Abstract
Nud1p, a protein homologous to the mammalian centrosome and midbody component Centriolin, is a component of the budding yeast spindle pole body (SPB), with roles in anchorage of microtubules and regulation of the mitotic exit network during vegetative growth. Here we analyze the function of Nud1p during yeast meiosis. We find that a nud1-2 temperature-sensitive mutant has two meiosis-related defects that reflect genetically distinct functions of Nud1p. First, the mutation affects spore formation due to its late function during spore maturation. Second, and most important, the mutant loses its ability to distinguish between the ages of the four spindle pole bodies, which normally determine which SPB would be preferentially included in the mature spores. This affects the regulation of genome inheritance in starved meiotic cells and leads to the formation of random dyads instead of non-sister dyads under these conditions. Both functions of Nud1p are connected to the ability of Spc72p to bind to the outer plaque and half-bridge (via Kar1p) of the SPB.
Keywords: Centriolin, meiosis, Nud1p, spindle polarity, yeast
Introduction
Diploid cells of the yeast Saccharomyces cerevisiae cultured in the absence of nitrogen and in the presence of a non-fermentable carbon source undergo meiosis and form four haploid spores. Both meiotic divisions occur within a single, continuous nuclear envelope, in which the spindle pole bodies (SPBs), the functional equivalents of centrosomes in higher eukaryotes, are embedded (Moens and Rapport, 1971). Regulation of SPB function during meiosis is critical for spore formation. During sporulation, the primary role of the cytoplasmic face of each SPB, the outer plaque (OP), changes from the anchoring of cytoplasmic microtubules to the initiation of prospore membrane (PSM) formation. This leads to the encapsulation of the post meiotic nuclei into individual compartments inside the boundaries of the mother cell and to the formation of spores. Thereby the SPBs are required for PSM synthesis by assisting with the docking and fusion of secretory vesicles and continuous anchorage of the nucleus to the growing membrane during cellularization (Moens and Rapport, 1971; Davidow et al, 1980; Okamoto and Iino, 1982; Neiman, 1998; Knop and Strasser, 2000; Knop et al, 2005).
The functional shift of the SPB during sporulation results from a change in the molecular composition of the OP. Spc72p is a mitotic component of the OP that recruits the γ-tubulin complex to the SPB and is required for cytoplasmic microtubule nucleation and anchorage (Knop and Schiebel, 1998). Spc72p binds to two different sites at the SPB, to Nud1p at the OP and to Kar1p at the SPB half-bridge (Pereira et al, 1999; Gruneberg et al, 2000). The half-bridge is required for SPB duplication and organizes microtubules during karyogamy and in the G1 phase of the cell cycle (Byers and Goetsch, 1975; Pereira et al, 1999). These cytoplasmic microtubules are specifically required for nuclear congression during mating (Rose and Fink, 1987; Brachat et al, 1998; Pereira et al, 1999). In meiosis, cytoplasmic microtubules are observed until late meiosis I. The function of cytoplasmic microtubules in meiosis is unknown and neither is it known whether Spc72p interacts with Nud1p or with Kar1p or both.
Spc72p is present at the SPB throughout meiosis I and disappears with the onset of meiosis II (Knop and Strasser, 2000; Nickas et al, 2004). It is replaced by the three essential meiosis-specific components Mpc54p, Mpc70p/Spo21p and Spo74p along with the non-essential protein Ady4p (Knop and Strasser, 2000; Bajgier et al, 2001; Nickas et al, 2003). These proteins form the meiotic plaque (MP). It has been proposed that Mpc54p, Mpc70p and Spo74p make up a paracrystalline protein matrix (Taxis et al, 2005), analogous to the central crystal of the SPB, which is composed of Spc42p (Bullitt et al, 1997). Cnm67p, a coiled-coil protein (Schaerer et al, 2001), and Nud1p (Adams and Kilmartin, 1999; Gruneberg et al, 2000) are constitutive components of the SPBs. Both proteins are thought to bring the meiosis-specific components of the MP into proximity to each other and anchor them to the SPB (Knop and Strasser, 2000; Bajgier et al, 2001; Nickas et al, 2003).
SPBs are duplicated twice during meiosis in a conservative manner. Thus, the four meiotic SPBs can be categorized into three different generations: the oldest SPB (present upon entry of the cell into meiosis), second oldest (formed during meiosis I) and two new SPBs (formed during meiosis II). S. cerevisiae is able to limit the number of formed spores to the amount of available external energy resources (e.g. acetate) (Davidow et al, 1980). It has been shown that MPs are assembled in higher probability on newer SPBs, in the case that less than four MPs are formed (Nickas et al, 2004; Taxis et al, 2005). The number of MPs that are formed, which determines the number of spores that are formed, is regulated via the amounts of MP components available. This allows a cell to precisely specify the number of spores it can afford to form. This system ensures that in asci containing only two spores (dyads), the spores contain genomes that derive from different meiosis II spindles (non-sister genomes). Because of the relatively tight genetic linkage of the mating type locus (MAT) to a centromere, the two spores in a dyad are mostly of opposite mating type (MATa and MATα) and able to mate with each other upon germination. This provides a selective advantage associated with preservation of heterozygosity (Taxis et al, 2005).
SPB inheritance is also subject to control in mitotically dividing yeast cells, where it has been shown that the old SPB is preferentially inherited into the bud (Pereira et al, 2001). Also in Schizosaccharomyces pombe, inheritance of SPBs in mitosis is subject to regulation (Grallert et al, 2004). Different fates have furthermore been reported for centrosomes and centrioles of different generations in higher organisms (Piel et al, 2001; Uetake et al, 2002). At present, the molecular basis for discrimination of SPBs from different generations is not clear, nor is it known whether the molecular mechanism is evolutionary conserved.
Nud1p is the only core component of the SPB, apart from calmodulin/Cmd1p, with recognizable homologies to proteins from other species. It is a constitutive component of the OP of the SPB in mitosis and meiosis (Adams and Kilmartin, 1999; Knop and Strasser, 2000). It shares approximately 120 aa of homology with the human protein Centriolin, which functions in cytoplasmic microtubule organization and in late stages of cytokinesis (Gromley et al, 2003). Recently, it was found that the Nud1p homology domain of Centriolin interacts with Sec15, a member of the exocyst complex, and that Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory vesicle-mediated abscission of the cells at the end of cytokinesis (Gromley et al, 2005). Cdc11p, the Nud1p homolog in the yeast S. pombe, is a scaffold protein of the SPB and its N-terminal domain is essential for the proper function of the septation initiation network (SIN; Krapp et al, 2004). In S. cerevisiae, Nud1p functions as a structural component of the SPB that binds the γ-tubulin receptor protein Spc72p to the OP. Furthermore, Nud1p is required in mitotic exit where it functions as a scaffold for Bub2p and Bfa1p that constitute a dual component GAP complex for the small GTPase Tem1p (Gruneberg et al, 2000; Pereira et al, 2001).
In this work, we report that Nud1p has different roles during meiosis, one during spore wall maturation and another during the selection of specific SPBs for the assembly of MPs, such that only non-sister dyads will form. Our results indicate that this process requires the binding of Spc72p to the SPB. We furthermore provide indication that two different mechanisms act to specify differences between all four SPBs present in cells in meiosis II.
