Abstract
The U2 and U6 snRNAs contribute to the catalysis of intron removal while U5 snRNA loop 1 holds the exons for ligation during pre-mRNA splicing. It is unclear how different exons are positioned precisely with U5 loop 1. Here, we investigate the role of U2 and U6 in positioning the exons with U5 loop 1. Reconstitution in vitro of spliceosomes with mutations in U2 allows U5–pre-mRNA interactions before the first step of splicing. However, insertion in U2 helix Ia disrupts U5–exon interactions with the intron lariat-3′ exon splicing intermediate. Conversely, U6 helix Ia insertions prevent U5–pre-mRNA interactions before the first step of splicing. In vivo, synthetic lethal interactions have been identified between U2 insertion and U5 loop 1 insertion mutants. Additionally, analysis of U2 insertion mutants in vivo reveals that they influence the efficiency, but not the accuracy of splicing. Our data suggest that U2 aligns the exons with U5 loop 1 for ligation during the second step of pre-mRNA splicing.
Keywords: pre-mRNA splicing, spliceosome, U2 snRNA, U5 snRNA, yeast
Introduction
Transcription in eukaryotic cells produces pre-messenger RNAs (pre-mRNAs) that contain intron regions that are removed by the process of pre-mRNA splicing. Accuracy of splicing is critical for production of functional mRNAs and subsequent synthesis of proteins that define and control cell behaviour. Pre-mRNAs contain conserved sequences found within the introns that define the splice sites and are involved in the splicing reaction. The yeast Saccharomyces cerevisiae (S. cerevisiae) has been employed extensively to study pre-mRNA splicing (Rymond and Rosbash, 1992). The availability of the sequence of the yeast genome has allowed the analysis of the conserved sequences found within the introns required for yeast splicing (Ares et al, 1999; Lopez and Séraphin, 1999). The splice sites are defined by conserved sequences at the 5′ and 3′ ends of the introns termed the 5′ and 3′ splice sites. Within the intron there is a conserved sequence, the branch point, which contributes to intron recognition and catalysis of intron removal.
Removal of introns from pre-mRNA and correct ligation of coding exons is catalyzed by the spliceosome, a large RNA–protein complex. The spliceosome is composed of five small nuclear ribonucleoprotein particles (snRNPs) (U1, U2, U4, U5 and U6 snRNPs) as well as numerous non-snRNP protein splicing factors (reviewed in Will and Lührmann, 2001). The snRNPs contain small nuclear RNAs (snRNAs) that interact with one another, and with the pre-mRNA, such that the splice sites are recognized and the snRNPs catalyze removal of the intron regions (reviewed in Nilsen, 1998). Introns are removed in a two-step transesterification reaction that produces characteristic intermediates and products. In the first step, a 2′ hydroxyl of a conserved adenosine within the branch point sequence attacks the 5′ splice site producing a 5′ exon intermediate. Simultaneously, the 5′ end of the intron is bonded to the adenosine of the branch point producing a looped molecule, called a lariat, to produce the intron lariat-3′ exon intermediate. In the second step, cleavage at the 3′ splice site produces spliced out intron-lariat and ligated mRNA product.
The snRNAs interact with pre-mRNA in an ordered sequence to identify the splice sites (reviewed in Moore et al, 1993; Nilsen, 1998). This process begins with the interaction of U1 with the 5′ splice site and U2 with the branch site. The U4 and U6 snRNAs are extensively base-paired and join the spliceosome with the U5 snRNA. Once all the snRNAs are present, a complex set of RNA rearrangements occurs to form the active spliceosome competent for the two catalytic steps of splicing. Specifically, U1 interaction at the 5′ splice site is exchanged for U6 at the intron side of the 5′ splice site and U5 loop 1 at the exon side of the 5′ splice site. In addition, interaction of U6 with U4 is disrupted allowing U6 to establish a mutually exclusive base-pairing interaction with conserved intron sequences at the 5′ splice site and with U2. The U2/U6 base-pairing forms helix Ia and Ib that are proposed to be part of the catalytic site of the spliceosome (Madhani and Guthrie, 1992, 1994). A recent structural study of the U2/U6 interaction proposes that U2 and U6 may form intramolecular contacts instead of helix Ib prior to the first step of splicing (Sashital et al, 2004). A phosphate at U80 of the S. cerevisiae U6 snRNA has been shown to contribute to catalysis at the 5′ splice site during the first step of splicing (Yean et al, 2000). Following the first step of splicing, U5 loop 1 maintains an interaction with the 5′ exon intermediate while forming a new interaction with the exon of the intron lariat-3′ exon intermediate. These U5–exon interactions hold the exon ends in the correct orientation for ligation during the second step of splicing (Newman and Norman, 1992; Sontheimer and Steitz, 1993; Newman et al, 1995; O'Keefe et al, 1996; O'Keefe and Newman, 1998) and are critical for the second step of splicing in the yeast S. cerevisiae (O'Keefe and Newman, 1998). Tertiary interactions between U2 A25 and U6 G52 (Madhani and Guthrie, 1994; Valadkhan and Manley, 2000) and an interaction of the U2 snRNA position U23 with the exon of the intron lariat-3′ exon intermediate (Newman et al, 1995) both occur before the second step of splicing.
Inherently, all exon sequences in pre-mRNAs are different as they will code for different protein products. To date, however, it is not known how U5 loop 1 is directed precisely to the hundreds of different exons at the splice sites. We hypothesized that the other snRNAs in the spliceosome may direct the exons to U5 loop 1. The U2 and U6 snRNAs were likely candidates as they both interact with the highly conserved intron sequences that define the splice sites and U2 interacts directly with the exon of the intron lariat-3′ exon intermediate following the first step of splicing (Newman et al, 1995). In yeast, a number of positions in U2 (G26 and helix Ia insertions) and U6 (A51, G52, C58 and A59) when mutated inhibit the second step of splicing in vitro (Fabrizio and Abelson, 1990; McPheeters and Abelson, 1992). Inhibition of the second step of splicing is also observed when exons are misaligned by changing the size of U5 loop 1 (O'Keefe and Newman, 1998). In addition, genetic analysis of some of these positions in U6 revealed that, while they base-pair with the U2 snRNA to form helix I, they may have an additional role in splicing (Madhani and Guthrie, 1992, 1994; Hilliker and Staley, 2004).
To address whether mutations in the U2 and U6 snRNAs, which block the second step of splicing, can influence the interaction of U5 loop 1 with exon sequences during splicing, we have reconstituted a number of U2 and U6 mutants in vitro and monitored their affects on U5–exon interactions. It was found that point mutations in U2 and U6 snRNA that block the second step of splicing do not affect the interaction of U5 loop 1 with exon sequences. However, a five-nucleotide insertion mutant of U2 in helix Ia allowed U5 loop 1 interactions with the pre-mRNA and 5′ exon intermediate but prevented the interaction of U5 with the intron lariat-3′ exon splicing intermediate. In contrast, insertion mutants of U6 in helix Ia prevented the interaction of U5 with the pre-mRNA. In vivo, synthetic lethal genetic interactions between U5 loop 1 insertion and U2 insertion mutants were identified. Furthermore, analysis in vivo revealed that U2 insertion mutants influenced the efficiency of the two steps of splicing but not the accuracy of splicing. Overall, these data suggest that the U2 snRNA is required for aligning the exons with U5 loop 1 for ligation during the second catalytic step of pre-mRNA splicing. The ability of mutants in U2 to influence the interaction of exons with U5 provides insight into how the spliceosome can bring together different exon sequences to produce functional mRNA following intron removal.
