment was reduced by almost 100-fold, as determined by real time qRT–PCR (Supplementary data). In addition, the hamster 14-3-3σ gene is not expressed in CHO cells as shown by its absence by immunoblot analysis (Figure 5A) and real-time qRT–PCR (Supplementary data). The localization of AUF1 in the presence or absence of 14-3-3σ could therefore be examined in a controlled manner in this system. CHO-14-3-3σ cells were treated with Dox or mock treated for 3 days and stained for endogenous AUF1 using an antibody that detects all four isoforms. Human and hamster AUF1 proteins are well conserved and are both efficiently recognized by antibodies to the human AUF1 proteins (Sarkar et al, 2003a, 2003b). Without 14-3-3σ expression (+Dox), the majority of AUF1 was localized to the nucleus, as expected (Figure 5B, left panel, +Dox). When 14-3-3σ was expressed, a significant cytoplasmic accumulation of AUF1 was observed (Figure 5B, left panel, −Dox), evidenced by the extranuclear spillover of FITC signal into the cytoplasm. The altered localization of AUF1 was not due to a global effect on protein distribution caused by 14-3-3σ expression, as shown by retention of nuclear replication protein A (RPA) under the same conditions (Figure 5B, right panel). Since 14-3-3σ preferentially binds to p37 AUF1 in vitro, we determined whether the increased cytoplasmic accumulation of AUF1 in the presence of 14-3-3σ overexpression is primarily due to relocalization of the p37 isoform. The cytoplasmic fractions and nuclear fractions of CHO-14-3-3σ cells, with or without 14-3-3σ expression (−/+ Dox) were isolated and proteins detected by immunoblot analysis (Figure 5C). As AUF1 is present at low levels in the cytoplasm, six-fold more cytoplasmic than nuclear protein was used for analysis. Of the four AUF1 isoforms, the p45 and p37 proteins were clearly resolved by SDS–PAGE, whereas the p40 and p42 proteins typically overlap. The p37 AUF1 protein showed a significant increase in cytoplasmic accumulation in the presence of 14-3-3σ, with a corresponding reduction in p37 AUF1 levels in the nucleus (Figure 5C). This observation correlates well with the finding that 14-3-3σ preferentially binds to the p37 isoform of AUF1. There was some decrease in the nuclear fraction of p45 AUF1, with a corresponding cytoplasmic increase, and a slight but similar change in the p40/p42 fraction. The slight change in AUF1 proteins likely reflects the poorly understood interactions and stoichiometry of association among the AUF1 isoforms (Sarkar et al, 2003a).
Figure 5.
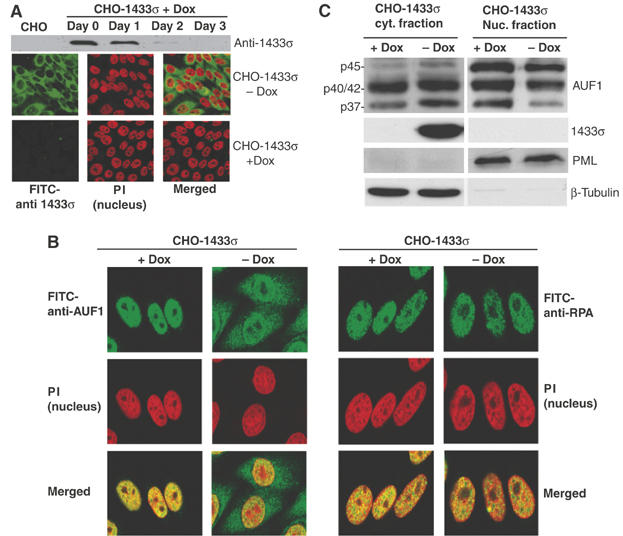
14-3-3σ expression promotes cytoplasmic accumulation of p37 AUF1. (A) A stable CHO cell line expressing 14-3-3σ under the control of a doxycycline (Dox)-suppressed promoter (pBSTR1-14-3-3σ) was developed. (Upper panel) Cells treated with Dox for up to 3 days to inhibit 14-3-3 expression were lysed and examined for 14-3-3σ protein levels by immunoblot analysis with 14-3-3σ antibody. (Lower panels) Cells were treated with Dox or mock treated for 3 days and examined by immunofluorescence microscopy with 14-3-3σ antibody followed by FITC-conjugated secondary antibody. Cell nuclei were visualized by staining with propidium iodide (PI). Representative individual panels of cells and panels with merged P.I./FITC staining are shown. (B) (Left panels) 14-3-3σ cells treated with Dox (+Dox) or mock treated (−Dox) under the same conditions as above, were fixed and stained for endogenous AUF1 with anti-AUF1 antibody followed by