Results
The nud1-2 mutation reduces spore formation in a KAR1-dependent manner
To examine the role of NUD1 in meiosis, we assayed the frequency of ascus formation in a temperature-sensitive nud1-2 mutant (Gruneberg et al, 2000). We chose this mutant allele of NUD1 because of its relatively low restrictive temperature of 34°C, at which vegetative cell division is completely prevented. Temperature-sensitive mutants with restrictive temperatures above 35°C are not useful for meiotic analysis, because meiosis in S. cerevisiae (as well as in many other species) is intrinsically temperature sensitive (Byers and Goetsch, 1982). When counting the number of cells that formed spores (asci) in the nud1-2 mutant at 34°C, we found that conduction of meiosis was not impaired (Figure 1A). Sporulating yeast cells often form asci containing a variable number of spores due to selective abortion of haploid genomes after meiosis II (Davidow et al, 1980; Okamoto and Iino, 1981). We therefore analyzed the number of spores present in the asci of the nud1-2 mutant compared to the wild-type strain. This revealed that asci containing fewer spores (one or two) were more frequent in the nud1-2 mutant at 34°C but not at 23°C (Figure 1B). This indicates that NUD1 is required to form wild-type number of spores per ascus. Nevertheless, the nud1-2 mutant is still able to form spores at 34°C, whereas vegetative cell division is completely abolished (data not shown), indicating that the functions of Nud1p that are impaired in the nud1-2 mutant are not essential for sporulation. We also tested germination of nud1-2 spores from sporulations at 23 or 34°C, and found no difference to the wild-type strain (data not shown).
Figure 1.
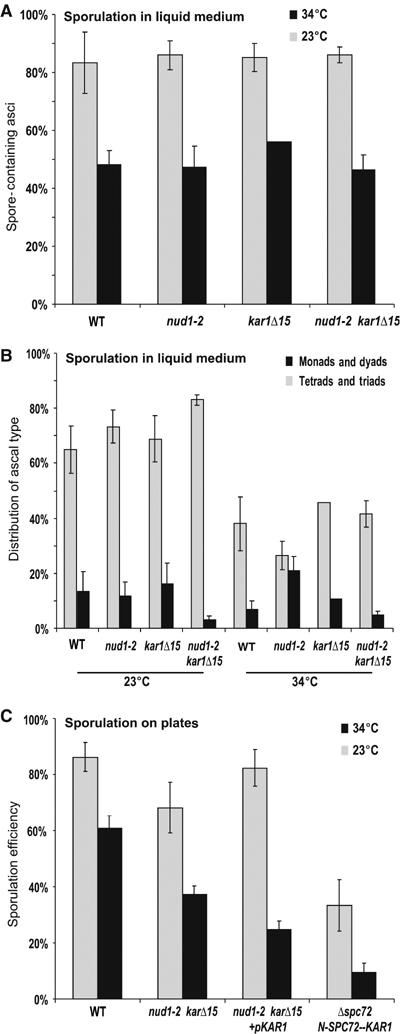
The nud1-2 mutation leads to reduced spore production under restrictive temperature in a KAR1-dependent manner. (A) Wild-type (WT), nud1-2, kar1Δ15 and nud1-2 kar1Δ15 double mutant cells (strains YKS32 (WT), YOG23, YOG52 and YOG66, respectively) were sporulated at the permissive (23°C) and restrictive (34°C) temperatures in standard acetate concentration (0.3%) for 1–2 days. At least 200 cells per strain and temperature were counted (4–13 experiments per strain; except only two for kar1Δ15 at 34°C). (B) Formation of ascus types with 1–2 or 3–4 spores in sporulated cultures of the strains from (A). At least 200 cells per strain and temperature were counted. (C) WT, nud1-2 kar1Δ15 double mutant cells, double mutant cells transformed with a KAR1 plasmid (pMR77) and a homozygous Δspc72 deletion strain expressing the fusion protein N-Spc72p-ΔN-Kar1p (YCT883-2) were sporulated at 23 and 34°C on sporulation medium plates for 3 days. The sporulation efficiency (the sum of spores divided by four times the sum of cells; 100% efficiency means 100% tetrads) was determined for at least 200 cells per strain and temperature. The means and standard errors from three to six experiments are shown.
Nud1p exhibits two reported functions during vegetative cell division. First, it is required for anchorage of cytoplasmic microtubules via the γ-tubulin receptor protein Spc72p to the OP of the SPB (Knop and Schiebel, 1998; Elliott et al, 1999; Gruneberg et al, 2000). Second, it serves as a scaffold for components that function in the mitotic exit network (MEN) (Gruneberg et al, 2000; Bardin and Amon, 2001; Luca et al, 2001).
In vegetative cells, not only the OP but also the half-bridge of the SPB binds Spc72p in order to organize cytoplasmic microtubules in a cell cycle-dependent manner. Binding of Spc72p to the half-bridge requires Kar1p but not Nud1p (Byers and Goetsch, 1975; Pereira et al, 1999). In order to investigate the consequences of Spc72p binding to the SPB on the abortion of spores, we combined the nud1-2 mutation with the kar1Δ15 mutation. kar1Δ15 encodes for an N-terminally deletion variant of Kar1p that is selectively impaired in binding of Spc72p but not in the other functions of Kar1p (e.g. in SPB duplication) (Vallen et al, 1992; Pereira et al, 1999). We found that the kar1Δ15 mutation did not significantly affect ascus (Figure 1A) or spore formation (Figure 1B). Surprisingly, spore formation at 34°C in the double mutant nud1-2 kar1Δ15 was comparable with wild-type levels (Figure 1B). This means that the kar1Δ15 mutation rescued the spore abortion phenotype of the nud1-2 mutation. Plasmid encoded KAR1 could restore spore abortion in the nud1-2 kar1Δ15 mutant, indicating the consistency of the result (Figure 1C). This suggests that the interaction of Spc72p with Kar1p, which is specifically affected by the karΔ15 mutation, leads to reduced spore formation in the nud1-2 mutant. To explore this further, we used a fusion of Spc72p to Kar1p (Spc72p1−272-Kar1p), which leads to constitutive localization of Spc72p at the half-bridge of the SPB. This fusion complements for loss of Spc72p at the OP of the SPB (Gruneberg et al, 2000). When we examined spore formation in the Δspc72 Spc72p1−272-Kar1p strain, we noticed that it exhibits significant lower sporulation efficiency than the wild type (Figure 1C). This is the opposite effect to the one seen in the kar1Δ15 mutant (Figure 1B), where Spc72p is not able to interact with the half-bridge. This indicates that Spc72p at the half-bridge may be responsible for the lower level of spore formation in the nud1-2 mutant.