Results
Mutations in U2 and U6 inhibit the second step or both steps of pre-mRNA splicing in vitro
A number of mutations and insertions flanking, and within, the helix Ia region formed by the U2 and U6 snRNAs are known to block the second step of splicing in vitro (Figure 1A). To determine whether these and other mutations in U2 and U6 influence the interaction of U5 loop 1 with exon sequences, we exploited the ability to deplete the U2 or U6 snRNA from whole-cell extracts of the yeast S. cerevisiae by RNase H targeting of the snRNA in the presence of complementary DNA oligonucleotides (Fabrizio et al, 1989; McPheeters et al, 1989). Splicing of a CYH2 based pre-mRNA in vitro is inhibited by depletion of U2 (Figure 1B, lane 2) or U6 (Figure 1C, lane 2) with complementary oligonucleotides. Addition of in vitro transcribed U2 or U6 snRNA to extracts depleted of U2 or U6 reconstitutes functional U2 or U6 snRNPs and both steps of pre-mRNA splicing in vitro (Figure 1B, lane 3, and C, lane 3; Fabrizio et al, 1989; McPheeters et al, 1989). However, insertion of one (U2 Ins1 28/29), three (U2 Ins3 28/29) or five (U2 Ins5 28/29) uridines between positions 28 and 29 in U2 helix Ia (Figure 1B, lanes 4–6) allowed the first step but progressively inhibited the second step of splicing (McPheeters and Abelson, 1992). In addition, U2 mutant G26C (Figure 1B, lane 7), U6 mutant A51U and U6 mutant A59U (Figure 1C, lanes 7 and 8) also inhibited the second step of splicing (Fabrizio and Abelson, 1990; McPheeters and Abelson, 1992). Alternatively, insertion of one (U6 Ins1 55/56), three (U6 Ins3 55/56) or five (U6 Ins5 55/56) uridines between positions 55 and 56 in U6 helix Ia, opposite the U2 insertions, progressively inhibited both steps of splicing (Figure 1C, lanes 4–6). Taken together, these data indicate that there is asymmetry in the effects on splicing of insertion mutations in the U2 and U6 strands of helix Ia. Phenotypic asymmetry resulting from alterations in interacting U2 and U6 nucleotides of the helix I region has been observed previously in vivo (Madhani and Guthrie, 1992, 1994).
Figure 1.
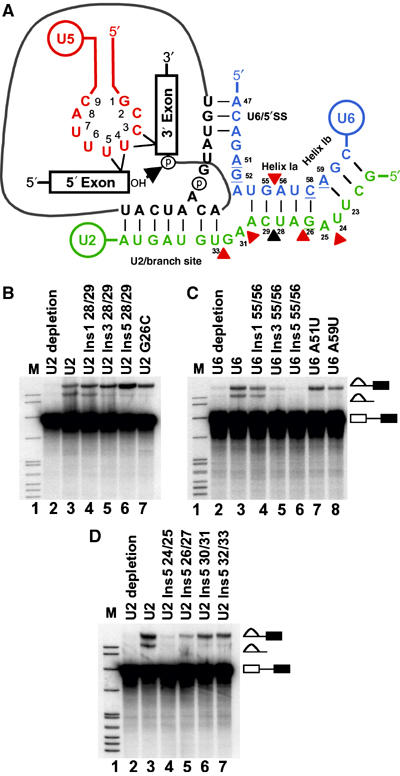
In vitro splicing in the presence of U2 and U6 snRNA mutations. (A) RNA–RNA interactions prior to the second catalytic step of pre-mRNA splicing in S. cerevisiae. Nucleotides in U5 snRNA loop 1 (red) numbered 1–9. Nucleotides in U2 (green) and U6 (blue) that block the second step of splicing when mutated are underlined (Fabrizio and Abelson, 1990; McPheeters and Abelson, 1992). Position where insertions in U2 progressively block the second step of splicing is identified with a black triangle (McPheeters and Abelson, 1992). Positions of insertions in U2 and U6 that are unique to this work are indicated by red triangles. (B, D) Uniformly labeled CYH2 based pre-mRNA used to monitor in vitro splicing in U2-depleted extract reconstituted with specified RNAs. The splicing intermediates and products indicated on the right. The 5′ exon intermediates and mRNA products are not shown. (C) Uniformly labeled CYH2 based pre-mRNA used to monitor in vitro splicing in U6-depleted extract reconstituted with specified RNAs. The splicing intermediates and products indicated on the right. The 5′ exon intermediates and mRNA products are not shown.
As U2 helix Ia insertion mutants between positions 28/29 blocked only the second step of splicing, it was of interest to determine how insertion mutants in different regions of U2 influenced splicing. Insertion mutants containing five uridines were produced in distinct locations on either side of positions 28/29. These mutants comprised an insertion in U2 helix Ia (U2 Ins5 26/27), an insertion in the bulge between U2 helix Ia and Ib (U2 Ins5 24/25) and two separate insertions between U2 helix Ia and the branch site binding sequence (U2 Ins5 30/31 and U2 Ins5 32/33) (Figure 1A). Addition of in vitro-transcribed U2 mutants U2 Ins5 30/31 and U2 Ins5 32/33 to U2-depleted extract allowed the first step of splicing but inhibited the second step of splicing (Figure 1D, lanes 6 and 7). Mutant U2 Ins5 26/27 in helix Ia also displayed a second step block but with slight reduction in the first step of splicing (Figure 1D, lane 5). In contrast, mutant U2 Ins5 24/25, in the bulge between U2 helix Ia and Ib, inhibited both steps of splicing (Figure 1D, lane 4). All U2 and U6 mutants displayed similar in vitro splicing phenotypes with a pre-mRNA derived from the ACT1 gene (Supplementary Figure S1). Thus, mutations within U2 or U6 can either block the second step or block both steps of pre-mRNA splicing in vitro.
