Abstract
Mutations in the adenomatous polyposis coli or β-catenin gene lead to cytosolic accumulation of β-catenin and, subsequently, to increased transcriptional activity of the β-catenin–T cell-factor/lymphoid-enhancer-factor complex. This process seems to play an essential role in the development of most colorectal carcinomas. To identify genes activated by β-catenin overexpression, we used colorectal cell lines for transfection with the β-catenin gene and searched for genes differentially expressed in the transfectants. There are four genes affected by β-catenin overexpression; three overexpressed genes code for two components of the AP-1 transcription complex, c-jun and fra-1, and for the urokinase-type plasminogen activator receptor (uPAR), whose transcription is activated by AP-1. The direct interaction of the β-catenin–T cell-factor/lymphoid-enhancer-factor complex with the promoter region of c-jun and fra-1 was shown in a gel shift assay. The concomitant increase in β-catenin expression and the amount of uPAR was confirmed in primary colon carcinomas and their liver metastases at both the mRNA and the protein levels. High expression of β-catenin in transfectants, as well as in additionally analyzed colorectal cell lines, was associated with decreased expression of ZO-1, which is involved in epithelial polarization. Thus, accumulation of β-catenin indirectly affects the expression of uPAR in vitro and in vivo. Together with the other alterations, β-catenin accumulation may contribute to the development and progression of colon carcinoma both by dedifferentiation and through proteolytic activity.
In normal colonic mucosa, β-catenin is involved in cell–cell adhesion. Together with α- and γ-catenin, β-catenin binds to the cytoplasmatic domain of the transmembrane protein E cadherin, and this complex is necessary for adhesion to occur (1). The adenomatous polyposis coli protein (APC) competes with E cadherin for binding to the internal repeats of β-catenin, which are like those encoded by the armadillo gene in Drosophila (2–4). In normal epithelial cells, both components of the APC/β-catenin complex are phosphorylated by a serine–threonine kinase, GSK3β, which results in β-catenin effectively being degraded in proteosomes (5, 6). Thus, in healthy colonic cells, levels of free cytosolic β-catenin are low. During embryogenesis and activation of the wnt signaling pathway, phosphorylation by GSK3β is inhibited, and cytosolic β-catenin accumulates (7). β-Catenin, present in the cytosol at high levels, binds to cytosolic T cell-factor/lymphoid-enhancer-factor (Tcf/Lef) proteins, and the resulting complex is shuttled to the nucleus. This bipartite complex, in which Tcf/Lef proteins provide a DNA-binding domain and β-catenin presents a potential transactivation domain, plays a key role as a transcription factor in the wingless/wnt signaling pathway (8, 9).
The APC gene is mutated in up to 80% of sporadic colorectal carcinomas (10, 11), usually resulting in a truncated protein. This truncated APC protein is unable to form a complex with β-catenin, and eventually the protein becomes unable to trigger β-catenin degradation (5). Indeed, nuclei of APC−/− colon carcinoma cells contain a stable, transcriptionally active β-catenin–Tcf4 complex (12). Reintroduction of wild-type APC to these cells removes β-catenin from this complex and abrogates transcriptional activity (12). In colorectal carcinoma cells with intact APC genes, CTNNB1 mutations of β-catenin itself were shown to contribute to the accumulation of β-catenin and the activation of the wingless/wnt pathway (13), although CTNNB1 mutations are also found in mutant APC carcinomas (14). These data indicate that either APC or β-catenin mutations may mimic the activation of the wingless/wnt signaling pathway in up to 80% of human colorectal carcinomas. It was suggested, therefore, that activation of this pathway is the main initial event in colorectal tumorigenesis (11, 15). Recently, c-myc was identified as a target of the APC pathway in HT 29 cells (16). Other genetic targets of this signaling pathway and their contribution to the neoplastic process have not yet been characterized.
The purpose of the present work is to identify target genes of the β-catenin–Tcf/Lef complex and to analyze the potential role of the encoded proteins in the development of human colorectal carcinomas. We transfected two colorectal cell lines, which have low endogenous β-catenin and high Tcf4 expression, with a plasmid containing the complete coding sequence of β-catenin. We then searched for altered gene expression by differential hybridization. The expression of these identified target genes was tested in other colorectal cell lines and in patient tissue samples.
Expression of transcription factors fra-1 and c-jun, members of the AP-1 complex, and of urokinase-type plasminogen activator receptor (uPAR), known to be activated through AP-1, was increased after transfection. Simultaneously, the expression of the zonula occludens-1 (ZO-1) gene, coding for a member of the MAGUK protein family that is related to β-catenin, was suppressed. We conclude that activation of the β-catenin–Tcf/Lef complex might facilitate loss of epithelial polarization and enhance proteolytic activity in human colorectal carcinomas.
MATERIALS AND METHODS
Cell Lines.