FITC-conjugated secondary antibody. Nuclei were visualized with PI. (Right panels) As a control, nuclear protein RPA, was stained with RPA antibody followed by FITC-conjugated secondary antibody. Representative panels of cells from at least three independent analyses are shown. (C) CHO-14-3-3σ cells were treated with Dox for 3 days or mock treated, collected and fractionated into cytoplasmic and nuclear fractions. Proteins were resolved by SDS–PAGE and detected by immunoblot analysis with AUF1 antibody and ECL. β-Microtubulin, a cytoplasmic protein, and PML, a nuclear protein, were used as controls for equal loading and fractionation efficacy. Representative results of at least three independent experiments are shown.
14-3-3σ promotes p37 AUF1-mediated ARE-mRNA decay
Although AUF1 proteins localize to a greater extent in the nucleus, they do shuttle and the rapid turnover of short-lived ARE-mRNAs is a cytoplasmic event associated with their cytoplasmic presence. Since binding of 14-3-3σ to p37 AUF1 promotes cytoplasmic accumulation of p37 AUF1, we asked whether this interaction facilitates a more rapid cytoplasmic turnover of ARE-mRNAs. To address this issue, we utilized an ARE-mRNA reporter assay system (Sarkar et al, 2003b). In this assay, CHO-14-3-3σ cells with and without 14-3-3σ expression (−/+ Dox) were transfected with a plasmid encoding LacZ reporter mRNA containing an ARE in its 3′UTR derived from the GM-CSF 3′UTR (LacZ-ARE) or a control stable LacZ reporter without an ARE. Cells were cultivated for 3 days in the presence or absence of Dox, then transfected and total RNA extracted 24 h post transfection. The steady-state levels of LacZ and LacZ-ARE mRNAs were determined by Northern blot analysis (Figure 6A). A stable GFP mRNA was co-expressed from a co-transfected plasmid as a control for transfection efficiency and RNA loading in Northern blot studies. In addition, LacZ-ARE and LacZ mRNAs were quantified by densitometry and by quantitative real-time RT–PCR relative to GFP mRNA levels, and data presented normalized to the levels of mRNA in the absence of 14-3-3σ induction (+Dox, Figure 6A and B). The levels of stable (control) LacZ mRNA present in cells with and without 14-3-3σ expression were identical, indicating that 14-3-3σ expression had no effect on non-ARE-mRNA steady-state levels. In contrast, steady-state levels of LacZ-ARE mRNA were reduced by three-fold in cells expressing 14-3-3σ (Figure 6A), as determined by Northern blot analysis and densitometry, and ∼2.5-fold by qRT–PCR (Figure 6B). Expression of very low levels of reporter ARE-RNAs in these cells did not alter the increased mRNA decay mediated by 14-3-3σ protein (data not shown). Thus, the promotion of ARE-mRNA decay by 14-3-3σ is not related to reporter ARE-mRNA levels. As expected, the steady-state LacZ-ARE mRNA level was five- to seven-fold lower than that of LacZ mRNA without the ARE, regardless of 14-3-3σ expression (Figure 6A and B, left panel), due to the destabilizing effect of the ARE (Sarkar et al, 2003b). The reduction in LacZ-ARE mRNA level due to 14-3-3σ expression was next shown to be mediated through AUF1. siRNAs developed against AUF1 were transfected into cells and found to mediate strong silencing of AUF1 expression (Figure 6C). Silencing of endogenous AUF1 protein expression by more than 90% (Figure 6C, left panel) abolished the ability of 14-3-3σ to promote LacZ-ARE mRNA decay, whereas a control siRNA that does not target AUF1 expression had no effect (Figure 6C, right panel).