Crosslinking of U5 loop 1 to the pre-mRNA before the first step of splicing is disrupted by insertion mutants in U6, but not U2
To monitor the interaction of U5 snRNA loop 1 with the 5′ exon of the pre-mRNA and 5′ exon intermediate in the presence of U2 or U6 mutants, a CYH2 based pre-mRNA containing a single crosslinkable 4-thio-uridine (4-thioU) in the 5′ exon three nucleotides from the 5′ splice site position (−3) and a single 32P between positions (−1) and (−2) was employed. Addition of this pre-mRNA to in vitro splicing reactions and irradiation with long-wave UV allows one to monitor U5–pre-mRNA interactions before the first step of splicing and U5–5′ exon intermediate interactions following the first step of splicing (Newman et al, 1995; O'Keefe et al, 1996; O'Keefe and Newman, 1998; Alvi et al, 2001). To visualise U5–pre-mRNA and U5–5′ exon intermediate crosslinks, total RNA isolated from UV-irradiated splicing reactions was hybridized with a biotinylated oligonucleotide complementary to U5 and U5-containing molecules were specifically captured with streptavidin paramagnetic particles. Captured RNA was then eluted from the particles and separated by gel electrophoresis. By utilizing this technique a radioactive band will only be present in the gel when there is a direct crosslink between the unlabeled U5 snRNA and the labeled pre-mRNA or splicing intermediate. In UV-irradiated splicing reactions containing wild-type U2 (Figure 2A and C, lane 3) or U6 (Figure 2B and D, lane 3), U5 biotinylated oligonucleotide captured four crosslinked species. RNase H analysis of these crosslinks revealed that they were the short and long forms of U5 crosslinked to the pre-mRNA and 5′ exon intermediate (Figure 3A). No U5–pre-mRNA or U5–5′ exon intermediate crosslinks were captured in an extract depleted of U2 (Figure 2A, lane 2) or U6 snRNA (Figure 2B, lane 2). Reconstitution with U2 insertion mutants (Figures 2A, lanes 4–6, and C, lanes 4–7) and U2 mutant G26C (Figure 2A, lane 7) still allowed formation of U5–pre-mRNA crosslinks. It is interesting to note that U2 mutant U2 Ins5 24/25, which blocked both steps of splicing, still allowed U5–pre-mRNA interactions (Figure 2C, lane 4). U6 mutants A59U, A51U and U6 Ins1 55/56 (Figure 2B, lanes 4 and 5, and D, lane 4) still allowed formation of U5–pre-mRNA crosslinks with increased U5–pre-mRNA cross–link efficiency of mutant A59U. However, reconstitution with U6 Ins3 55/56 and U6 Ins5 55/56 mutants, which blocked the first step of splicing, did not allow U5–pre-mRNA crosslinks (Figure 2D, lanes 5 and 6). The block of U5–pre-mRNA interactions with U6 Ins3 55/56 and U6 Ins5 55/56 may result from the mutations or the inability of the mutants to assemble into functional snRNPs (see Discussion). In either case, as these mutants block the first step of splicing, do not allow U5–pre-mRNA interactions and do not allow the formation of splicing intermediates their affects on U5–exon interactions can not be investigated further.
Figure 2.
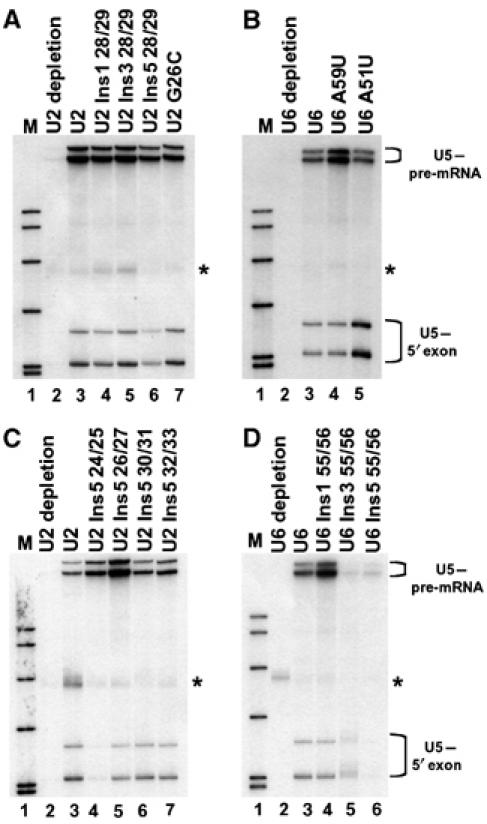
Isolated crosslinks of U5 snRNA with the pre-mRNA and 5′ exon intermediate in the presence of U2 and U6 mutations. U5 snRNA was selected from reconstituted splicing reactions with a biotinylated oligonucleotide complementary to U5 and streptavidin paramagnetic particles. (A, C) Selection of U5 snRNA from U2-depleted extract reconstituted with specified RNAs and CYH2 pre-mRNA containing 4-thioU at (−3) in the 5′ exon. Crosslinking of the two forms of U5 to the pre-mRNA and 5′ exon intermediate indicated at right. (B, D) Selection of U5 snRNA from U6-depleted extract reconstituted with specified RNAs and CYH2 pre-mRNA containing 4-thioU at (−3) in the 5′ exon. Crosslinking of the two forms of U5 to the pre-mRNA and 5′ exon intermediate indicated at right. Asterisks indicate background pre-mRNA captured with streptavidin paramagnetic particles.
Figure 3.
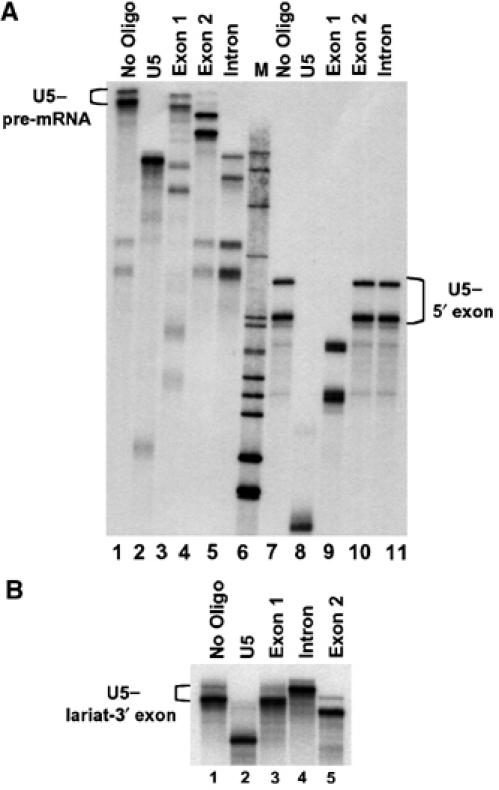
RNase H analysis of U5 snRNA-exon crosslinks. (A) RNase H analysis of isolated U5–pre-mRNA and U5-5′ exon intermediate crosslinks with specified oligonucleotides. (B) RNase H analysis of isolated U5–intron lariat-3′ exon intermediate crosslinks with specified oligonucleotides. In lane 4 the mobility of the crosslinks is decreased following RNase H treatment as the intron-lariat is broken to form a branched RNA. Additional bands seen below the major crosslinks are a combination of degradation products that appear as a result of the incubation under the RNase H reaction conditions and specific bands resulting from oligonucleotide directed RNase H cleavage.
The interaction of U5 loop 1 with the 5′ exon splicing intermediate produced following the first step of splicing can be monitored with the pre-mRNA containing a single 4-thioU in the 5′ exon at position (−3). Reconstitution with U6 mutants A59U, A51U and U6 Ins1 55/56, which allowed U5–pre-mRNA crosslinks, also allowed U5–5′ exon intermediate crosslinks with increased crosslink efficiency of mutant A51U (Figure 2B, lanes 4 and 5, and D, lane 4). Reconstitution with six of the U2 insertion mutations (Figure 2A, lanes 4–6, and C, lanes 5–7) and U2 mutant G26C (Figure 2A, lane 7) still allowed formation of U5–5′ exon splicing intermediate interactions. In contrast, the insertion mutant U2 Ins5 24/25, in the bulge between U2 helix Ia and Ib, prevented the U5–5′ exon intermediate interaction (Figure 2C, lane 4). However, as mutant U2 Ins5 24/25 did not allow the first step of splicing (Figure 1D, lane 4) there is no 5′ exon intermediate produced that could crosslink to U5.