The nud1-2 mutation leads to kar115-dependent defects in spore wall maturation
Reduced spore formation may reflect either a deficiency during initiation and formation of the PSMs or during the maturation of the spore wall. We examined the reduced spore formation phenotype in the nud1-2 strain, using immunofluorescence microscopy of PSMs in meiotic cells. We counted the total number of PSMs that were present in the cells at different time points during a sporulation time-course experiment (Figure 2A) and compared this with the frequencies of spores formed at the end of sporulation (after 36 h) (Figure 2B). This experimental approach counts slightly more PSMs than spores for the wild-type strain. This is owing to a systematic error that is made in the quantification of PSM formation, because meiotic progression of sporulating cells is not perfectly synchronous, and cells forming more PSMs (three or four) are usually performing sporulation faster than the ones forming less PSMs (one or two) (Taxis et al, 2005). The comparison revealed that at 34°C the nud1-2 mutant formed approximately 15–20% fewer spores, when compared with the wild type, the kar1Δ15 and the nud1-2 kar1Δ15 mutants (Figure 2C). However, the number of PSMs formed did not show significant differences. This suggests that the spore abortion phenotype in the nud1-2 mutant at 34°C is due to a late defect, after the formation of PSMs.
Figure 2.
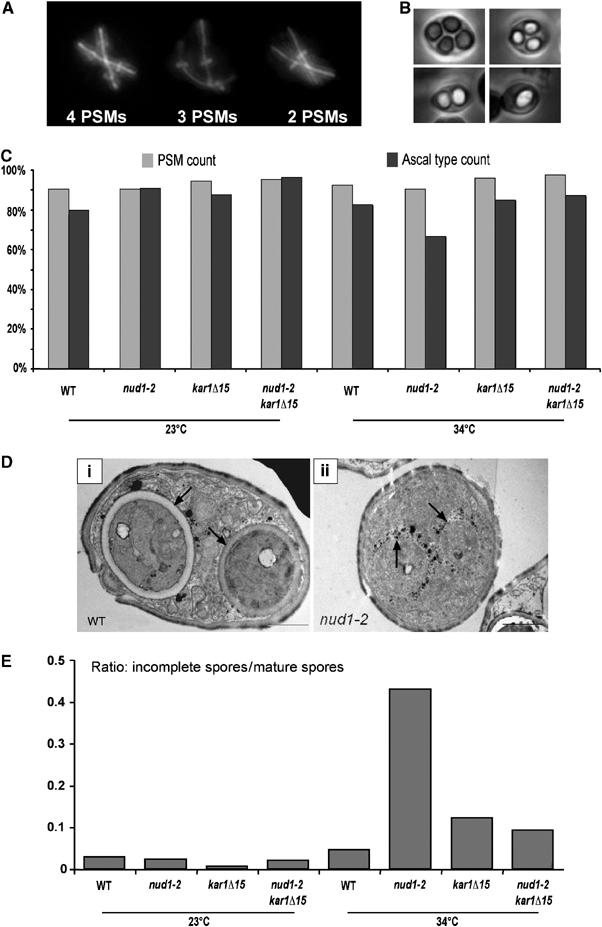
The nud1-2 mutation leads to PSM loss at the restrictive temperature. (A) Colocalization of Ady3, tubulin and DNA in cells in meiosis II using immunofluorescence microscopy. The figure shows examples of cells with different numbers of PSMs. Ady3 localizes to the leading edge of the PSM; hence, a bar, vertically aligned to a meiosis II spindle, formed by the flattened ring-like opening of the PSM indicates a forming PSM. (B) Examples of asci containing 1–4 spores. Phase-contrast microscopy was used to obtain these pictures. (C) Wild-type (WT), nud1-2, kar1Δ15 and nud1-2 kar1Δ15 mutant cells (strains YKS32 (WT), YOG23, YOG52 and YOG66, respectively) were sporulated at the permissive (23°C) and restrictive (34°C) temperatures in liquid cultures. Aliquots of cells were taken after 5, 6, 7, 8 and 9 h following induction of meiosis. The number of PSMs per cell in anaphase of meiosis II (two elongated spindles) was determined (PSM count) (∼150 cells per strain and temperature were counted). After 36 h, the number of spores per ascus (monads, dyads, triad and tetrads) was assayed (spore count). By integration of the PSM count over sporulation time, the percent of PSM per ascus was determined (100% means that 100% of the cells produced four PSMs). The same was carried out for the spore count (∼200 cells per strain and temperature were counted). One experiment was carried out for each strain at each temperature, except for nud1-2, in which case the average of two experiments is shown (4% difference between the two experiments). (D) Electron microscopy of examples of sporulated cells from the WT and the nud1-2 strains used in (C). The section of the WT cell shows two mature spores, whereas the nud1-2 section shows two immature spores. Arrows point to the spore walls of the WT and the mutant spores. The immature spores are surrounded by black dots of unclear origin (they might consist of vesicular structures that contain dityrosin, a component of the outermost layer of the spore wall; Coluccio et al, 2004). (E) Quantification of spore maturation in sporulated cells using electron microscopy (24 h after induction of sporulation). For each strain, the ratio of immature versus mature spores as visible by electron microscopy (150–200 spore containing cells per strain) is shown.
To analyze directly the spore formation defect in the nud1-2 mutant, we used electron microscopy to investigate the cells after completion of sporulation (24 h after induction). In the nud1-2 mutant, at 34°C, we detected an increased number of asci containing immature spores in comparison to wild type, kar1Δ15 and nud1-2 kar1Δ15 mutants (Figure 2D and E). At 23°C, no significant difference between the different strains was observed (Figure 2E). This indicates that the nud1-2 mutant exhibits a partial defect during maturation of PSMs to spores at 34°C, a late step in spore formation, and that this defect can be rescued by the kar1Δ15 mutation, in which the binding of Spc72p to the half-bridge is abolished.
The nud1-2 mutation leads to random encapsulation of genomes present in the two spores of dyads
It has previously been shown that the haploid spores in dyads contain genomes that originate from a different meiosis II spindle. This is owing to regulation of SPB inheritance. Thereby, in dyads, only one SPB per meiosis II spindle, namely the new one formed during meiosis II, becomes involved in spore formation (Davidow et al, 1980; Okamoto and Iino, 1981; Nickas et al, 2004). As the chromatids present in the two spores are thus of non-sister origin in centromere-linked regions (each derived from a different homologous chromosome), the dyads are called non-sister dyads. We examined the fidelity of formation of non-sister dyads in the nud1-2, kar1Δ15 and nud1-2 kar1Δ15 homozygous strains. For this, we used spore-autonomously expressed red and green fluorescent proteins (FPs) that are expressed from loci next to the centromeres of the two homologous chromosomes V (CENV-FP assay; illustrated in Figure 3A). This assay enables quantitative measurement of non-sister dyad formation using fluorescence microscopy (Figure 3B). We found 4% of dyads to be sister dyads in the wild-type strain and only slightly elevated levels in the kar1Δ15 mutant. In the nud1-2 and nud1-2 kar1Δ15 mutants, the frequency of sister dyads was almost exactly 33% (Figure 3C). This value is expected when regulation of genome inheritance into the spores in dyads is completely abolished. No difference between the two temperatures used (23 and 34°C) was observed. At 34°C, the production of random dyads in the nud1-2 strain may be caused by abortion of PSMs. However, the randomization of genome inheritance in dyads is also seen in the nud1-2 strains at 23°C. In addition, the kar1Δ15 mutation does rescue spore abortion at 34°C, but not faithful non-sister dyad formation (Figures 1B, 2 and 3C). These results strongly indicate that nud1-2 is impaired in faithful regulation of SPB inheritance, and that this function is genetically distinct from the function of Nud1p in promoting spore formation.