Figure 6.
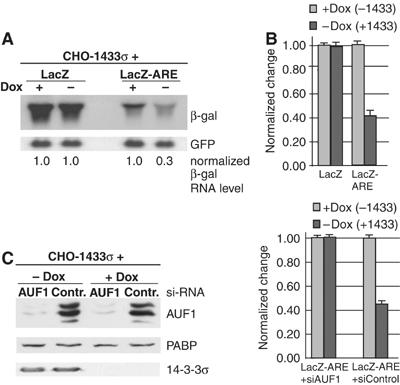
Overexpression of 14-3-3σ reduces steady-state reporter levels of ARE-mRNAs. (A) CHO-14-3-3σ cells were treated with Dox for 3 days or mock treated, then transfected with vectors expressing a stable LacZ reporter mRNA (pCH110) or a LacZ-ARE reporter mRNA containing the ARE sequence derived from the GM-CSF 3′UTR. At 24 h post-transfection, total RNA was extracted and the level of β-gal mRNA remaining was detected by Northern blot analysis. As a control, a plasmid encoding GFP from a stable mRNA was co-transfected and detected with a GFP probe to ensure equal transfection efficiency. Autoradiograms were quantified by densitometry and values normalized to 1.0 for β-gal mRNA relative to the sample lacking 14-3-3σ induction (+Dox). (B) Total RNA extracted in (A) was further treated with DNaseI and quantified by real-time qRT–PCR using primers designed to amplify the middle region of the LacZ reporter mRNA. The data presented are averaged from three independent experiments and normalized as above. (C) CHO-14-3-3σ cells treated with Dox, or mock treated, were transfected with siRNA targeting AUF1, or a control siRNA, followed by transfection of LacZ-ARE reporters. The level of AUF1 silencing was verified by immunoblot analysis using AUF1 antibody. The level of LacZ-ARE mRNA remaining 24 h post-transfection was determined as in (B).
The reduction in the steady-state levels of LacZ-ARE mRNA with 14-3-3σ expression was found to result from accelerated mRNA decay. CHO-14-3-3σ cells with and without 14-3-3σ expression (−/+ Dox) were transfected as described above, and treated with actinomycin D (Act D) to block new transcription. Total RNA was extracted and examined by Northern blot to determine mean relative rates of mRNA decay (Figure 7). The LacZ-ARE mRNA reporter exhibited an average half-life of 3.5 h without 14-3-3σ expression (+Dox), and 1.2–1.5 h with 14-3-3σ expression (Figure 7). As expected, there was no change in the stability of the control LacZ mRNA, which displayed a half-life of 12–15 h (data not shown). Therefore, these data confirm that 14-3-3σ expression selectively decreases the stability of ARE-mRNAs, in association with increased cytoplasmic localization of p37 AUF1.
Figure 7.
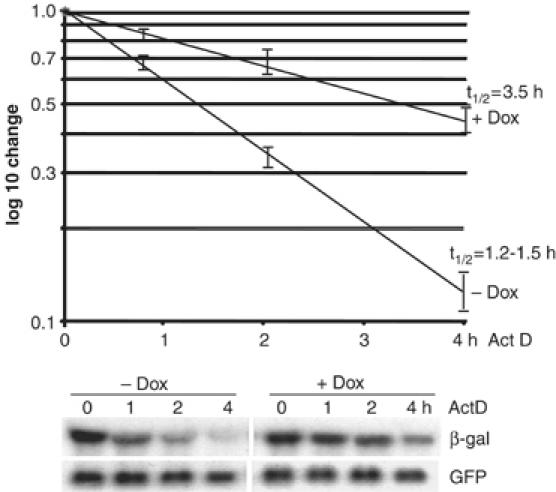
Overexpression of 14-3-3σ reduces ARE-mRNA half-life. CHO-14-3-3σ cells were treated with Dox for 3 days or mock treated, then cotransfected with plasmids expressing the LacZ-ARE or stable LacZ reporter and GFP mRNAs. Cells were treated with Act D for up to 4 h. Total RNA was extracted and analyzed by Northern blot analysis using β-gal and GFP probes. A representative Northern blot from three independent experiments is shown. mRNA levels were quantified by densitometry and mRNA half-lives (t1/2) determined from a log10 semilog plot, normalized to GFP and the initial levels of the β-gal mRNA.