Human colonic adenocarcinoma cell lines HT 29, COLO 205, SW480, SW620, and LS 174T were obtained from the American Type Culture Collection, and cell line CCO7 was obtained from the Imperial Cancer Research Fund (London). Cell lines CSC-1 and CD-NCM425 were derived from normal colonocytes as described (17). All cell lines were cultured under standard conditions.
Patient Tissue.
Tissue samples from primary colorectal carcinomas, their liver metastases, and specimens of normal colonic mucosa (taken from tissue 10 cm away from the tumor) were obtained at the time of surgery from six patients with International Union Against Cancer stage IV colorectal carcinomas. Total RNA and whole cell lysates were prepared immediately, as described (18).
Northern Blotting.
Total RNA was extracted from the tissue samples with guanidinium thiocyanate followed by centrifugation in CsCl as described (18); total RNA was extracted from cell lines with RNAzol (Biotex Laboratories, Houston), according to the recommendations of the manufacturer. Poly(A)-RNA was isolated from 500 μg of total RNA with DynaBeads (Dynal, Oslo), according to the recommendations of the manufacturer. For Northern blotting, 25 μg of total RNA was denatured in dimethyl sulfoxide/glyoxal buffer (18) and run on a 1% agarose gel. The RNA was transferred onto Nytran membranes (Schleicher and Schüll, Dassel, Germany) overnight by capillary force. After prehybridization, hybridization was carried out for 1–2 h at 68°C in QuickHyb (Stratagene) solution with a 32P-labeled probe. Membranes were washed at 60°C, according to the background signal. After autoradiography, the band intensities were evaluated densitometrically and normalized to β-actin expression on the same blot. Specific cDNA sequences of β-catenin (base pairs −18–524), Tcf4 (base pairs 600–900), c-jun (base pairs 1,441–1,622), fra-1 (base pairs 147–888), uPAR (base pairs 776–1,106), and ZO-1 (base pairs 3,329–4,143) were amplified by reverse transcription–PCR, isolated, and used as probes. For β-actin detection, a full-length β-actin cDNA was used as a probe.
Immunobloting.
Whole cell lysates were prepared from approximately 5 × 106 cultured cells in lysis buffer (1% Triton X-100/20 mM Tris⋅HCl/140 mM NaCl/1.5 mM MgCl2/1 mM EGTA/10% glycerol/10 μg/ml DNase/1 mM NaVO3/50 mM NaF/1 mM phenylmethylsulfonyl fluoride/1 mM benzamidine/10 μg/ml aprotinin/10 μg/ml leupeptin/10 μg/ml pepstatin). Whole cell lysates also were prepared from tissue samples after pulverization of approximately 1 g (wet weight) of snap-frozen tissue. After centrifugation, 10–100 μg of the supernatant protein was separated on an SDS/3–10% polyacrylamide gel and blotted onto poly(vinylidene difluoride) membranes. Membranes were incubated overnight with 5% dry milk in 1× PBS at 4°C and then for 1 h with specific primary antibodies (1 μg/ml). The membranes then were incubated for 1 h with a second antibody conjugated to peroxidase (Dako), followed by detection with the enhanced chemiluminescence system (Amersham Pharmacia). The specific antibodies used were anti-human β-catenin mAb 7D11 (Alexis Deutschland, Grünberg, Germany), anti-human APC mAb FE-9 (Oncogene Science), anti-human uPAR rabbit polyclonal antibody 399R (American Diagnostica, Greenwich, CT), anti-human ZO-1 rabbit polyclonal antibody 61–7300 (Zymed), and anti-human β-actin mouse mAb 5441 (Sigma).
β-Catenin Transfection.
Cell lines CCO7 and CD-NCM425 were transfected with full-length β-catenin cDNA containing the complete coding region and fragments of 5′ and 3′ untranslated regions. This cDNA was isolated from the pBAT expression vector (19) by KpnI/SalI restriction and cloned into the pcDNA 3.1 vector. The resulting pcDNAβ-415 plasmid, which was kindly provided by J. Behrens (Max-Dell Brück-Centrium, Berlin), was transiently transfected into CCO7 and CD-NCM425 cells by using Lipofectin (Life Technologies, Palo Alto, CA; ref. 20). Briefly, 5 × 106 cells were cultured until cells were 70% confluent; 6 μg of DNA was diluted in 100 μl of OPTI-MEM (Life Technologies), and 24 μg of Lipofectin was diluted in 100 μl of OPTI-MEM and incubated for 1 h at room temperature before both solutions were pooled and incubated for 30 min at room temperature. Next, 5 ml of OPTI-MEM was added, and the solution was added to the cells and incubated for 5 h at 37°C. Then, 5 ml of DMEM containing 20% fetal calf serum was added, and, after 12 h, serum was replaced by DMEM containing 10% fetal calf serum. Control cells were treated with Lipofectin alone. Cells were harvested 48 h after transfection, and whole cell lysates and RNA were isolated.
cDNA-Array Hybridization.