Figure 3.
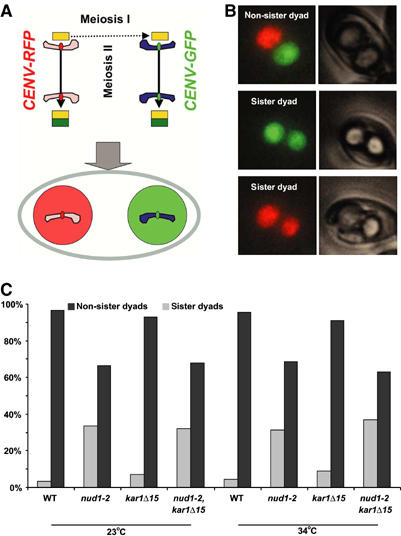
The nud1-2 mutation leads to random encapsulation of genomes in dyads. (A) Cartoon illustrating the CENV-FP assay (see text). (B) Samples of non-sister and sister dyads. Merged fluorescence pictures and phase-contrast images are shown. (C) Wild-type (WT) nud1-2, kar1Δ15 and nud1-2, kar1Δ15 double mutant cells (strains YCT716, YOG26, YOG72 and YOG73, respectively) were sporulated at the permissive (23°C) and restrictive (34°C) temperatures. Dyad formation was induced by the presence of limited amounts of acetate (0.01%) in the sporulation medium. The segregation of centromere-linked spore markers (CENV-linked FP markers, either heterozygote CENV-GFP/CENV-RedStar or heterozygote CENV-GFP/no marker, each under the control of a spore autonomous promoter) was assayed in approximately 200 dyads per strain and temperature. Different labelling of the spores in dyads (red/green or green/dark) indicates the presence of non-sister genomes (genomes segregated in meiosis I). Random incorporation of genomes in dyads would give rise to 2/3 dyads of spores with different labelling (non-sister dyads) and 1/3 with equal labelling (sister dyads). The picture shows examples for the three different possibilities of dyads from the WT strain containing CENV-GFP/CENV-RedStar (phase-contrast pictures and fluorescence images are shown). The error for this experiment (estimated in independent experiments and by ‘dyad' dissection) was found to be less than 10%.
The nud1-2 mutation leads to randomization of SPB selection during MP assembly
The results so far indicate that randomization of genome inheritance in dyads in the nud1-2 strain is not caused by abortion of spore formation on the level of PSMs or spore maturation. Therefore, randomization of genome inheritance must originate from a defect earlier in the process of spore formation. The assembly of a functional MP is the earliest directly observable feature of SPBs that form a PSM. The most likely defect would be randomization of MPs assembly such that older SPBs from the same meiosis II spindle rather than only new ones become also involved in spore formation in dyads. To address this possibility, we used the previously established reporter assay that allows the differentiation of SPBs based on their age by using a slow-maturing red fluorescent protein (RFP) fused to the integral SPB component Spc42p (Pereira et al, 2001; Nickas et al, 2004; Taxis et al, 2005). The t1/2 maturation rate of the chromophore of the particular RFP used (RedStar) (Knop et al, 2002) is approximately 3–5 h and thus matches the time it requires for a cell to progress through meiosis (approximately 5–7 h). This allowed us to distinguish between SPBs from the three different generations present in a cell in meiosis II, based on the brightness of the SPB signal (Figure 4A, see cartoon). Assembly of MPs was visualized using Mpc54p-YFP or GFP at 23 and 34°C (Figure 4B and C).
Figure 4.
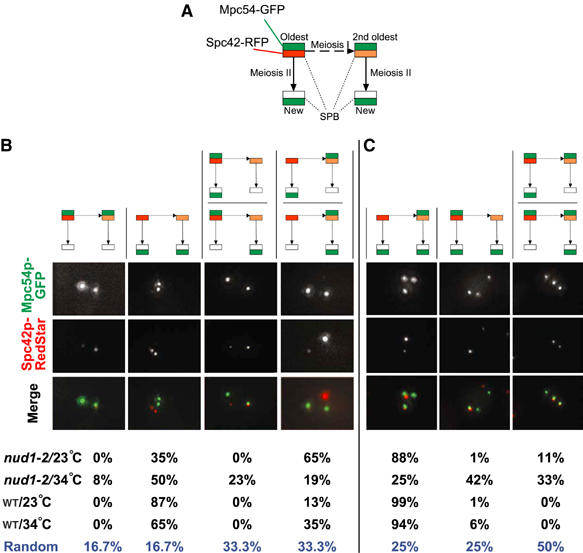
The nud1-2 mutation leads to randomization of MPs assembly. Wild-type (WT) and nud1-2 strains harboring MPC54-GFP (nud1-2) or MPC54-YFP (WT) and SPC42-RedStar (strains YCT806-1 and YOG84) were sporulated at the permissive (23°C) and restrictive (34°C) temperatures. The configuration of MPs assembly on old and new SPBs was determined by imaging green and red fluorescence in cells in meiosis II. The cartoon (A) outlines the relationship of the SPBs and the FP labels used to distinguish SPBs from three different generations. Arrows indicate duplication of the SPBs during meiosis I (dashed arrow) and meiosis II (normal arrow). MPs were visualized using Mpc54p-GFP (green signals); old and new SPBs were discriminated based on the signal brightness of a fluorescent timer (RedStar/RFP; Knop et al, 2002) fused to the integral SPB component Spc42p (Donaldson and Kilmartin, 1996). This allows correlating the age of SPBs with the assembly of MPs. The oldest SPB is marked with the brightest RFP signal, and the second oldest SPB with the second brightest RFP signal. Statistical evaluation of ∼30 cells showing two MPs (B) and of ∼100 cells showing three MPs (C). The fluorescence images in (B) and (C) provide examples for the different possible constellations. The associated cartoons illustrate the different categories that the examples belong to and that can be distinguished. Formation of cells with two and three MPs was induced by the presence of limited amounts of acetate (0.01%) in the sporulation medium.
We first investigated cells with two MPs. In wild type at 23°C, we observed a fidelity of 87% for assembly of the MPs preferentially at the new SPBs, whereas in 13% of the cases one new and the second oldest SPB contained an MP (Figure 4B). Interestingly, the corresponding values were 65 and 35% at 34°C. We also performed this experiment using in addition Spc110p-CFP to assess the position of all SPBs. This allows distinguishing situations where two SPBs are aligned behind each other. In this strain, the fidelity for the involvement of the second oldest SPB instead of its associated new SPB is 4% at 30°C (Taxis et al, 2005). At 34°C, we found again a temperature dependency for the distinction between the second oldest and its associated new SPB in this strain (data not shown).
As the fidelity of non-sister dyad formation in the wild-type strain is not compromised at 34°C at all (Figure 3), the logical explanation for the above observation is that the distinction between the second oldest SPB and its direct daughter is compromised under elevated temperatures, whereas the daughter SPB of the oldest one is still the most likely one to assemble an MP.