Discussion
Although it has been established that AUF1 family members bind to ARE sequences (DeMaria et al, 1997; Sirenko et al, 1997) and facilitate rapid degradation of ARE-mRNAs (Brewer, 1991; Sarkar et al, 2003a, 2003b), the molecular mechanisms by which the different AUF1 proteins function in this process are still poorly understood. The aim of the current study was the identification of novel AUF1-interacting proteins and elucidation of their function. We utilized recovery of proteins from whole-cell lysates that bound unmodified recombinant His-p37 AUF1 protein in vitro. This approach also identified known AUF1 binding proteins, such as Hsp70 and PABP (data not shown). The 30 kDa p37 AUF1 interacting protein detected, 14-3-3σ, was confirmed to be authentic in vitro and in vivo, and functionally relevant in regulating AUF1 cytoplasmic accumulation and promoting ARE-mRNA decay.
The 14-3-3 family of proteins are closely related acidic proteins that play critical roles in many physiological events of the cell, including signal-transduction pathways that control cell-cycle checkpoints, kinase activation and apoptosis among other processes (Xing et al, 2000; Yang et al, 2003). In general, the 14-3-3 proteins act as molecular scaffolds and chaperones. The different biological actions of the various 14-3-3 proteins are thought to depend on interactions with their binding partners, many of which are nuclear or mitochondrial proteins, some of which are relocalized in the cytoplasm (Muslin and Xing, 2000). More than one hundred 14-3-3 protein binding partners have been identified to date.
The 14-3-3σ protein, which we have now identified as a p37 AUF1 interacting protein is a unique and least well-conserved member of the 14-3-3 family. 14-3-3σ is also known as HME1 (Human Mammary Epithelial marker 1), because it is expressed at high levels in cells of epithelial origin. In contrast to the other 14-3-3 isoforms, 14-3-3σ has been shown to act as a tumor suppressor due to its ability to inhibit G2/M progression in a p53-regulated manner (Hermeking et al, 1997; Yang et al, 2003). Disruption of 14-3-3σ expression blocks cytoplasmic sequestration of cyclin B1/cdc2, which accounts in part for its tumor suppression activity (Chan et al, 1999). It is interesting to note that the ARE-binding protein TTP, another protein shown to accelerate ARE-mRNA decay, binds to 14-3-3 family members, which increases its cytoplasmic accumulation (Johnson et al, 2002). The binding of TTP to 14-3-3η requires phosphorylation of the 14-3-3η binding site on TTP (serine 178) (Johnson et al, 2002). Similarly, BRF1, a member of the Tis11 protein family which is related to TTP, binds to the 14-3-3β isoform in a phosphorylation-dependent manner (Schmidlin et al, 2004). In contrast, the interaction between p37 AUF1 and 14-3-3σ is unlikely to be phosphorylation-dependent because the p37 AUF1 used in our assay was a recombinant protein purified from Escherichia coli, and therefore lacks phosphorylation. In preliminary studies, addition of phosphatases to the interaction assays also had no effect on binding (data not shown). In addition, unlike TTP, which possesses phosphopeptides with sequences similar to that of two consensus 14-3-3 binding motifs (Yaffe et al, 1997), the 20 amino-acid sequence in the CTD of p37 AUF1 which is required for 14-3-3σ binding does not possess homology to other known consensus 14-3-3 binding motifs. This finding is not surprising since a number of 14-3-3 binding partners identified to date also do not require phosphorylation (Andrews et al, 1998). Our finding that both 14-3-3σ and 14-3-3η interact with p37 AUF1 in vitro suggests that AUF1 proteins also belong to the class of 14-3-3 binding partners that lack a requirement for phosphorylation.