Mutation in U2 prevents U5 loop 1 crosslinking to the intron lariat-3′ exon intermediate
To investigate how U5 loop 1 interacted with the intron lariat-3′ exon intermediate in the presence of U2 and U6 mutants, a CYH2 based pre-mRNA containing a single crosslinkable 4-thioU in the 3′ exon two nucleotides from the 3′ splice site position (+2) and a single 32P at the 3′ splice site was utilized. In UV-irradiated reconstitution reactions containing wild-type U2 (Figure 4A and C, lane 2) or U6 (Figure 4B and D, lane 2), U5 biotinylated oligonucleotide captured a doublet of crosslinked species. RNase H analysis of the doublet revealed that these crosslinked species were the two forms of U5 crosslinked to the intron lariat-3′ exon intermediate (Figure 3B). No U5–intron lariat-3′ exon intermediate crosslinks were captured in an extract depleted of U2 (Figure 4A and C, lane 1) or U6 snRNA (Figure 4B and D, lane 1). Reconstitution with U6 mutants A59U, A51U (Figure 4B, lanes 3 and 4) and U6 Ins1 55/56 (Figure 4D, lane 3) still allowed formation of U5–intron lariat-3′ exon crosslinks but with varying intensity. Reconstitution with U2 mutants G26C (Figure 4A, lane 3), U2 Ins1 28/29, U2 Ins3 28/29 (Figure 4C, lanes 3 and 4), U2 Ins5 26/27, U2 Ins5 30/31 and U2 Ins5 32/33 (Figure 4E, lanes 3–5) still allowed formation of U5–intron lariat-3′ exon crosslinks. Reactions containing mutant U2 Ins5 24/25 (Figure 4E, lane 2) prevented the U5–intron lariat-3′ exon intermediate interaction. However, as mutant U2 Ins5 24/25 did not allow the first step of splicing (Figure 1D, lane 4), there is no intron lariat-3′ exon intermediate produced that could crosslink to U5. Interestingly, reactions containing mutant U2 Ins5 28/29, which allowed the first step of splicing, prevented the U5–intron lariat-3′ exon intermediate interaction (Figure 4C, lane 5). This indicates that the helix Ia region of U2 is required for the association of the intron lariat-3′ exon intermediate with the U5 snRNA loop 1. A summary of both in vitro splicing and U5–exon interactions in the presence of U2 and U6 snRNA mutants is presented in Table I.
Figure 4.
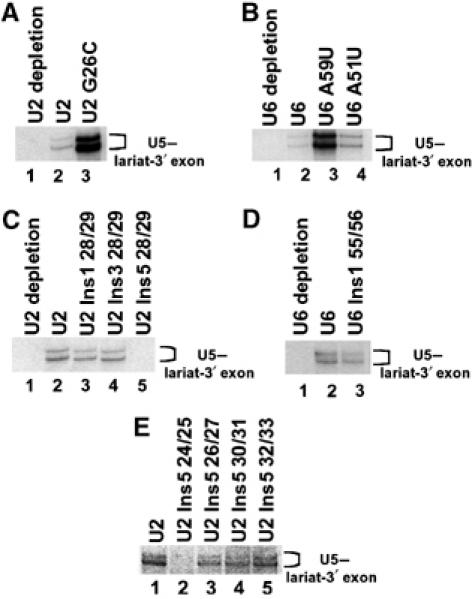
Isolated crosslinks of U5 snRNA with the intron lariat-3′ exon intermediate in the presence of U2 and U6 snRNA mutations. U5 snRNA was selected from reconstituted splicing reactions with a biotinylated oligonucleotide complementary to U5 and streptavidin paramagnetic particles. (A, C, E) Selection of U5 snRNA from U2-depleted extract reconstituted with specified RNAs and CYH2 pre-mRNA containing 4-thioU at (+2) in the 3′ exon. Crosslinking of the two forms of U5 to the intron lariat-3′ exon intermediate indicated at right. (B, D) Selection of U5 snRNA from U6-depleted extract reconstituted with specified RNAs and CYH2 pre-mRNA containing 4-thioU at (+2) in the 3′ exon. Crosslinking of the two forms of U5 to the intron lariat-3′ exon intermediate indicated at right.
Table 1.
Summary of in vitro splicing and crosslinking
| U5 crosslinking to |
||||
|---|---|---|---|---|
| Splicing | Pre-mRNA | 5′ exon | Lariat-3′ exon | |
| U2 | WT | + | + | + |
| U2 Ins1 28/29 | Slight step 2 block | + | + | + |
| U2 Ins3 28/29 | Step 2 block | + | + | + |
| U2 Ins5 28/29 | Step 2 block | + | + | − |
| U2 Ins5 24/25 | Step 1 block | + | − | − |
| U2 Ins5 26/27 | Slight step 1/2 block | + | + | + |
| U2 Ins5 30/31 | Step 2 block | + | + | + |
| U2 Ins5 32/33 | Step 2 block | + | + | + |
| U2 G26C | Step 2 block | + | + | + |
| U6 | WT | + | + | + |
| U6 Ins1 55/56 | WT | + | + | + |
| U6 A51U | Step 2 block | + | + | + |
| U6 A59U | Step 2 block | + | + | + |
| Data for this table were taken from Figures 1, 2 and 4. | ||||
In circumstances where U5–exon crosslinks were still present with mutations in the U2 or U6 snRNAs, it was important to determine whether the crosslinks were to the same position on U5 loop 1 as compared to wild-type. Selected U5–pre-mRNA and U5–5′ exon intermediate crosslinks were mapped by primer extension to establish the site of attachment of the 5′ exon to U5 loop 1. It was found that in the presence of U2 G26C, U2 Ins5 28/29, U6 A51U and U6 A59U mutants, the site of attachment of the 5′ exon in the pre-mRNA and 5′ exon intermediate were to the same positions in U5 loop 1 as a wild-type extract (Supplementary Figure S2). Selected U5–intron lariat-3′ exon intermediate crosslinks were also mapped by primer extension. It was found that the site of attachment of the 3′ exon of the intron lariat-3′ exon intermediate was to the same positions in U5 loop 1 in the presence of U2 G26C, U6 A59U and A51U mutants as a wild-type extract (Supplementary Figure S2).
Synthetic lethal interactions between U5 loop 1 insertion and U2 insertion mutants
As both U2 and U6 can influence U5–exon interactions, this implies a possible interaction between U2, and/or U6, with U5 loop 1. To test this hypothesis, we investigated U2 and U6 interactions with U5 loop 1 in vivo. As a genetic interaction had already been identified between the U5 loop 1 and U2 positions in helix I (Xu et al, 1998), we decided to investigate how the size of U5 loop 1 related to insertion mutants in U2. U2 single-nucleotide insertion mutants at positions 24/25 and 26/27 were lethal with wild-type U5 (Table II and Supplementary Figure S3). In contrast, U2 single-nucleotide insertions at positions 28/29, 30/31 and 32/33, within helix Ia and between helix Ia and the branch site binding sequence were viable with wild-type U5 or U5 loop 1 single nucleotide deletions U5 DelG1 and U5 DelC2 (Table II and Supplementary Figure S3). However, these U2 single-nucleotide insertions were synthetically lethal with U5 loop 1 single-nucleotide insertions U5 Ins1/2 and U5 Ins2/3 (Table II and Supplementary Figure S3). It appears, therefore, that an insertion in U2 is compatible with a deletion in U5 loop 1, whereas an insertion in U2 is not compatible with an insertion in U5 loop 1. This suggests that the spacing of U2 between the branch site binding sequence and helix Ia is related to the size of U5 loop 1.
Table 2.