In the nud1-2 strain, a much higher degree of randomization was observed. At 23°C, only 35% of the cells involved the two new SPBs for MP assembly, whereas in 65%, the second oldest SPB and one new SPB were involved. As the nud1-2 mutant shows compromised non-sister dyad formation at 23°C, this suggests that cells are no longer able to distinguish between the two new SPBs in relation to their origin from the oldest or second oldest SPB. Under elevated temperature, the distinction between the two old SPBs was also compromised (Figure 4B). We intended to perform this experiment also with the additional Spc110p-CFP marker as well. However, nud1-2 combined with Spc42p-RFP and Spc110p-CFP (or Spc29p-CFP or Cnm67p-CFP) was inviable (data not shown).
When looking at cells with three MPs, the results were similar (Figure 4C). Wild-type cells show a clear preference not to assemble MPs on the oldest SPB. In this case also, a weak temperature dependency was observed. The nud1-2 strain exhibited significant randomization of SPB selection for MP assembly already at 23°C. This was enhanced further at 34°C.
Together, these results suggest that the nud1-2 mutant is compromised in directing MP assembly to specific (new) SPBs, whereas MP assembly per se is not compromised. This explains the defect in SPB inheritance and preference in the mutant and provides a reason for the abnormal random dyad formation in starvation conditions.
Simulation of spore number control in the nud1-2 kar1Δ15 mutant
We recently established a mathematical model for spore number control, which is based on molecular feedback mechanisms for MP assembly. The simulation was fitted to sporulation profiles that were obtained when spore formation was assayed as a function of acetate provided to populations of sporulating cells (Taxis et al, 2005).
To apply the simulation to the analysis of the nud1-2 mutant, we recorded sporulation profiles of mutant and wild-type populations. We used the double mutant nud1-2 karΔ15 in order to prevent abortion of spore formation after their assembly has been initiated. Fitting of the simulation to the two sporulation profiles (Figures 5A and B; for experimental details, see Materials and methods) revealed that the mutant's profile required different values for the parameters that describe the SPBs' individual ability to assemble an MP as compared to the wild type. The highest value for the mutant, corresponding to the first SPB that acquires an MP, was lower than the corresponding value of the wild type, whereas the other values were higher (Figure 5C). This indicates that the mutant exhibits less pronounced differences between the preferences of the individual SPBs to assemble MPs. Initiation of MP formation is thought to be a stochastic event, which depends on the likelihood of the different SPBs to initiate MP formation (Taxis et al, 2005). Thus, less pronounced differences between the SPBs would give rise to a more promiscuous SPB selection for MP assembly and thus explain the defects in SPB inheritance seen in this mutant.
Figure 5.
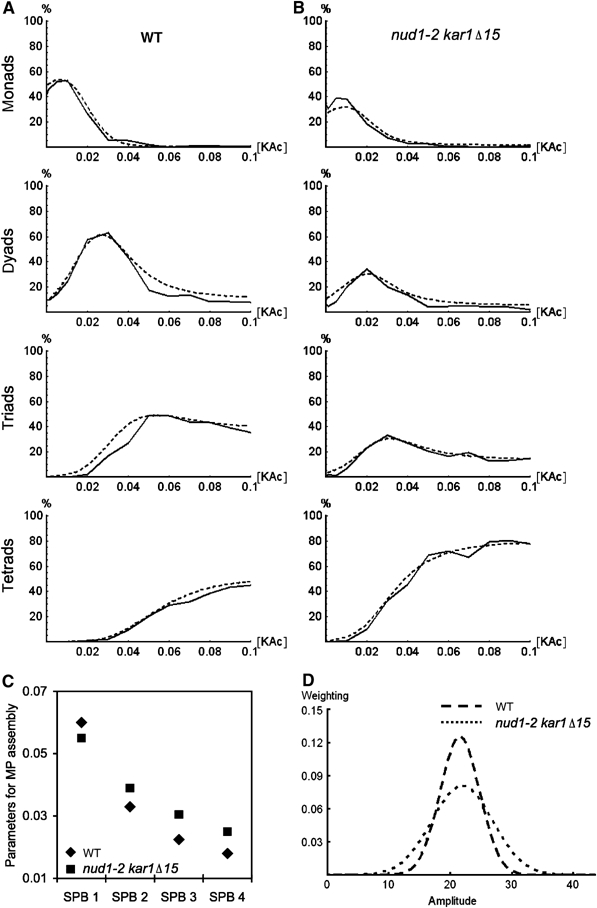
Analysis of the nud1-2 kar1Δ15 mutant sporulation profile using computer simulation. (A) Sporulation profile of the wild-type (WT) strain (YKS32; solid lines) and the simulated sporulation profile (dashed lines). (B) As (A), but for the nud1-2 kar1Δ15 mutant (YOG66). Note that the profiles were recorded at 30°C, in order to match previously established conditions for the recording of sporulation profiles. (C) Graph showing the four parameters that were derived by fitting the model to the experimental data and that describe the ability of four SPBs to promote MP assembly; for the WT and the nud1-2 kar1Δ15 double mutant. (D) Graph showing the Gaussian distributions that resulted from fitting the simulation. These distributions describe the variation of MP component production level in the yeast population.
The simulation furthermore considers population statistics in order to describe population-based sporulation profiles. The center (aav) of the Gaussian distribution that describes the variation of MP component production level in the yeast population was the same for both strains (Figure 5D). This result indicates that the mutant cells produce on average the same amounts of MP components in response to external acetate as wild-type cells. This was confirmed qualitatively using immunoblotting (data not shown). However, optimal fitting of the simulation to the nud1-2 kar1Δ15 mutant required a larger width of the Gaussian distribution (Figure 5D). This can be explained by the fact that the nud1-2 kar1Δ15 mutant exhibits a slight vegetative growth defect at 23°C, which gives rise to a less homogenous cell population as a starting culture used for the assessment of the sporulation profile. This heterogeneity influences the synchronicity of meiotic progression of the cells in the population and thus affects the range of acetate concentrations (but not the average acetate concentration) the cells in the population are exposed to (Taxis et al, 2005).
As a result, the mathematical analysis of the sporulation profiles indicates that the nud1-2 kar1Δ15 mutant is impaired in faithful establishment of SPB differences that are required for non-sister dyad formation.
Spc72p is required for SPB inheritance
The SPB inheritance defect seen for the nud1-2 mutants could be owing to a function of Nud1p required specifically for this process, or owing to more subtle effects of the mutant allele that are sufficient to disturb SPB inheritance but not its other essential functions. One essential function of Nud1p is binding of Spc72p, which in turn is required for cytoplasmic microtubule formation and organization (Knop and Schiebel, 1998; Gruneberg et al, 2000).
To test how sensitive SPB inheritance is with respect to amounts of Spc72p that are present at the SPBs, we analyzed SPB selection for MP formation in a diploid cell that was deleted for one copy of SPC72 (Δspc72/SPC72 strain).