The CTD of AUF1 was previously shown to play a significant role in regulating the subcellular distribution of AUF1 proteins (Sarkar et al, 2003a). An intact CTD from p37 and p40 AUF1 isoforms was found to possess nuclear localizing activity, whereas disruption of the CTD by the insertion of exon 7, emulating the p42 and p45 proteins, resulted in cytoplasmic AUF1 accumulation (Sarkar et al, 2003a). This region overlaps both the core 12 amino-acid NLS within the CTD, as well as the site of exon 7 insertion (Figure 4C). In essence, binding of 14-3-3σ and insertion of exon 7 have the same effect on p37 AUF1: both events disrupt the activity of the NLS (Figure 4) and lead to p37 AUF1 cytoplasmic accumulation. It has been well documented that other 14-3-3 proteins employ this same type of mechanism to regulate the intracellular distribution of their respective binding partners. For example, a 14-3-3 protein inhibits nuclear import of cdc25c in human cells (Graves et al, 2001) by interfering with the cdc25 NLS, which is closely associated with the 14-3-3 binding site. Similarly, mammalian cdc25b possesses an NLS that is closely associated with the 14-3-3 binding site (Davezac et al, 2000; Mils et al, 2000).
A key finding of this study is that increased accumulation of p37 AUF1 in the cytoplasm occurs upon 14-3-3σ expression and binding, and corresponds to a reduction in both the steady-state level and the half-life of reporter ARE-mRNAs (Figures 6 and 7). It is important to bear in mind that the level of ectopic 14-3-3σ protein expression in the engineered CHO cells is not very high, and is comparable to endogenous levels of 14-3-3σ expression in HeLa cells (data not shown). Higher expression levels stalled cell cycling and led to cell death, consistent with the known properties of 14-3-3σ (Hermeking et al, 1997; Chan et al, 1999; data not shown). Consequently, the 2.5- to three-fold reduction in ARE-mRNA stability promoted by 14-3-3σ expression is quite significant. The regulation of ARE-mRNA decay facilitated by AUF1 is believed to be complex and likely involves a number of factors in addition to 14-3-3σ. 14-3-3σ is probably only one of these factors, and other factors which are likely rate-limiting have yet to be identified.
Materials and methods
Plasmids
GST-14-3-3σ, GST-14-3-3η and HA-14-3-3σ plasmids were provided by Michael Yaffe. Plasmid GST-14-3-3ζ was provided by Helen Piwnica-Worms. The pBSTR1-1433σ construct was provided by PCR amplification of 14-3-3σ cDNA to generate a NotI site upstream of the AUG codon, and a HindIII site downstream of the termination codon. The PCR product digested with NotI and HindIII was ligated into pBSTR1 (Paulus et al, 1996). Construction of plasmid pET23b-p37 containing full-length/ΔN78/ΔC45 (His-p37 full-length/ΔN78/ΔC45) was described previously (Sarkar et al, 2003a). AUF1 truncation mutants pET23b-p37 ΔC35, ΔC15, and ΔC15PM were generated as described above, by PCR amplification of p37 AUF1 cDNA with a forward primer that introduced a BamH1 site, and a reverse primer that introduced a XhoI site. In the case of ΔC15PM, the reverse primer contains two nucleotide substitutions to give rise to R → G and D → G amino-acid mutations. Primer sequences are available upon request. All constructs were validated by DNA sequence analysis. The pCH110-ARE (LacZ-ARE) reporter containing the 62 bp ARE from the GM-CSF mRNA 3′UTR has been described previously (Laroia et al, 1999).
Recombinant protein purification
All recombinant proteins were expressed in E. coli strain BL21 codon plus bacteria. GST-14-3-3 proteins were purified using glutathione-Sepharose 4B resin (Amersham Biosciences), according to the manufacturer's instructions. His-p37 AUF1 and related truncation and point mutants were purified using Ni2+-NTA resin (Qiagen), following the manufacturer's instructions. Purified proteins were tested for purity by gel electrophoresis and Coomassie blue staining and concentrations determined by spectrometry compared to bovine serum albumin (BSA) standards.