Differential gene expression was investigated by using Atlas Array membranes (CLONTECH). Briefly, in the first step, 1 μg of mRNA of each cell line was reverse-transcribed to [α-32P]dATP-labeled cDNA. The resulting cDNA probes were hybridized separately overnight to the Atlas Array membranes, on which 588 human cDNA fragments from known genes related to different biological processes are applied in duplicate. Plasmid and bacteriophage DNAs are included as negative controls to confirm hybridization specificity, along with nine housekeeping cDNAs as positive controls for normalizing mRNA abundance. After a high-stringency wash, the hybridization patterns were quantified by phosphorimaging and then analyzed by autoradiography. The signal intensity of each double spot was scanned and normalized to the expression of all nine housekeeping genes. Finally, the mean value of these nine expression ratios was calculated and taken as the actual expression of the gene. Differences in gene expression in the β-catenin-transfected cells of more than 2.5-fold were verified by Northern blot analysis.
Electrophoretic Mobility-Shift Assay.
We generated synthetic duplex oligonucleotides encompassing positions 231–250 of the human fra-1 promoter (accession no. D16365; 5′-AGAATCCCTTTAGCTCAGGA-3′) and positions 261–280 of the flanking, evolutionarily conserved region of the c-jun promoter (accession no. X59744; 5′-GAGAGCCCTTTGAGACACCT-3′) respectively. Each contained homologous sequence to the reported Tcf/Lef binding motif (21). Nuclear extracts from SW480 cells, which showed the highest endogenous β-catenin and Tcf4 expression of our cell lines, were prepared as described (22). For binding reaction, 2 ng of [α-32P]dCTP-labeled oligonucleotide was incubated with 1.2 μg of nuclear extract, 100 ng of poly(dI-dC), and 10% glycerol in 20 μl of binding buffer (15 mM Hepes, pH 7.9/60 mM KCl/1 mM DTT/1 mM EDTA). For supershift analysis, 0.75 μg of anti-human β-catenin murine mAb (Transduction Laboratories, Lexington, KY) was added. The DNA–protein complexes were separated on a 6% polyacrylamide gel, and, after drying, the gel was exposed on a phosphorimager plate.
Statistical Evaluation.
The differences in β-catenin and uPAR expression in patient tissue samples were analyzed with the t test, and the correlation between β-catenin and uPAR expression was calculated by using the standard correlation coefficient (r).
RESULTS
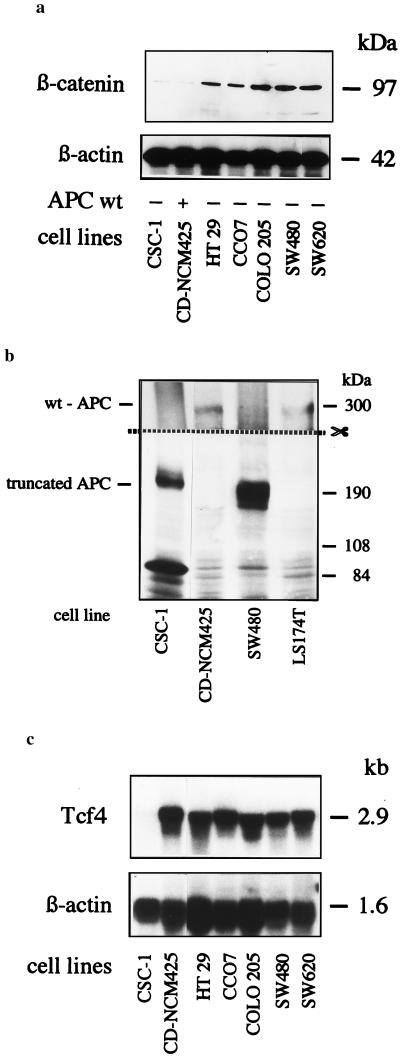
We selected seven cell lines with strong differences in their endogenous β-catenin protein expression (Fig. 1a). Of these, five cell lines are known to have mutated APC (14). APC from the two remaining cell lines was analyzed by Western blotting and found to be truncated in the CSC-1 cell line and apparently normal in the CD-NCM425 cell line (Fig. 1b). All cell lines, except for CSC-1, strongly expressed Tcf4 mRNA (Fig. 1c). Cell lines CD-NCM425 (apparently wild-type APC) and CCO7 (APC mutated; ref. 14), both expressing high levels of Tcf4 and low levels of β-catenin, were chosen for transfection.
Figure 1.
Analysis of β-catenin expression, APC status, and Tcf4 expression in the cell lines used. (a) Variable expression of β-catenin. Whole cell lysates were separated on a 6% polyacrylamide gel, blotted, and developed with anti-β-catenin mAb 7D11 or, for control of the amount of protein per lane, with anti-β-actin mAb 5441. (b) Cell line CD-NCM425 and LS 174T have APC with the expected molecular mass, whereas, in CSC-1 and SW480, the APC is truncated. Immunoblots from whole cell lysates were developed with FE9 mAb directed against the NH2-terminal region of APC. The upper part of the membrane was exposed 10-fold longer than the lower part. Cell lines SW480 and LS 174T were blotted in parallel as controls for truncated and intact APC, respectively. (c) Tcf4 is expresed strongly in all cell lines except CSC-1; 25 μg of total RNA per lane was hybridized with probes for Tcf4 or for β-actin for control of amounts of RNA.