This time, we used the previously established three FP reporter assay (Spc110p-CFP, Spc42p-RedStar and Mpc54p-YFP) (Taxis et al, 2005). We found a significant alteration in the frequencies of involvement of the second oldest SPB in MP formation, for situations with one MP or two MPs (Figure 6A). When we analyzed the fidelity of non-sister formation in the Δspc72/SPC72 strain using the CENV-FP assay (Figure 3A and B), we found no alteration in the frequency of sister dyad formation (Figure 6B).
Figure 6.
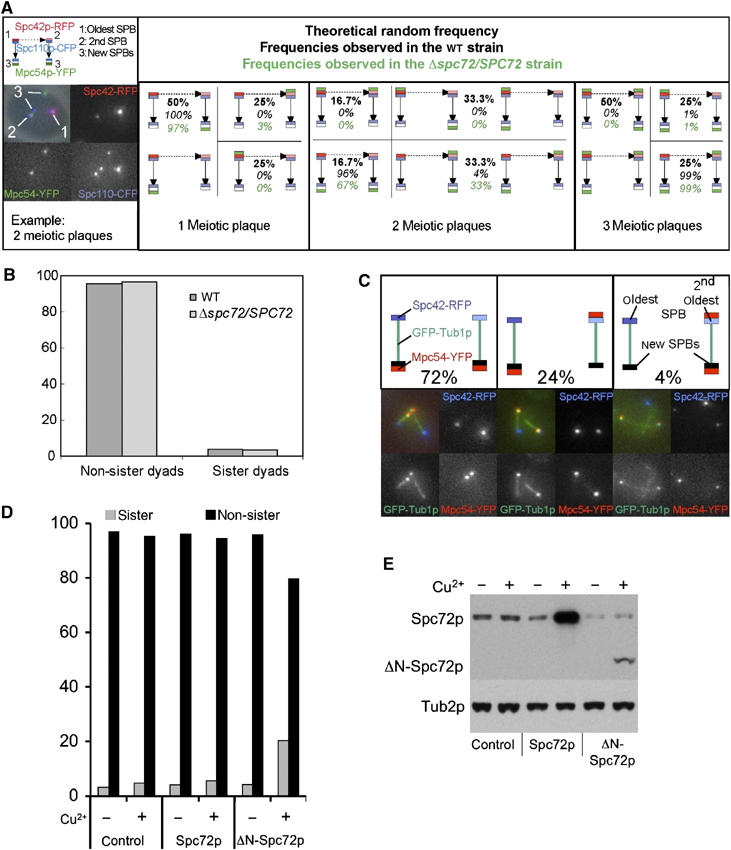
Spc72p is required for spindle pole body selection during sporulation. (A) SPB selection during sporulation in wild-type (WT; YCT806) and Δspc72/SPC72 (YMK834) cells. SPBs of three different generations are present in meiosis II. Arrows indicate the relationship between the SPBs (duplication of the SPBs during meiosis I (dashed arrow) and meiosis II (normal arrow)). The pictograms show all theoretically possible constellations for the situation with one, two or three MPs per cell. Pictograms for constellations that are indistinguishable by this method are grouped together. The data were recorded using strains harboring FP reporter as described (see text). The values for the WT strain (italic numbers) are identical to the ones previously published (Taxis et al, 2005). The theoretical random distribution is indicated in bold numbers, and the distribution found in the Δspc72/SPC72 cells, by italic green numbers. (B) Genetic analysis of non-sister dyad formation. Dyads from WT and Δspc72/SPC72 cells were analyzed for formation of non-sister versus sister dyads as before (Figure 3B), but using only one spore autonomous reporter (green/dark spores in non-sister dyads instead of green/red spores) (strains YCT716 and YCT871, respectively). (C) The neighbor relationship of the new SPBs influences MP formation in Δspc72/SPC72 cells in the case of two MPs. MPs were visualized using Mpc54p-YFP (red signals). GFP-Tub1p was used to visualize the spindles (green signals) and Spc42-RedStar to discriminate the age of the SPBs (blue signals). The pictograms show the distribution of the MPs in the case of two MPs. The percentage values indicate the distribution of the MPs in Δspc72/SPC72 cells (YCT917). (D) Frequency of formation of non-sister and sister dyads in strains overexpressing Spc72p, ΔN-Spc72p or control cells. Overexpression of Spc72p or ΔN-Spc72p was achieved by integration of a CUP1 promoter in front of one copy of the SPC72 gene in a diploid (strain YCT716) strain by using PCR targeting (Janke et al, 2004), either in front of codon 1 or 177 (ΔN-Spc72p) of the gene (resulting strains YCT1106 and YCT1107). Non-sister spores' formation was assayed using the colored spores assay (Figure 3B) in cultures sporulated in the presence of 0.01% KAc. Induction of the CUP1 promoter was achieved by addition of 1 μM CuSO4 to the sporulation medium. (E) Overexpression of Spc72p or ΔN-Spc72p in the experiment shown in (D) was monitored by Western blotting 4 h after induction of meiosis. Note that the anti-Spc72p antibody recognizes only the N-terminus of the protein (Knop and Schiebel, 1998); therefore, the signal of the ΔN-Spc72p appears much weaker because it lacks a fraction of the recognized epitopes.
This is consistent with the idea that in this strain the two SPBs from the spindle that is formed by the second oldest SPB are affected with respect to SPB selection for MP formation, but not the other two SPBs. This resembles the situation of the wild-type strain at elevated temperature (34°C) (Figure 4) and suggests again that the two new SPBs formed during meiosis II are different from each other in terms of their preference to assemble an MP.
To address the difference between the two new SPBs in the Δspc72/SPC72 strain directly, we used GFP-tubulin labelling in combination with Spc42p-RedStar and Mpc54p-YFP to reveal the connectivity between the four SPBs in meiosis II. For situations with two MPs, we found that 96% of all cells assembled one MP on the new SPB that is connected to the oldest SPB, whereas the second MP had only a preference of 76% to become formed on the second new SPB. In the other 24% of the cases, this second MP was found on the second oldest SPB (Figure 6C). This supports the notion that the likelihood of the two new SPBs to form an MP depends on the age of the SPB next to which they were formed during SPB duplication.
In order to address directly the requirement of Spc72p for regulation of SPB inheritance, we overexpressed ΔN-Spc72p in cells undergoing sporulation. In vegetative cells, this has been shown to interfere with Spc72p function and to cause depletion of astral microtubules from the cytoplasmic face of the SPB (Knop and Schiebel, 1998). For sporulation, we found that this did not affect sporulation efficiency at all (data not shown), but the faithful formation of non-sister dyads was severely impaired (Figure 6D and E).
Together, these findings argue for a function of Spc72p in the regulation of SPB inheritance in meiosis.