Synthetic lethal interactions between U5 loop 1, U2 and U6
| U5 | U5 Del G1 | U5 Ins1 1/2 | U5 Del C2 | U5 Ins1 2/3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| U2 | + | + | + | + | + | ||||||
| U6 | + | + | + | + | + | ||||||
| U2 Ins1 24/25 | − | − | − | − | − | ||||||
| U2 Ins1 26/27 | − | − | − | − | − | ||||||
| U2 Ins1 28/29 | + | + | − | + | − | ||||||
| U2 Ins1 30/31 | + | + | +/− | + | − | ||||||
| U2 Ins1 32/33 | + | + | − | + | − | ||||||
| U6 Ins1 55/56 | + | − | − | − | − | ||||||
| U5 loop 1 insertion (Ins1) mutants contain one uridine between the loop 1 position numbers indicated and U2 or U6 insertion (Ins1) mutants contain one uridine between position numbers indicated. | 5-FOA plates were scored after 3 days at 30°C. +, normal growth; −, no growth; +/−, slow growth. | ||||||||||
We next investigated a U6 single-nucleotide insertion (U6 Ins 55/56) in helix Ia with both U5 loop 1 insertion and deletion mutants. We found that U6 mutant Ins1 55/56 was viable with wild-type U5 but synthetically lethal with both U5 single-nucleotide insertions and deletions (Table II and Supplementary Figure S3). This is the first evidence of a genetic interaction between the U5 and U6 snRNAs. Additionally, as a U6 insertion opposite a U2 insertion in helix Ia displays distinct genetic interactions with U5 loop 1 insertions and deletions, this again reinforces the idea that there is asymmetry in the affects of mutation in helix Ia (Madhani and Guthrie, 1992, 1994).
Insertion mutants in U2 affect the efficiency of splicing in vivo
As insertion mutants in U2 influenced the interaction of the intron lariat-3′ exon intermediate with U5 loop 1 and were synthetic lethal with U5 loop 1 insertion mutants, it is predicted that insertion mutants in U2 might also influence the efficiency and specificity of the second step of splicing. To test this prediction, we constructed yeast strains containing viable U2 single nucleotide insertion mutations U2 Ins1 28/29, U2 Ins1 30/31 or U2 Ins1 32/33 as the sole source of U2 in the strains. These strains were transformed with a reporter plasmid harboring the ACT1 5′ exon, intron and a small portion of 3′ exon fused in frame to a luciferase reporter gene containing either a wild-type 5′ splice site or a G1 to C 5′ splice site mutation (Figure 5A). The G1 to C mutation at the 5′ splice site blocks the second step of splicing preventing production of mRNA, but also has reduced efficiency of the first step of splicing leading to an accumulation of pre-mRNA (Vijayraghavan et al, 1989; Lesser and Guthrie, 1993). Protein extract and total RNA prepared from these strains were utilized to assay splicing in vivo by luciferase assay and primer extension, respectively. Each of the U2 single nucleotide insertion mutations U2 Ins1 28/29, U2 Ins1 30/31 and U2 Ins1 32/33 inhibited splicing of the luciferase reporter gene to at least 50% of the level of wild-type U2 (Figure 5B). Negligible luciferase activity was detected with the G1 to C containing reporter construct (Figure 5B). Primer extension analysis with a primer complementary to the ACT1 intron revealed an accumulation of both pre-mRNA and intron lariat-3′ exon intermediate with each U2 single nucleotide insertion mutant (Figure 5C). This is indicative of decreased efficiency of both the first and second steps of splicing, which is revealed more clearly by comparing the levels of pre-mRNA and intron lariat-3′ exon intermediates between the wild-type and mutants through quantitation of the primer extension products (Figure 5D). To investigate the levels of mRNA produced from the reporter constructs and the accuracy of exon ligation in the presence of U2 insertion mutants, RT–PCR analysis was performed. Primers were utilized that produced a small PCR product for the mRNA and PCR products were separated on a sequencing gel to detect any changes in the accuracy of splicing. Cells with U2 insertion mutants contained mRNA that migrated at the same position in the gel as mRNA from cells with wild-type U2 from the reporter containing the wild-type 5′ splice site (Figure 5E). Thus, U2 insertion mutations influence the efficiency, but not the accuracy, of splicing in vivo.
Figure 5.
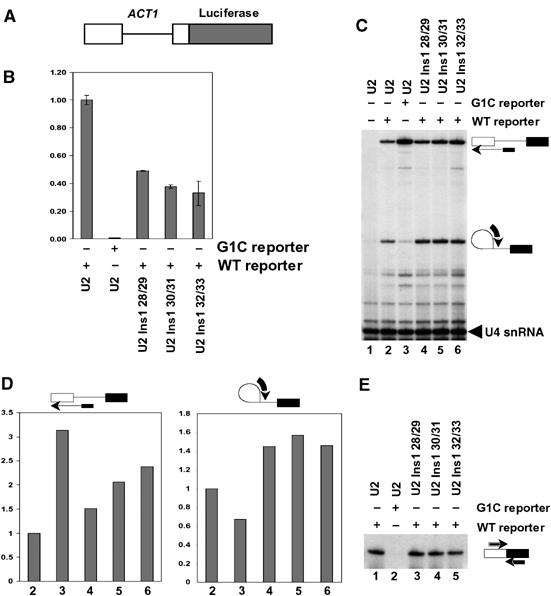
In vivo analysis of pre-mRNA splicing in the presence of U2 snRNA insertion mutations. (A) Schematic of ACT1-luciferase reporter pre-mRNA utilized to assay pre-mRNA splicing in vivo. (B) Quantitation of luciferase activity in the presence of wild-type U2 or the indicated U2 insertion mutants. Data are shown relative to the wild-type reporter in the presence of wild-type U2, which has been set at 1.00. Mean values and standard deviations from three experiments are shown. (C) Primer extension analysis with a primer specific for the ACT1 intron of RNA isolated from cells with the indicated U2 snRNA and reporter construct. Primer extension products for pre-mRNA and lariat intron-3′ exon intermediate are indicated on the right. A primer specific for the U4 snRNA was included as a loading control. (D) Quantitation by phosphorimaging of the results presented in (C). Data are shown relative to the wild-type reporter in the presence of wild-type U2, which has been set at 1. (E) RT–PCR analysis of mRNA from cells containing the indicated U2 snRNA and reporter construct with primers specific to the 5′ and 3′ exons of the reporter construct.
Discussion
Coordinated alignment of the two exon ends for ligation during pre-mRNA splicing is an essential step in the production of functional mRNA. In this study, we have investigated how the U2 and U6 snRNAs influence the alignment of the exon ends with U5 snRNA loop 1 for ligation during the second step of splicing. We found that mutation in U2 helix Ia allowed the interaction of U5 loop 1 with the pre-mRNA but prevented the interaction of U5 loop 1 with the intron lariat-3′ exon intermediate in vitro. In contrast, U6 helix Ia insertion mutants prevented the interaction of U5 loop 1 with the pre-mRNA. In vivo, genetic interactions were identified between U2 and U5 that were related to the size of U5 loop 1. The first genetic interactions between U5 and U6 were also identified with these interactions displaying differences to the U2/U5 genetic interactions. Furthermore, analysis of splicing in the presence of U2 insertion mutants in vivo revealed that these mutants influenced the efficiency, but not the accuracy, of splicing. Taken together, these data suggest that the U2 snRNA is required for the alignment of the exon ends with U5 loop 1 for the second catalytic step of splicing.