Gene-Expression Analysis.
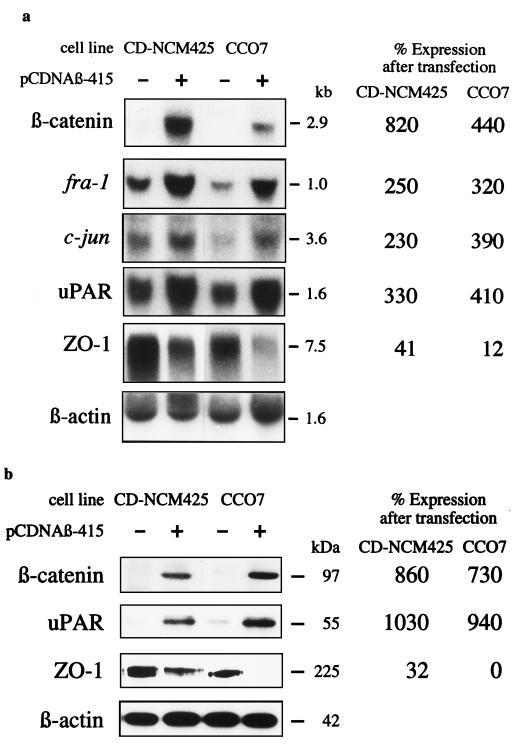
Transfection of both cell lines with pcDNAβ-415 containing the coding region of the β-catenin gene resulted in overexpression of both β-catenin mRNA and protein (Fig. 2). Hybridization of Atlas Array membranes with radioactively labeled cDNA of both β-catenin-transfected and mock-transfected cells identified four differentially expressed genes. These differences in gene expression were confirmed in both cell lines by Northern blot analyses of control and transfected cells (Fig. 2a). The expression of the protooncogenes c-jun and fra-1 and, additionally, of uPAR mRNA increased 2- to 4-fold in the transfectants. On the other hand, mRNA expression of the tight-junction-associated ZO-1 was lowered 2- to 10-fold in the two cell lines after transfection with β-catenin. These data clearly show that β-catenin overexpression is directly or indirectly affecting transcription of the four identified genes. The expression of uPAR protein (23), determined by Western blotting, was increased 10-fold, and the expression of ZO-1 protein (24) decreased substantially in both cell lines after β-catenin transfection (Fig. 2b).
Figure 2.
Effect of β-catenin transfection on target gene expression. (a) Total RNA (25 μg per lane) from CD-NCM425 and CCO7 cells transfected with pcDNAβ-415 (+) or from CD-NCM425 and CCO7 cells that were mock transfected (−) was hybridized with probes for the genes that were found to be expressed differentially by the Atlas Array. Hybridization with the β-actin probe was performed to control the amounts of RNA applied per lane. (b) Expression of the uPAR and ZO-1 protein in the transfected cell lines. Immunoblots from whole cell lysates were developed with anti-β-catenin mAb 7D11, anti-uPAR rabbit IgG 399R, anti-ZO-1 rabbit IgG 61–7300, or, for control of the amount of protein per lane, anti-β-actin mAb 5441. (Expression differences in a and b are indicated as the percentage of expression in transfected cells related to mock transfected cells after normalization to the β-actin expression on the same blot.)
Electrophoretic Mobility-Shift Assay.
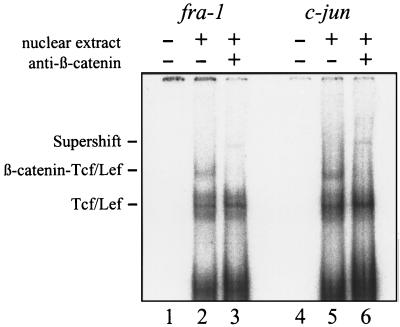
By comparing sequences, we recognized potential Tcf/Lef binding sites in the promoter regions of fra-1 and c-jun, with homologies to the reported Tcf/Lef binding motif (21). To prove directly whether the β-catenin–Tcf/Lef complex interacts with the promoter of fra-1 and c-jun genes, the binding of the complex to each promoter region was tested in an electrophoretic mobility-shift assay by using nuclear extracts from SW480 cells. The observed supershift indicates that the promoter regions of c-jun and fra-1 are the direct targets of the β-catenin–Tcf/Lef complex (Fig. 3).
Figure 3.
Promoter regions of fra-1 and c-jun are direct targets of the β-catenin–Tcf/Lef complex. Gel retardation assays were performed on nuclear extracts from SW480 cells with radioactively labeled oligonucleotides from the indicated promoter regions containing Tcf/Lef binding motifs. Anti-β-catenin mAb (0.75 μg) was added to the samples in lanes 3 and 6. No nuclear extracts were added in lanes 1 and 4.