Cell culture and reagents
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% bovine calf serum and gentamicin at 50 μg/ml. Chinese hamster ovary (CHO) cells were maintained in DMEM with 10% fetal BSA and 50 μg/ml gentamicin. For mRNA steady-state and half-life analysis, cells were subcultured for 18 h prior to transfection with Lipofectamine Plus (Invitrogen), at 1 μg DNA per 106 cells, according to the manufacturer's instructions. A CHO-1433σ stable expression cell line was generated by transfecting CHO cells with plasmid pBSTR1-14-3-3σ followed by selection with 8 μg/ml puromycin (Calbiochem) at 2-day post-transfection for 4 weeks. Colonies of cells were then selected and screened for 14-3-3σ protein expression. To inhibit 14-3-3σ expression, 2 μg/ml doxycycline (Dox, Calbiochem) was added to the media for 3 days before cells were used for experiments.
His-p37 AUF1 isolation and mass spectrometry
Whole cell HeLa lysates were obtained by resuspending PBS-washed cells in lysis buffer (0.5% NP40, 150 mM NaCl, 20 mM HEPES, pH 7.5, 4 mM Na-orthovanadate, 1 mM NaF, 25 mM β-glycerophosphate, 1:100 protease inhibitor (Sigma), and 100 μg/ml RNaseA), with the addition of 40 mM imidazole. Cell suspensions were incubated on ice for 15 min, and disrupted with periodic vortexing. Lysate was then cleared by microcentrifugation at 4°C for 10 min. For protein isolation, 5 mg of lysate were precleared with Ni2+-NTA resin for 6 h at 4°C. Simultaneously, 20 μg of His-p37 AUF1 was coupled to 50 μl of Ni2+-NTA resin at 4°C. The NTA–His-p37 AUF1 complex was then added to pre-cleared HeLa lysate and incubated overnight at 4°C with shaking. NTA resin–protein complex was washed three times with washing buffer (0.5% NP40, 150 mM NaCl, 20 mM HEPES, pH 7.5) and the pellet was resuspended in 50 μl of 2 × loading buffer. His-p37 AUF1-associating proteins were separated by electrophoresis in 12% SDS–PAGE, and visualized by Silver Staining (Invitrogen) according to the manufacturer's instructions. In-gel proteolysis and Q-TOF mass-spectrometry analysis was performed by the NYU Protein Analysis Facility.
Immunoprecipitation and in vitro binding assays
For co-immunoprecipitation experiments using HeLa lysate, 800 μg of lysate was mixed with 2 μg of 14-3-3σ antibody, or Flag antibody as a negative control, at 4°C overnight. The following day, 20 μl of washed protein G resin (Santa Cruz) was added to both samples at 4°C with shaking for 2 h. Resin was then washed three times with washing buffer and resuspended in 2 × loading dye. For in vitro binding assays, recombinant GST-14-3-3 proteins and His-p37 AUF1 full-length or truncation mutants were mixed in binding buffer (10 mM HEPES pH 7.5, 5 mM Mg-acetate, 100 mM K-acetate, 0.02% NP-40) at 4°C for 2 h. Pre-washed GSH resin or NTA resin were then added to the mix and incubated at 4°C with shaking for 1 h. Proteins bound to resin were washed three times with binding buffer, and re-suspended in 2 × loading dye. Samples were resolved by SDS–PAGE and visualized by immunoblotting.
Immunofluorescence
Cells grown on coverslips were washed three times with cold PBS, fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100 and treated with 100 μg/ml RNaseA to remove cellular RNA. Cells were then blocked in 3% BSA. Primary antibodies were diluted in 3% BSA and incubated for 2 h at room temperature. 14-3-3 antibody (Upstate Biotechnology) and RPA antibody (Sigma) were used at 0.5 μg/ml, and AUF1 antibody was used at 1:100 dilution of the stock. After washing with PBS, cells were incubated at room temperature for 1 h with secondary anti-mouse or anti-rabbit antibody conjugated to fluorescein isothiocyanate (FITC) (Jackson ImmunoResearch). Cells were washed again, and mounted using Vectashield medium containing propidium iodide (Vector Laboratories). Samples were examined and photographed using a Zeiss LSM510 Meta confocal microscope.