It is well established that U2 base pairs with U6 to form helix Ia and Ib. However, there is evidence of asymmetry in the affects of mutations in the U2 and U6 regions of helix I (Madhani and Guthrie, 1992, 1994). We have found that insertion mutants at opposite positions in helix Ia displayed asymmetry in their influence on splicing and U5–exon interactions. U2 helix Ia insertion mutants at position 28/29 allowed the first step of splicing whereas U6 helix Ia insertion mutants at position 55/56, opposite the U2 insertions, progressively blocked both steps of splicing. In addition, there is also a difference in the synthetic lethal interactions of U2 and U6 helix Ia insertion mutants with U5 loop 1 insertion and deletion mutants. There are two possible explanations for these results. First, the helix Ia region of U2 may be tolerant of insertions or may allow alternative base-pairings that maintain the integrity of helix Ia. On the other hand, our results could suggest that helix Ia does not form prior to the first step of splicing.
The idea of alternative RNA interactions to U2/U6 helix I is not new. During yeast spliceosome assembly, when U4 is released from U6, it has been proposed that U6 may form an intramolecular stem-loop (ISL) before interacting with U2 to form helix I (Fortner et al, 1994). In yeast, helix Ib is essential only when helix II is disrupted (Field and Friesen, 1996). However, the helix Ib structure has been shown to be important for 5′ splice site selection in yeast (Luukkonen and Séraphin, 1998b). In the mammalian spliceosome, U2/U6 helix Ib does not form and the nucleotides of this region participate in intramolecular base-pairing (Sun and Manley, 1995). A model for the formation of the catalytic core of the minor U12-dependent spliceosome proposes two assembly pathways for helix I (Frilander and Steitz, 2001). In one pathway, the U6atac remains associated with the U4atac precluding formation of helix I with the U12 snRNA. In an alternative pathway the U6atac forms helix Ia with the U12 snRNA and the U6atac region of helix Ib remains associated with the U4atac.
Recently, a structural study of the U2/U6 interaction was used to propose a model where, prior to the first step of splicing, helix Ib is not formed and sequences encompassing helix Ib are included in extended intramolecular helices U6 ISL and U2 stem I (Figure 6A) (Sashital et al, 2004). Following the first step of splicing, U2 and U6 would then form the previously proposed helix Ia and Ib required for the second step of splicing (Figure 6B) (Madhani and Guthrie, 1992). While our data do not support or refute this model, it does suggest that there is asymmetry in the region of helix Ia between the U2 and U6 snRNAs. It will be interesting to determine any additional interactions that the residues of U2/U6 in the helix Ia region might participate in prior to the first step of splicing in yeast.
Figure 6.
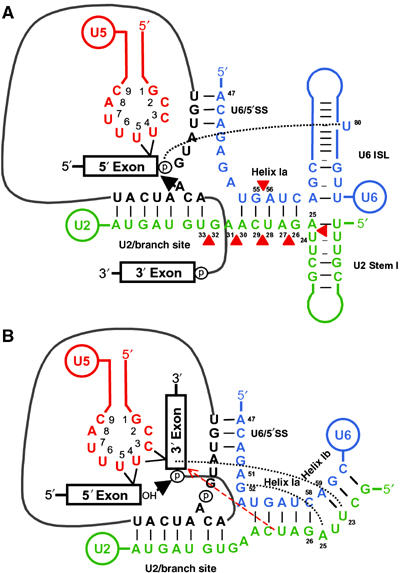
Models of RNA–RNA interactions in the yeast spliceosome required for the first and second steps of pre-mRNA splicing. (A) Intermolecular interactions are depicted prior to the first step of splicing between U5 loop 1 and the 5′ exon, U2 and pre-mRNA at the branch site (U2/branch site) and U6 and pre-mRNA at the 5′ splice site (U6/5′SS). Intramolecular stem loops proposed for U2 (U2 stem I) and U6 (U6 ISL) are indicated (Sashital et al, 2004). An arrow identifies the branch site adenosine attack of the 5′ splice site phosphate. Positions of insertions in U2 and U6 are indicated by red triangles. Black dotted line identifies the potential interaction of the metal binding site at U6 U80 required for 5′ splice site cleavage (Yean et al, 2000). (B) Intermolecular interactions are depicted prior to the second step of splicing between U5 loop 1 and the 5′ and 3′ exons, U2 and intron lariat-3′ exon intermediate at the branch site (U2/Branch site), U6 and intron lariat-3′ exon intermediate at the 5′ splice site (U6/5′SS), and between U2 and U6 to form helix Ia and Ib. A black arrow identifies the 5′ exon attack of the 3′ splice site phosphate. Black dotted lines identify the tertiary interaction of U6 G52 with U2 A25 (Madhani and Guthrie, 1994; Valadkhan and Manley, 2000) and crosslink of the 3′ exon of the intron lariat-3′ exon intermediate to U2 position U23 (Newman et al, 1995). A red dashed line with arrow represents potential interaction of the U2 snRNA with the splicing intermediate suggested from the influence of U2 mutant Ins5 28/29 on U5–lariat intron 3′-exon intermediate crosslinks.
We have not ascertained exactly how U6 insertion mutants in helix Ia inhibit the first step of splicing. U6 helix Ia insertions could disrupt the proposed interaction of the metal binding site at U6 U80 with the 5′ splice site required for the first step of splicing (Yean et al, 2000). On the other hand, previous studies of mutations in the helix Ia region of U6, which also inhibit the first step of splicing, revealed an accumulation of A1 spliceosome complexes indicative of a defect before the rearrangements required for spliceosome activation (Ryan and Abelson, 2002). We have shown that the U6 Ins3 55/56 and Ins5 55/56 mutants inhibit U5–pre-mRNA interactions at the (−3) position upstream of the 5′ splice site. There are two distinct interactions of U5 with the pre-mRNA prior to the first step of splicing in yeast, an early interaction in the (−8) region that is not maintained following the first step of splicing and a later interaction at the (−1) to (−3) region that is maintained following the first step of splicing (Newman et al, 1995). It will be interesting to determine whether the early (−8) interaction of U5 with the pre-mRNA still occurs in the presence of the U6 Ins3 55/56 and Ins5 55/56 mutants. Nevertheless, there is the possibility that U6 Ins3 55/56 and Ins5 55/56 mutants may not assemble into snRNPs.
It was previously found that insertions between positions 28 and 29 of U2 in helix Ia block the second step of splicing (McPheeters and Abelson, 1992). In an extension of that work we have analyzed U5–exon interactions in the presence of one (U2 Ins1 28/29), three (U2 Ins3 28/29) or five (U2 Ins5 28/29) inserted uridines. In addition, we investigated five nucleotide insertions at different locations in and around the U2 helix Ia region. We found that pre-mRNA interactions with U5 loop 1 were present with all mutants, whereas intron lariat-3′ exon intermediate interactions with U5 loop 1 were inhibited in the presence of U2 mutant U2 Ins5 28/29. This suggests that the helix Ia region of U2 may be responsible for aligning the 3′ exon with U5 loop 1 (Figure 6B). The U2/U6 helix Ia region has been implicated in 3′ splice site selection (Chang and McPheeters, 2000). As the (+1) position in the exon of the intron lariat-3′ exon intermediate can be crosslinked to U2 nucleotide U23 and U5 loop 1 following the first step of splicing (Newman et al, 1995), this places U2/U6 helix I, U5 snRNA loop 1 and the 3′ exon in close proximity during the second step of splicing. In vivo, synthetic lethal interactions we have identified between U2 and U5 point to a possible relationship between the size of U5 loop 1 and the spacing within U2 from the region of helix Ia to the branch point binding region. In addition, U2 insertion mutants affect the efficiency of both steps of splicing in vivo. It remains to be determined whether there is any direct interaction between U2 and U5 loop 1.