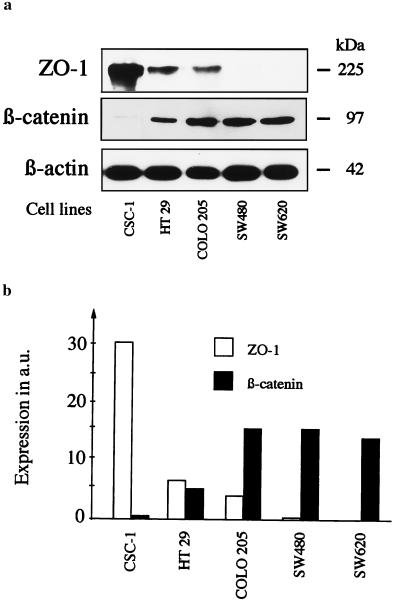
To examine whether the accumulation of β-catenin protein contributes to the suppression of ZO-1 in colorectal carcinoma cells and not only in the transfected lines, we analyzed the ZO-1 expression in five additional cell lines with significant differences in endogenous β-catenin levels. These cell lines showed an inverse relationship between β-catenin and ZO-1 protein expression (Fig. 4). Thus, expression of ZO-1 in colorectal cancer cell lines is decreased not only after exogenous induction of β-catenin but also seems to depend on endogenous β-catenin expression levels.
Figure 4.
Suppression of ZO-1 protein expression in cell lines with high endogenous β-catenin expression. (a) Immunoblots from whole cell lysates developed with anti-ZO-1 rabbit IgG 61–7300, anti-β-catenin mAb 7D11, or, for control of the amount of protein per lane, anti-β-actin mAb 5441. (b) Expression of ZO-1 and β-catenin protein in the analyzed cell lines, after normalization to β-actin expression (a.u., arbitrary units). The expression of ZO-1 was inversely related to β-catenin expression.
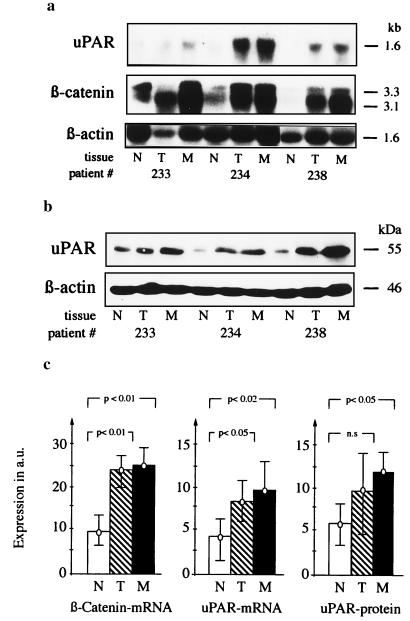
The expression of uPAR, a proteinase receptor that has been reported to contribute to invasive growth of carcinomas of different organs (25–27), was analyzed at mRNA and protein levels in tissue samples from six patients with International Union Against Cancer stage IV colorectal carcinomas. In the same samples, β-catenin mRNA levels were also determined. The expression of the β-catenin-specific bands at 3.1 and 3.3 kb (28) was increased significantly in primary carcinomas and their liver metastases, as shown previously (29). Correspondingly, uPAR was overexpressed in the same neoplastic tissue samples (Fig. 5). The significant increase of uPAR expression at the transcriptional (r = 0.8; P < 0.001) and the translational (r = 0.5; P < 0.05) level correlated with the increase in β-catenin mRNA expression in the analyzed tissue samples.
Figure 5.
uPAR and β-catenin expression levels are increased concomitantly in primary carcinomas and their liver metastases. (a) Total RNA (25 μg per lane) from tissue samples of normal colonic mucosa (N), primary tumor (T), and metastases (M) were hybridized with probes for uPAR, β-catenin, or β-actin. (b) Immunoblot of whole cell lysates, developed with anti-uPAR rabbit polyclonal antibody 399R or with anti-β-actin mAb 5441. Representative blots from three of six analyzed patients are shown in a and b. (c) Expression of β-catenin mRNA, uPAR mRNA, and uPAR protein in tissue samples from N, T, and M of six analyzed patients after normalization to β-actin expression on the same blots. Bars indicate two standard deviations.
DISCUSSION
The activation of the wnt-pathway and its immediate triggering—the increase of cytosolic concentration of β-catenin—can be caused by alterations at the mRNA, protein, or signal-transduction levels. β-Catenin mRNA is overexpressed in many colorectal carcinoma cells and tissues (29), indicating that its transcription is enhanced in neoplastic tissue. At the protein level, it is likely that the inhibition of β-catenin degradation is caused mostly by mutations in the APC or the β-catenin genes themselves. It is also possible that mutations in other regulatory proteins, such as conductin, are involved (30). APC is mutated both in patients with familial adenomatous polyposis and in up to 80% of sporadic colorectal carcinomas (10, 11). The mutations in this gene are probably the initial step in this pathway (31) of colorectal tumorigenesis (10, 11, 31). The triggering of the accumulation of cytosolic β-catenin by mutations of the APC gene can be replaced by β-catenin CTNNB1 mutations, detectable in several colorectal carcinoma cell lines and in about 50% of sporadic colorectal carcinomas with wild-type APC (11, 13, 14, 32). Another potential target protein whose mutations may affect cytosolic levels of β-catenin is conductin, the recently characterized protein that seems to regulate β-catenin degradation independently of APC (30). Finally, E cadherin and α-catenin have been shown to antagonize the wnt-signal (33, 34). The loss of these two proteins during the progression of colorectal cancer (35–38) may enhance the β-catenin signal further in neoplastic cells.