The nud1-2 and the kar1Δ15 mutations lead to mislocalization of Spc72p from the SPBs during meiosis I
The results so far suggest that recruitment of Spc72p is required for faithful spore number control and the establishment of differences between the SPBs. Therefore, we analyzed the abundance of Spc72p at the SPBs in the mutants by using immunofluorescence microscopy. In wild-type meiosis, Spc72p is present before and during meiosis I, whereas in cells with four SPBs (meiosis II), no Spc72p can be detected at the SPBs anymore. In contrast, Nud1p is found on all SPBs (Knop et al, 1999; Nickas et al, 2004). We found that in wild-type and also in Δspc72/SPC72 cells, ∼70% of the SPBs present in meiosis I (cells with one elongated spindle) show a clear staining for Spc72p. For kar1Δ15, only ∼30% of meiosis I SPBs show such staining. In the nud1-2 mutant and in the nud1-2 kar1Δ15 double mutant, there is almost no Spc72p staining (0–15%) (Figure 7). This means that Spc72p is binding to the SPBs in meiosis I at the OP (via Nud1p) and to the half-bridge (via Kar1p). These results are consistent with the idea that Spc72p is involved in the mechanism that generates the age bias of the SPBs that underlies SPB inheritance and control of MP assembly during meiosis such that the cells are able to regulate precisely genome inheritance as well as the number of spores to be assembled (spore number control).
Figure 7.
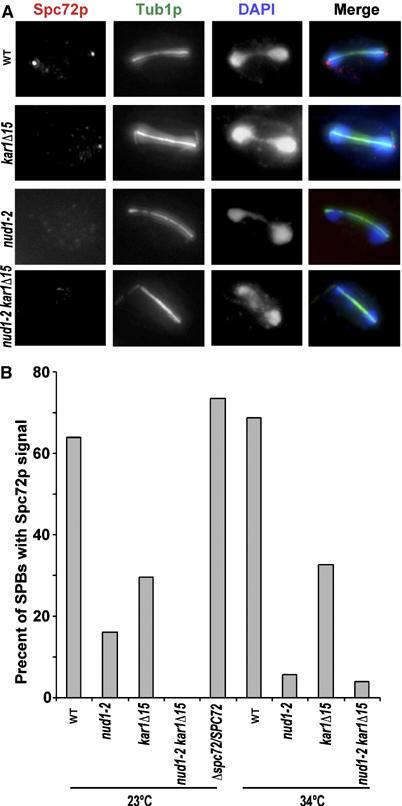
Spc72p binds via Nud1p and Kar1p to the SPBs in meiosis I. (A) Immunofluorescence microscopy of sporulating cells (at 23 and 34°C) 4 h after induction of meiosis and detection of Spc72p, tubulin and DNA. The picture shows examples from cells in meiosis I (34°C). Strains used are wild type (WT; YKS32), YOG23, YOG52 and YMK828; genotypes are indicated in the figure. (B) Quantification of the number of Spc72p foci that can be detected at the SPBs. Approximately 40 cells in anaphase of meiosis I (one elongated spindle) were evaluated per strain and temperature.
Discussion
Spindle polarity is an intrinsic characteristic of cell division in yeast and in other organisms such as Drosophila, Caenorhabditis elegans, starfish oocytes or mammalian cells (Pereira et al, 2001; Smeets and Segal, 2002; Uetake et al, 2002; Kusch et al, 2003). In yeast mitosis, spindle polarity is generated by two intertwined mechanisms. The first is a built-in spindle pole asymmetry, which sets temporal asymmetry to limit cytoplasmic microtubules to a single SPB before spindle pole separation. The second mechanism uses the established asymmetry to capture these microtubules at the bud cell-cortex. In mitotic S. cerevisiae cells, spindle asymmetry is essential for spindle positioning along the mother-cell bud axis enabling proper mitosis and chromosome movement to the bud. This system produces an invariant pattern of SPB inheritance in which the ‘old' SPB is destined to the bud (Pereira et al, 2001). The molecular basis for the built-in spindle asymmetry is presently unknown. Interestingly, the built-in spindle pole asymmetry, and thus the faithful inheritance of the old SPB into the bud, is per se not essential for mitotic spindle positioning, as other mechanisms enable recruitment of factors to one specific pole independently of the age of the SPBs (Pereira et al, 2001; Kusch et al, 2003; Maekawa et al, 2003).
In contrast, the preferential inheritance of new SPBs in meiosis is essential to enable faithful non-sister dyad formation and to ensure that the two spores in most dyads are of opposite mating types (owing to the linkage of the MAT locus to the centromere on chromosome III; Hawthorne and Mortimer, 1960). This enables within-ascus (dyad) mating upon germination. Such mating is required to maximize transmission of heterozygosities following meiosis and to account for possible loss of genomes due to associated haploid lethal mutations. Non-sister dyad formation is thus associated with a population genetic advantage (Taxis et al, 2005; Zakharov, 2005) and theoretical models have been generated that predict the evolution of mechanisms for intratetrad mating (Johnson et al, 2005).
In meiosis, spindle asymmetry is used to distinguish between the old SPBs and the SPBs appearing in meiosis II (Nickas et al, 2004; Taxis et al, 2005). Our results strongly suggest that each SPB has a different probability to assemble an MP, which in turn enables the selective formation of PSMs at specific SPBs and thereby precise spore number control. The probability for each SPB to assemble an MP depends on two factors. Newer SPBs that have experienced fewer cell cycles have a higher probability. In addition, the ‘quality' of an SPB also seems to depend on the age of the SPB next to which it was formed (neighborhood relationship): an SPB formed next to an old SPB (two cell cycles or more) is ‘better' than the one formed next to a more recent one (one cell cycle).
Previously, we have shown that the SPBs from different generations have different probabilities for MP assembly, which gives rise to three different types of SPBs (Taxis et al, 2005). However, in our simulation of the mechanism of MP assembly and spore number control, we used four different values, one for each individual SPB, to reproduce the measured experimental situation. Interestingly, we now find that all four SPBs are in fact different. This reinforces the value of this simulation for the understanding of the principles of spore number control.
A comparable system of spindle polarity pattern was observed previously for SPBs in vegetative S. pombe cells, where the number of experienced cell cycles and the neighborhood relationship together determine the localization of the NIMA-related kinase Fin1 to specific SPBs over consecutive rounds of cell division (Grallert et al, 2004). Fin1 seems to function in the SIN network, and to restrain SIN activity at the SPBs where it is bound. We tested the homologous gene in S. cerevisiae, KIN3, for its role in non-sister dyad formation and spore number control, but could not find any defect associated with its deletion (A Benjak and M Knop, unpublished observation).
Nud1p is a MEN component and is responsible for the localization of the spindle checkpoint proteins Bub2p and Bfa1p, as well as other MEN components to the SPB during mitosis (Gruneberg et al, 2000; Pereira et al, 2001; Segal and Bloom, 2001; Smeets and Segal, 2002). This may suggest that SPB selection in meiosis and faithful spore number control could involve this function of Nud1p. However, Δbub2 or Δbfa1 mutants produce wild-type level of non-sister dyads, and spore number control is unaffected (O Gordon and M Knop, unpublished results). This argues against a role for Nud1p binding of MEN components during selection of SPBs for MP assembly.
How could Nud1p then work in meiosis to establish faithful discrimination between all four SPBs? We can imagine two alternative models. In one scenario, Spc72p specifically occupies Nud1p binding sites for MP components. This would then direct MP assembly preferentially to newer SPBs. Several observations argue against this model. First, it does not explain the difference between the two new SPBs, because we and others never observed Spc72p on these SPBs, even using live cell imaging (Knop and Strasser, 2000; Nickas et al, 2004). Second, fusion of Spc72p to Kar1p in the homozygous Δspc72 background leads to a dominant-negative phenotype with respect to spore formation, albeit this strain is completely devoid of Spc72p at the OP, the place where the MP assembles. Third, MP assembly is not blocked at all in the nud1-2 mutant under restrictive conditions, whereas Spc72p binding is impaired (Figure 7). This indicates that Spc72p and the MP components bind in different ways to the SPBs and thus direct competition for binding sites is not very likely.