Antibodies
Polyclonal AUF1 antibody has been described previously (Sarkar et al, 2003a). Monoclonal 14-3-3σ and 14-3-3γ antibodies were purchased from Upstate Biotechnology. Monoclonal 14-3-3β/ζ antibody was a gift from Ed Ziff. Antibodies against PABP and GST were from AbCam and Santa Cruz, respectively. PML and β-microtubulin antibodies were both from Sigma.
RNA extraction, Northern blot, steady-state and half-life analysis and quantification
At 24 h post-transfection, cells were washed once with cold PBS and lysed in Trizol reagent (Invitrogen). Total RNA extraction was carried out according to the manufacturer's instructions. For Northern blot analysis, RNA was separated by denaturing formaldehyde gels, transferred onto GeneScreen membrane and hybridized to labeled probes. For quantitative real-time RT–PCR analysis, total RNA was treated to remove residue DNA contaminants using the TURBO DNAfree kit (Ambion) and reverse transcribed followed by PCR amplification using SYBR Green Quantitative RT–PCR kit (Sigma) and a Roche LightCycler.
RNAi expression
Interfering RNAs were delivered directly by transfection of siRNAs. Double-stranded siRNAs were designed following the procedure described by Elbashir et al (2001) based on a reported sequence (Raineri et al, 2004). Lyophylized siRNA duplexes were synthesized (Qiagen Corp. HPP grade) dissolved in siRNA suspension buffer (100 mM potassium acetate, 30 mM HEPES–KOH, 2 mM magnesium acetate, pH 7.4) to a final concentration of 1 μg/μl and stored at −20°C until use. Universal negative (non-silencing) control siRNA was purchased from Qiagen. Cells were transfected with 5 μg of siRNA in 10 cm plates, 24 h after plating at 20% density, using Oligofectimine reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. siRNA transfections were repeated 24 h later, the next day cells were used for LacZ-ARE reporter assays.
Supplementary Material
Supplementary Figure 1
Legend to Supplementary Figure 1
Acknowledgments
We thank Michael Yaffe and Helen Piwnica-Worms for 14-3-3σ plasmids and Werner Paulus for pBPSTR1 plasmid. Cheng He was a recipient of a postgraduate fellowship from the Natural Sciences and Engineering Research Council (NSERC) of Canada. This work was supported by NIH Grant GM 60428 to RJS.
References
- Andrews RK, Harris SJ, McNally T, Berndt MC (1998) Binding of purified 14-3-3 zeta signaling protein to discrete amino acid sequences within the cytoplasmic domain of the platelet membrane glycoprotein Ib-IX-V complex. Biochemistry 37: 638–647 [DOI] [PubMed] [Google Scholar]
- Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS (2001) ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res 29: 246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G (1991) An A+U rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol 11: 2460–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D, Moorhead GB (2005) 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE 2005: re10. [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ (1998) Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ (2000) Evidence that tristetraprolin is a physiological regulator of granulocyte–macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95: 1891–1899 [PubMed] [Google Scholar]
- Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B (1999) 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature 401: 616–620 [DOI] [PubMed] [Google Scholar]
- Davezac N, Baldin V, Gabrielli B, Forrest A, Theis-Febvre N, Yashida M, Ducommun B (2000) Regulation of CDC25B phosphatases subcellular localization. Oncogene 19: 2179–2185 [DOI] [PubMed] [Google Scholar]
- DeMaria CT, Sun Y, Long L, Wagner BJ, Brewer G (1997) Structural determinants in AUF1 required for high affinity binding to A+U-rich elements. J Biol Chem 272: 27635–27643 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Fan XC, Steitz JA (1998) Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 17: 3448–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY (2004) A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell 14: 571–583 [DOI] [PubMed] [Google Scholar]
- Graves PR, Lovly CM, Uy GL, Piwnica-Worms H (2001) Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 protein binding. Oncogene 20: 1839–1851 [DOI] [PubMed] [Google Scholar]
- Guhaniyogi J, Brewer G (2001) Regulation of mRNA stability in mammalian cells. Gene 265: 11–23 [DOI] [PubMed] [Google Scholar]
- Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B (1997) 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell 1: 3–11 [DOI] [PubMed] [Google Scholar]