Collectively, these data suggest that β-catenin–Tcf/Lef signaling underlies this pathway of colorectal carcinogenesis and can be triggered by a variety of cellular alterations. Cytosolic β-catenin can bind to different members of the Tcf/Lef family in neoplastic colorectal epithelial cells; complexes with Tcf4 (10), Tcf1 (39), and Lef-1 (40) have been shown to be transcriptionally active. Their target genes, however, were known only in Xenopus embryos. These targets are the homeobox genes siamois (41), nr3 (42), and Xtwn (43), which encode proteins involved in Xenopus dorsal-axis formation. Recently, one target gene was identified in HT 29 cells by using serial analysis of gene expression. The expression of the protooncogene c-myc was shown to be repressed by the reintroduction of wild-type APC and activated by the accumulation of β-catenin through Tcf4 binding sites in the c-myc promoter (16).
For this study, we applied a different approach and identified four other target genes of the β-catenin–Tcf/Lef complex in human colorectal carcinoma cells. Expression of the protooncogenes c-jun and fra-1, both of which are components of the AP-1 transcription factor complex (44), was increased severalfold after transfection. It has been reported that GSK3β activation, which antagonizes wingless/wnt signaling, inhibits c-jun activity in vitro (45). We show here the direct interaction of the β-catenin–Tcf/Lef complex with the promoter regions of both components of the AP-1 complex. AP-1, which is known to be activated also by the Jun-amino-terminal kinase/stress-activated protein kinase mitogen-activated protein kinase pathway (46), has been shown to increase the expression of uPAR in the colorectal cancer cell line RKO (47). The present work elaborates on this observation in terms of upstream and downstream events. Indeed, the overexpression of uPAR as a consequence of β-catenin accumulation may have important biological effects. uPAR is known to facilitate extracellular-matrix proteolysis by accelerating plasmin formation at the cell surface (48). Both the proteolytic activity and the motility-enhancing role (49) of uPAR may be responsible for its potential roles in physiological processes, such as wound healing (50) and invasive growth, as well as metastasis formation of human carcinomas of different organs, including the colorectum (25–27, 51). In agreement with these roles, we observed a concomitant increase in β-catenin and uPAR expression in tumors and metastases. This increase in uPAR expression in colorectal carcinoma cells after β-catenin–Tcf/Lef activation might enhance their proteolytic activity and thus be instrumental in invasive growth and metastasis formation. It is perhaps surprising that APC mutations, which are known to be an early event in colorectal carcinogenesis, may trigger indirectly the expression of a proteinase receptor, relevant for tumor progression. Because the level of free cytosolic β-catenin is in a dynamic equilibrium, regulated by a number of factors (see ref. 52 for a review), an alteration in only one of these might be necessary, but not sufficient, to activate the oncogenic properties of the β-catenin–Tcf/Lef complex.
In addition to uPAR overexpression, we found low ZO-1 expression after β-catenin transfection. The tight-junction-associated protein ZO-1 has been reported to colocalize with E cadherin at the lateral plasma membrane and to coimmunoprecipitate with β-catenin (53). Catenins participate in the mobilization of ZO-1 from the cytosol to the cell surface early in the development of tight junctions (53), which are structural markers of polarized epithelial cells. Neoplastic transformation may block tight-junction assembly (54). Indeed, in well differentiated gastrointestinal adenocarcinomas, ZO-1 is expressed at the apical cell border as in the normal epithelium, whereas in poorly differentiated adenocarcinomas, ZO-1 is frequently lost (55). The present data suggest that activation of the β-catenin–Tcf/Lef pathway in colorectal epithelial cells suppresses the expression of ZO-1 through a hitherto unknown mechanism, which may contribute to a loss of epithelial polarization in neoplastic cells. This suppression of an “adhesive partner” after activation of β-catenin signaling suggests that these two functions of β-catenin might be intertwined in epithelial cells.