The point mutants U2 G26C, U6 A51U and U6 A59U, all of which block the second step of splicing, allowed the crosslinking of U5 loop 1 to the 5′ and 3′ exons. The defect associated with these mutants, therefore, remains to be determined. Primer extension analysis of U5–exon interactions in the presence of these three point mutations revealed that the orientation of U5 loop 1 with the 5′ and 3′ exons is unchanged. The fact that U5–exon interactions are unchanged suggests that there is no major rearrangement of the spliceosome by these mutants and that there may be some other subtle defect that blocks the second step of splicing. One interesting observation was that the U5–intron lariat-3′ exon crosslink intensity appeared stronger with mutants U2 G26C and U6 A59U suggesting some difference within the spliceosome. In fact, U6 position A59 has been proposed to play a role during splicing that is distinct from its function in base-pairing with U2 (Madhani and Guthrie, 1992, 1994; Hilliker and Staley, 2004). This role was proposed to be the binding of a critical nucleotide or essential metal ion (Hilliker and Staley, 2004). Genetic and crosslinking studies have placed the U6 A51/G52 region at the 5′ splice site suggesting a role in the first step of splicing (Kim and Abelson, 1996; Luukkonen and Séraphin, 1998a). Ultimately, it will be important to determine how these point mutants in U2 and U6 block the second step of splicing.
The correct alignment of the exon ends for ligation to form functional mRNA is a critical step in pre-mRNA splicing. All exon sequences in pre-mRNAs are different as they code for different protein products. It was unclear how all the different exon ends were aligned precisely with U5 loop 1 during pre-mRNA splicing. We have shown that the U2 snRNA is required for aligning the exon ends with U5 loop 1 during splicing. It remains to be determined whether this alignment is through direct interactions between U2 and U5 loop 1 or indirect interactions. As pre-mRNA splicing is almost certainly catalyzed by the snRNAs, it is hoped that the network of interactions between the U2, U5 and U6 snRNAs that form the active spliceosome can be determined.
Materials and methods
Pre-mRNA, U2 and U6 snRNA
Body-labeled CYH2 and ACT1 pre-mRNA were produced by in vitro transcription as previously described (O'Keefe et al, 1996; Alvi et al, 2001). Modified CYH2 pre-mRNAs were produced by RNA ligation of one chemically synthesized RNA containing 4-thioU (Dharmacon) with one in vitro-transcribed RNA based on published methods (O'Keefe and Newman, 1998). Chemically synthesized RNAs were the following sequences with X representing 4-thioU: Exon 1 (−3), 5′-GACUAGAAAGCACAGAGGUCACGUCUXA; Exon 2 (+2), 5′-CXGGUAAGGGUCGUAUCGGUAAGCACAGAAAG CACCCCGGUGGUAG. Control RNAs contained uridine in place of 4-thioU. In vitro-transcribed RNAs were made from PCR templates by incorporating a T7 promoter into the forward primer. The transcription of 3′ RNA for the ligation of exon 1 (−3) pre-mRNA was primed with UpG (Sigma) to allow end labeling with 32P. The wild-type U2 snRNA gene in pRS316 (m871) was used for oligomutagenesis to produce U2 insertion and G26C mutations. These plasmids were used as templates for PCR with a forward primer containing a T7 promoter. The wild-type U6 snRNA gene in pBluescript (m737) was used for oligomutagenesis to produce U6 insertion, A51U and A59U mutations. These plasmids were used as templates for PCR with a forward primer containing a T7 promoter and a back primer with a recognition sequence for the VS ribozyme. In vitro transcription and purification of U2 and U6 snRNAs from PCR products was performed as described (Alvi et al, 2001).
In vitro splicing and crosslinking
Splicing extract was prepared from yeast strain SC261 or BJ2168 as described (Alvi et al, 2001). Depletion of U2 or U6 snRNA by targeted oligonucleotide directed RNase H cleavage from extracts (Fabrizio et al, 1989; McPheeters et al, 1989) was performed at 30°C for 30 min in a reaction containing 50% extract in 1 × splicing buffer (60 mM potassium phosphate pH 7.0, 3% PEG 8000, 2.5 mM MgCl2, 2 mM ATP) with depletion oligonucleotides complementary to the U2 or U6 snRNA (1 μM U6KO36-50, 5′-CTGTATTGTTTCAAA or 0.45 μM U2KO, 5′-CAGATACTACACTTG). U6 antisense oligo (U6A36-50, 5′-TTTGAAACAATACAG) was added to U6-depleted extract at a final concentration of 1.25 μM to block any remaining U6 depletion oligo (K Derry and RJ Lin, personal communication). Reconstitution was performed at 23°C for 5 min by addition of 125 nM wild-type or mutant in vitro-transcribed snRNA. Splicing was initiated by addition of approximately 2 nM body-labeled or ligated pre-mRNA in splicing buffer to produce a reaction with all components in 1 × splicing buffer with 40% extract. The size of reactions ranged from 5 to 2000 μL. UV crosslinking, recovery of RNA from UV-irradiated splicing reactions, RNase H analysis of crosslinks and electrophoresis were as previously described (Newman et al, 1995; Alvi et al, 2001).
Synthetic lethal screens
The yeast strain YROK4 (Mat α; ura3-52; trp1Δ63; leu2Δ1; his3Δ200; GAL2; snr7::kanMX6; snr6::kanMX6; pRS416-U5/U6) and YROK5 (Mat α; ura3-52; trp1Δ63; leu2Δ1; GAL2; snr7::kanMX6; snr20::kanMX6; pRS416-U5/U2) were constructed by mating haploid strains containing chromosomal U5 and U2 or U6 gene replacements complemented by pRS416 plasmids with the respective wild-type genes. Diploids were selected on 5-FOA then transformed with pRS416-U5/U2 or pRS416-U5/U6. Selected diploids were sporulated and haploid progeny identified that contained both U5 and U2 or U6 replacements and the complementing plasmid. Synthetic lethality was tested by co-transforming U5 and U2 or U6 plasmids, then growing on 5-FOA at 30°C for 3 days which selects against the URA3-marked pRS416-U5/U2 or pRS416-U5/U6 plasmid. The U5 wild-type and mutant plasmids were TRP1-marked and constructed as previously described (O'Keefe and Newman, 1998). The U2 wild-type and mutant plasmids were LEU2 marked and constructed as follows. The wild-type U2 snRNA gene in pRS316 (m871) was used for oligomutagenesis to produce U2 insertion and G26C mutations. The different U2 genes were then cut out and ligated into pRS415. The U6 wild-type and mutant plasmids were HIS3-marked and constructed as follows. The wild-type U6 snRNA gene in pBluescript (m737) was cut out and ligated into pRS413 to produce pRS413-U6. U6 mutants were produced by in vitro mutagenesis of pRS413-U6.