- Johnson BA, Stehn JR, Yaffe MB, Blackwell TK (2002) Cytoplasmic localization of tristetraprolin Involves 14-3-3-dependent and -independent mechanisms. J Biol Chem 277: 18029–18036 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG (1999) Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev 13: 1067–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol 19: 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G, Cuesta R, Brewer G, Schneider RJ (1999) Control of mRNA decay by heat shock–ubiquitin–proteasome pathway. Science 284: 499–502 [DOI] [PubMed] [Google Scholar]
- Leffers H, Madsen P, Rasmussen HH, Honore B, Andersen AH, Walbum E, Vandekerckhove J, Celis JE (1993) Molecular cloning and expression of the transformation sensitive epithelial marker stratifin. A member of a protein family that has been involved in the protein kinase C signaling pathway. J Mol Biol 231: 982–998 [DOI] [PubMed] [Google Scholar]
- Loflin P, Chen CY, Shyu AB (1999) Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev 13: 1884–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mils V, Baldin V, Goubin F, Pinta I, Papin C, Waye M, Eychene A, Ducommun B (2000) Specific interaction between 14-3-3 isoforms and the human CDC25B phosphatase. Oncogene 19: 1257–1265 [DOI] [PubMed] [Google Scholar]
- Min H, Turck CW, Nikolic JM, Black DL (1997) A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev 11: 1023–1036 [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Xing H (2000) 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal 12: 703–709 [DOI] [PubMed] [Google Scholar]
- Myer VE, Fan XC, Steitz JA (1997) Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J 16: 2130–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus W, Baur I, Boyce FM, Breakefield XO, Reeves SA (1996) Self-contained, tetracycline-regulated retroviral vector system for gene delivery to mammalian cells. J Virol 70: 62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB (1998) RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J 17: 3461–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioli PA, Hamilton BJ, Connolly JE, Brewer G, Rigby WF (2002) Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. J Biol Chem 277: 35738–35745 [DOI] [PubMed] [Google Scholar]
- Raineri I, Wegmueller D, Gross B, Certa U, Moroni C (2004) Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res 32: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar B, Lu JY, Schneider RJ (2003a) Nuclear import and export functions in the different isoforms of the AUF1/hnRNP D protein family. J Biol Chem 278: 20700–20707 [DOI] [PubMed] [Google Scholar]
- Sarkar B, Xi Q, He C, Schneider R J (2003b) Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol Cell Biol 23: 6685–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin M, Lu M, Leuenberger SA, Stoecklin G, Mallaun M, Gross B, Gherzi R, Hess D, Hemmings BA, Moroni C (2004) The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J 23: 4760–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G, Kamen R (1986) A conserved AU sequence from the 3′ untranslated region of GM–CSF mRNA mediates selective mRNA degradation. Cell 46: 659–667 [DOI] [PubMed] [Google Scholar]
- Sirenko OI, Lofquist AK, DeMaria CT, Morris JS, Brewer G, Haskill JS (1997) Adhesion-dependent regulation of an A+U-rich element-binding activity associated with AUF1. Mol Cell Biol 17: 3898–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G (1998) Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics 48: 195–202 [DOI] [PubMed] [Google Scholar]
- Wilson GM, Sutphen K, Chuang K, Brewer G (2001) Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J Biol Chem 276: 8695–8704 [DOI] [PubMed] [Google Scholar]
- Xing H, Zhang S, Weinheimer C, Kovacs A, Muslin AJ (2000) 14-3-3 proteins block apoptosis and differentially regulate MAPK cascades. EMBO J 19: 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB (2002) How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett 513: 53–57 [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91: 961–971 [DOI] [PubMed] [Google Scholar]
- Yang HY, Wen YY, Chen CH, Lozano G, Lee MH (2003) 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Mol Cell Biol 23: 7096–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Winkler K, Yoshida M, Kornbluth S (1999) Maintenance of G2 arrest in the Xenopus oocyte: a role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J 18: 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Legend to Supplementary Figure 1