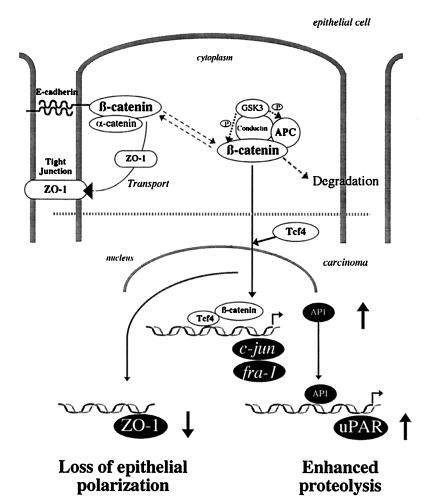
We have identified four target genes of the β-catenin–Tcf/Lef complex in human colorectal carcinomas. The expression of ZO-1, which is known to interact with β-catenin in tight-junction assembly, was decreased in the β-catenin transfectants. The transcription of c-jun and fra-1, members of the AP-1 transcription factor complex, was increased through direct interaction of the β-catenin–Tcf/Lef complex with the promoters of these genes. We have also shown that the transcription of uPAR, a proteinase that is known to contribute to invasive tumor growth in several carcinomas, is up-regulated indirectly by the accumulation of β-catenin (Fig. 6). The in vivo data presented here provide a direct link between β-catenin expression and the overproduction of the uPAR protein in human colon carcinomas and their metastases, thus supporting the in vitro data. Activation of the β-catenin–Tcf/Lef pathway in colorectal carcinoma cells is assumed to be an early event in carcinogenesis. Our results establish a molecular link between this major signaling pathway and the expression of proteins potentially facilitating subsequent tumor progression and metastasis.
Figure 6.
Genes targeted by the β-catenin–Tcf/Lef complex and the related upstream and downstream cellular processes.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (Project Ma-1989 1/1), Deutsche Krebshilfe (Project 10-1025-HaI), Sonnenfeld Stiftung, and the Berliner Krebsgesellschaft.
ABBREVIATIONS
- APC
adenomatous polyposis coli protein
- Lef
lymphoid enhancer factor
- Tcf
T cell factor
- uPAR
urokinase-type plasminogen activator receptor
References
- 1.Jou T S, Stewart D B, Stappert J, Nelson W J, Marrs J A. Proc Natl Acad Sci USA. 1995;92:5067–5071. doi: 10.1073/pnas.92.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su L K, Vogelstein B, Kinzler K W. Science. 1993;262:1731–1734. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 3.Hülsken J, Birchmeier W, Behrens J. J Cell Biol. 1994;127:2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubinfeld B, Souza B, Albert I, Munemitsu S, Polakis P. J Biol Chem. 1995;270:5549–5555. doi: 10.1074/jbc.270.10.5549. [DOI] [PubMed] [Google Scholar]
- 5.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi S, Rubinfeld B, Souza B, Polakis P, Wieschaus E, Levine A. Proc Natl Acad Sci USA. 1997;94:242–247. doi: 10.1073/pnas.94.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinck L, Nelson W J, Papkoff J. J Cell Biol. 1994;124:729–741. doi: 10.1083/jcb.124.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behrens J, von Kries J P, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 9.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Cell. 1997;88:788–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 10.Fearon E R, Vogelstein B. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 11.Bodmer W F. J R Coll Physicians Lond. 1996;31:82–89. [PMC free article] [PubMed] [Google Scholar]
- 12.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 13.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 14.Ilyas M, Tomlinson I P M, Rowan A, Pignatelli M, Bodmer W. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peifer M. Science. 1997;275:1752–1753. doi: 10.1126/science.275.5307.1752. [DOI] [PubMed] [Google Scholar]
- 16.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 17.Stauffer J S, Manzano L A, Balch G C, Merriman R L, Tanzer L R, Moyer P M. Am J Surg. 1995;169:190–195. doi: 10.1016/S0002-9610(99)80135-4. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Nagafuchi A, Takeichi M. EMBO J. 1988;7:3679–3684. doi: 10.1002/j.1460-2075.1988.tb03249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allshire R C. Proc Natl Acad Sci USA. 1990;87:4043–4047. doi: 10.1073/pnas.87.11.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giese K, Cox J, Grosschedl R. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 22.Newell C L, Deisseroth A B, Lopez-Berestein G. J Leukocyte Biol. 1994;56:27–35. doi: 10.1002/jlb.56.1.27. [DOI] [PubMed] [Google Scholar]
- 23.Carriero M V, Franco P, Del Vecchio S, Massa O, Botti G, D’Aiuto G, Stoppelli M P, Salvatore M. Cancer Res. 1994;54:5445–5454. [PubMed] [Google Scholar]