Luciferase assays, primer extension and RT–PCR
A reporter plasmid harboring the ACT1 5′ exon and intron fused to the luciferase coding sequence was constructed by PCR amplification from plasmid p283 (O'Keefe et al, 1996) of the ACT1 5′ exon, intron and 11 nucleotides of the 3′ exon with primers ActF-5′-CGTCTAGACTTTTAGATTTTTCACGCTTACTGCTTTTTTC and ActB-5′-CGGGATCCAGCAGCAACCTCTAAACATATAATATAGC containing recognition sequences for the restriction enzymes XbaI and BamHI (underlined). The digested PCR product was inserted into p416GPD (Mumberg et al, 1995) digested with XbaI and BamHI. The luciferase open reading frame was PCR amplified from plasmid pGL3 (Promega) with primers LucF-5′-CGGGATCCGAAGACGCCAAAAACATAAAG and LucB-5′-CGGAATTCTTACACGGCGATCTTTCCGC containing recognition sequences for the restriction enzymes BamHI and EcoRI (underlined). The digested PCR product coding for the luciferase gene was inserted in frame with the ACT1 gene in p416GPD digested with BamHI and EcoRI to produce plasmid p416GPD-Act-Luc and sequenced. A G1 to C 5′-splice site mutation was produced by in vitro mutagenesis of p416GPD-Act-Luc with oligonucleotide ActG1C-5′-CTAGAACATAGCAGAATCCAT to produce plasmid p416GPD-Act-LucG1C.
For luciferase assays cells were grown in SD-Ura at 30°C. One milliliter of culture was taken, washed in phosphate-buffered saline (PBS), resuspended in 300 μl cold passive cell lysis buffer (Promega) and lysed by vortexing with acid washed glass beads (Sigma) for 3 min. One microliter of a 1:50 dilution of lysate was added to 100 μl luciferase assay reagent (Promega) and luciferase activity detected with a luminometer (TD-20/20 Turner Designs). Concentrations of crude lysates were determined by the Bradford method (BioRad) and used to normalize luciferase activity values. Assays were carried out in triplicate and the mean normalized activity calculated. Values were then plotted relative to the wild-type U2 strain containing the wild-type reporter.
Primer extension was carried out with primers ActI-5′-CGTGGTTATTACAGATCAGTCA, U4-5′-GACGGTCTGGTTTATAATTAAATTTC and 10–20 μg total RNA as previously described (Dobbyn and O'Keefe, 2004). RT–PCR was carried out with primers ActE1-5′-TCCCAAGATCGAAAATTTACTG and LucRT-5′-CGCCGGGCCTTTCTTTAT as previously described (O'Keefe, 2002) except that PCR reactions were spiked with 32P-labeled LucRT primer and separated on a 6% denaturing sequencing gel.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Acknowledgments
We thank Andy Newman for U2 and U6 plasmids. Riz Alvi for construction of the U2/U5 knockout strain. Daniel Wallerstorfer for construction of the ACT1-luciferase reporter plasmid. Jean Beggs, Mike Grant, Steve Oliver, Andy Newman and the O'Keefe lab members for advice on the manuscript. This work was supported by grants from the BBSRC, The Wellcome Trust and The Royal Society.
References
- Alvi RK, Lund M, O'Keefe RT (2001) ATP-dependent interaction of yeast U5 snRNA loop 1 with the 5′ splice site. RNA 7: 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M, Grate L, Pauling MH (1999) A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA 5: 1138–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JS, McPheeters DS (2000) Identification of a U2/U6 helix Ia mutant that influences 3' splice site selection during nuclear pre-mRNA splicing. RNA 6: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbyn HC, O'Keefe RT (2004) Analysis of Snu13p mutations reveals differential interactions with the U4 snRNA and U3 snoRNA. RNA 10: 308–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J (1990) Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science 250: 404–409 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, McPheeters DS, Abelson J (1989) In vitro assembly of yeast U6 snRNP: a functional assay. Genes Dev 3: 2137–2150 [DOI] [PubMed] [Google Scholar]
- Field DJ, Friesen JD (1996) Functionally redundant interactions between U2 and U6 spliceosomal snRNAs. Genes Dev 10: 489–501 [DOI] [PubMed] [Google Scholar]
- Fortner DM, Troy RG, Brow DA (1994) A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes Dev 8: 221–233 [DOI] [PubMed] [Google Scholar]
- Frilander MJ, Steitz JA (2001) Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol Cell 7: 217–226 [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Staley JP (2004) Multiple functions for the invariant AGC triad of U6 snRNA. RNA 10: 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Abelson J (1996) Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA 2: 995–1010 [PMC free article] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C (1993) Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics 133: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez PJ, Séraphin B (1999) Genomic-scale quantitative analysis of yeast pre-mRNA splicing: implications for splice-site recognition. RNA 5: 1135–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen BG, Séraphin B (1998a) Genetic interaction between U6 snRNA and the first intron nucleotide in Saccharomyces cerevisiae. RNA 4: 167–180 [PMC free article] [PubMed] [Google Scholar]
- Luukkonen BGM, Séraphin B (1998b) A role for U2/U6 helix Ib in 5′ splice site selection. RNA 4: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C (1992) A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817 [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C (1994) Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev 8: 1071–1086 [DOI] [PubMed] [Google Scholar]
- McPheeters DS, Abelson J (1992) Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell 71: 819–831 [DOI] [PubMed] [Google Scholar]
- McPheeters DS, Fabrizio P, Abelson J (1989) In vitro reconstitution of functional yeast U2 snRNPs. Genes Dev 3: 2124–2136 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA (1993) Splicing of precursors to mRNA by the spliceosome. In The RNA World, Gesteland RF, Atkins JF (eds), pp 303–357. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Mumberg D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Newman AJ, Norman C (1992) U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68: 743–754 [DOI] [PubMed] [Google Scholar]
- Newman AJ, Teigelkamp S, Beggs JD (1995) snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA 1: 968–980 [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW (1998) RNA–RNA interactions in nuclear pre-mRNA splicing. In RNA Structure and Function, Simons RW, Grunberg-Manago M (eds), pp 279–307. Cold Spring Harbor, NY: Cold Spring Harbor Press [Google Scholar]
- O'Keefe RT (2002) Mutations in U5 snRNA loop 1 influence the splicing of different genes in vivo. Nucleic Acids Res 30: 5476–5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe RT, Newman AJ (1998) Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J 17: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe RT, Norman C, Newman AJ (1996) The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell 86: 679–689 [DOI] [PubMed] [Google Scholar]
- Ryan DE, Abelson J (2002) The conserved central domain of yeast U6 snRNA: importance of U2–U6 helix Ia in spliceosome assembly. RNA 8: 997–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymond BC, Rosbash M (1992) Yeast pre-mRNA splicing. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression, Broach JR, Pringle JR, Jones EW (eds), Vol. 2, pp 143–192. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE (2004) U2–U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol 11: 1237–1242 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA (1993) The U5 and U6 small nuclear RNAs as the active site components of the spliceosome. Science 262: 1989–1996 [DOI] [PubMed] [Google Scholar]
- Sun J-S, Manley JL (1995) A novel U2–U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev 9: 843–854 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Manley JL (2000) A tertiary interaction detected in a human U2–U6 snRNA complex assembled in vitro resembles a genetically proven interaction in yeast. RNA 6: 206–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U, Company M, Abelson J (1989) Isolation and characterization of pre-messenger RNA splicing mutants of Saccharomyces cerevisiae. Genes Dev 3: 1206–1216 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13: 290–301 [DOI] [PubMed] [Google Scholar]
- Xu D, Field DJ, Tang S-J, Moris A, Bobechko BP, Friesen JD (1998) Synthetic lethality of yeast slt mutations with U2 small nuclear RNA mutations suggests functional interactions between U2 and U5 snRNPs that are important for both steps of pre-mRNA splicing. Mol Cell Biol 18: 2055–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean S-L, Wuenschell G, Termini J, Lin R-J (2000) Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3