- 24.Anderson J M, Van Itallie C M, Peterson M D, Stevenson B R, Carew E A, Mooseker M S. J Cell Biol. 1989;109:1047–1056. doi: 10.1083/jcb.109.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crowley C W, Cohen R, Lucas B K, Liu G, Shuman M A, Levinson A D. Proc Natl Acad Sci USA. 1993;90:5021–5025. doi: 10.1073/pnas.90.11.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kariko K, Kuo A, Boyd D, Okada S, Cines D, Barnathan E. Cancer Res. 1993;53:3109–3117. [PubMed] [Google Scholar]
- 27.De Petro G, Tavian D, Copeta A, Portolani N, Giulini S M, Barlati S. Cancer Res. 1998;58:2234–2239. [PubMed] [Google Scholar]
- 28.Nollet F, Berx G, Molemans F, van Roy F. Genomics. 1996;32:413–424. doi: 10.1006/geno.1996.0136. [DOI] [PubMed] [Google Scholar]
- 29.Mann B, Gratchev A, Riede E, Schmidt-Wolf I, Trojanek B, Moyer M P, Buhr H J, Hanski C. J Mol Med. 1998;76:B16. (abstr.). [PubMed] [Google Scholar]
- 30.Behrens J, Jerchow B A, Würtele M, Grimm J, Asbrand C, Wirtz R, Kühl M, Wedlich D, Birchmeier W. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 31.Kinzler K W, Vogelstein B. Nature (London) 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 32.Sparks A, Morin P, Vogelstein B, Kinzler K W. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 33.Fagotto F, Funayama N, Glück U, Gumbiner B M. J Cell Biol. 1996;132:1105–1114. doi: 10.1083/jcb.132.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sehgal R N M, Gumbiner B M, Reichardt F L. J Cell Biol. 1997;139:1033–1046. doi: 10.1083/jcb.139.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabre M, de Herreros A G. J Cell Sci. 1993;106:513–522. doi: 10.1242/jcs.106.2.513. [DOI] [PubMed] [Google Scholar]
- 36.Hennig G, Behrens J, Truss M, Frisch S, Reichmann E, Birchmeier W. Oncogene. 1995;11:475–484. [PubMed] [Google Scholar]
- 37.Perl A K, Wilgenbus P, Dahl U, Semb H, Christofori G. Nature (London) 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 38.Vermeulen S J, Bruyneel E A, Bracke M E, DeBruyne G K, Vennekens K M, Vleminckx K L, Berx G J, van Roy F M, Mareel M M. Cancer Res. 1995;55:4722–4728. [PubMed] [Google Scholar]
- 39.Mayer K, Hieronymus T, Castrop J, Clevers H, Ballhausen W G. Int J Cancer. 1997;72:625–630. doi: 10.1002/(sici)1097-0215(19970807)72:4<625::aid-ijc13>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 40.Porfiri E, Rubinfeld B, Albert I, Hovanes K, Waterman M, Polakis P. Oncogene. 1997;15:2833–2839. doi: 10.1038/sj.onc.1201462. [DOI] [PubMed] [Google Scholar]
- 41.Brannon M, Kimelman D. Dev Biol. 1996;180:344–347. doi: 10.1006/dbio.1996.0306. [DOI] [PubMed] [Google Scholar]
- 42.Medina A, Wendler S R, Steinbeisser H. Int J Dev Biol. 1997;41:741–745. [PubMed] [Google Scholar]
- 43.Laurent M N, Blitz I L, Hashimoto C, Rothbacher U, Cho K W. Development (Cambridge, UK) 1997;124:4905–4916. doi: 10.1242/dev.124.23.4905. [DOI] [PubMed] [Google Scholar]
- 44.Valone D, Battista S, Pierantoni G M, Fedele M, Casalino L, Santoro M, Viglietto G, Fusco A, Verde P. EMBO J. 1997;16:5310–5321. doi: 10.1093/emboj/16.17.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutherland C, Leighton I A, Cohen P. Biochem J. 1993;296:15–19. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter T. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 47.Lengyel E, Wang H, Stepp E, Juarez J, Wang Y, Doe W, Pfarr C M, Boyd D. J Biol Chem. 1996;271:23176–23184. doi: 10.1074/jbc.271.38.23176. [DOI] [PubMed] [Google Scholar]
- 48.Ellis V, Behrendt N, Dano K. J Biol Chem. 1991;266:12752–12758. [PubMed] [Google Scholar]
- 49.Nguyen D H, Hussaini I M, Gonias S L. J Biol Chem. 1998;273:8502–8507. doi: 10.1074/jbc.273.14.8502. [DOI] [PubMed] [Google Scholar]
- 50.Romer J, Lund L R, Eriksen J, Pyke C, Kristensen P, Dano K. J Invest Dermatol. 1994;102:519–522. doi: 10.1111/1523-1747.ep12373187. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Skibber J, Juarez J, Boyd D. Int J Cancer. 1994;58:650–657. doi: 10.1002/ijc.2910580506. [DOI] [PubMed] [Google Scholar]
- 52.Dale T. Biochem J. 1998;329:209–223. doi: 10.1042/bj3290209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajaserkan A K, Hojo M, Huima T, Rodriguez-Boulan E. J Cell Biol. 1996;132:451–463. doi: 10.1083/jcb.132.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsukita S, Itoh M, Nagafuchi A, Yonemura S, Tsukita S. J Cell Biol. 1993;123:1049–1053. doi: 10.1083/jcb.123.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura Y, Shiozaki H, Hirao M, Maeno Y, Doki Y, Inoue M, Monden T, Akatsuta Y, Furuse M, Tsukita S, et al. Am J Pathol. 1997;151:45–54. [PMC free article] [PubMed] [Google Scholar]