An alternative model to explain the Nud1p-dependent SPB differences could be that Nud1p acts (probably via Spc72p) to recruit an activity to the SPBs, which is required to generate the differences between the SPBs. A possible way in which SPB differences could be generated would be differences in the phosphorylation ‘status' of the SPBs. These could regulate the affinity of proteins, for example, kinases and phosphatases, to individual SPBs. This explanation is in line with the observation of heterogeneous hyperphosphorylation of core SPB components, in particular Spc42p, Cnm67p and Nud1p itself (Donaldson and Kilmartin, 1996; Gruneberg et al, 2000; Schaerer et al, 2001; Jaspersen et al, 2004). However, at present, no experimental evidence that would strengthen such a model is available. Nevertheless, or may be because of its enigmatic nature, the SPB differences are a fascinating problem that deserves further investigation.
Does the role of Nud1p and Spc72p during non-sister spore formation involve astral microtubules? This question is suggested by the notion that non-sister dyad formation is impaired in the ΔN-Spc72p overexpression strain (Figure 6D), which is known to be impaired in astral microtubule binding in vegetative cells (Knop and Schiebel, 1998). In order to address the requirement of functional astral microtubules for this process, we investigated non-sister dyad formation in a Δkar9 mutant (Miller and Rose, 1998), but we noticed no difference with the wild-type strain. It could however well be that microtubules are required to recruit activity to the SPB that acts during generation of the SPB age bias. Further investigation will be required to solve this issue.
How could SPB differences regulate MP assembly? From our sporulation profiles with the nud1-2 kar1Δ15 (Figures 5D) and also the Δspc72/SPC72 (data not shown) strains, it seems that impaired differences between SPBs cause not only the randomization of SPB selection, but also the formation of more spores. Thus, a likely explanation would be that SPBs recruit activity that is inhibitory for MP assembly. Such activity must exist, because MP assembly, but not binding of MP components to the SPBs, is cell cycle regulated (Taxis et al, 2005). A variable amount of inhibitory activity on different SPBs is also consistent with the notion that increased gene dosage of MP components is able to overcome MP assembly on less ‘good' SPBs under conditions where wild-type cells would not assemble MPs on such SPBs (Nickas et al, 2004; Taxis et al, 2005). In the future, it will be a challenge to unravel the activities that encompass cell cycle regulation and SPB specificity of MP assembly.
Spore wall formation and maturation in S. cerevisiae is viewed as a model system for the de novo formation of a complex extracellular matrix (Coluccio et al, 2004). Spore wall development starts with PSM closure following deposition of spore wall material between the two membrane layers of the PSM. We found that the temperature-sensitive mutation nud1-2 leads to partial spore wall defects (Figure 2). The molecular basis of this defectis not clear, neither do we understand why the deletion of the Spc72p binding domain of Kar1p remedies this phenotype. One possibility is, that the SPB has a late function in spore biogenesis that requires Nud1p and somehow also the half-bridge of the SPB. To understand this in more detail, it will be important to screen for new nud1 alleles with meiosis-specific phenotypes. Another obvious question is whether Nud1p interacts with SNARES/exocytic machinery, as found for Centriolin. During initiation of PSM formation, Nud1p localization in close vicinity of highly localized vesicle fusion events may suggest so, but the absence of defects in initiation of PSM formation in the nud1-2 mutant, as well as the fact that the vesicles and Nud1p are still separated by the structure of the MP, may indicate that Nud1p does not play a direct role in vesicle fusion. However, further experiments are needed to resolve this issue completely.
In this paper, we report our findings regarding the involvement of Nud1p and Spc72p in controlling spore formation on selected SPBs during meiosis II. These two proteins exhibit an inhibitory effect on MP assembly and are important for the regulation of spore number with regard to energy availability. As Nud1p is conserved in other organisms, it might also fulfill functions in centrosome inheritance in higher eukaryotes.
Materials and methods
Yeast strains and plasmids
All strains used in this study were derived from SK1 strains NKY289 and NKY292 (Alani et al, 1987), and their relevant genotypes are listed in Table I. Manipulation of yeast strains using PCR targeting (tagging with FPs, marker insertions, gene deletions) was performed as described (Janke et al, 2004). For all other yeast strain construction, standard methods (Guthrie and Fink, 1991) were used. The plasmids used are also listed in Table I. For CENV-linked labelling of spores with an FP (CENV-FP assay), cassettes containing the spore autonomously regulated promoter of the gene YKL050c followed by either GFP or RedStar and a dominant selection marker (natNT1 or kanMX4; Janke et al, 2004) were integrated between ORFs YER001w and YER002w using PCR targeting in haploid cells (details and template plasmids are available upon request).
Table 1.
Yeast strains and plasmids
| Yeast strains | ||
|---|---|---|
| NKY289 | SK1 background; MATa ura3 lys2 ho∷hisG | Alani et al (1987) |
| NKY292 | SK1 background; MATα ura3 lys2 leu2∷hisG ho∷LYS | Alani et al (1987) |
| YKS32 | Diploid from NKY289 and NKY292 | Knop and Strasser (2000) |
| YOG23 | MATa/alpha nud1-2∷LEU2/nud1-2∷LEU2 Δnud1∷HIS3MX/Δnud1∷HIS3MX | This study |
| YOG52 | MATa/alpha kar1Δ15/kar1Δ15 | This study |
| YOG66 | MATa/alpha nud1-2∷LEU2/nud1-2∷LEU2 Δnud1∷HIS3MX/Δnud1∷HIS3MX kar1Δ15/kar1Δ15 | This study |
| YOG26 | MATa/alpha nud1-2∷LEU2/nud1-2∷LEU2 Δnud1∷HIS3MX/Δnud1∷HIS3MX MNN1∷PYKL050c∷GFP∷natNT2/MNN1∷PYKL050c∷RFP∷natNT2 | This study |
| YOG72 | MATa/alpha kar1Δ15/kar1Δ15 MNN1∷PYKL050c∷GFP∷natNT2/MNN1 | This study |
| YOG73 | MATa/alpha nud1-2∷LEU2/nud1-2∷LEU2 Δnud1∷HIS3MX/Δnud1∷HIS3MX kar1Δ15/kar1Δ15 MNN1∷PYKL050c∷GFP∷natNT2/MNN1 | This study |
| YOG84 | MATa/alpha nud1-2∷LEU2/nud1-2∷LEU2 Δnud1∷HIS3MX/Δnud1∷HIS3MX MPC54∷eGFP∷kanMX4/MPC54∷eGFP∷kanMX4 SPC42∷RFP∷natNT2/SPC42∷RFP∷natNT2 | This study |
| YCT806 | MATa/alpha MPC54∷YFP∷hphNT1/MPC54∷YFP∷hphNT1 SPC42∷RedStar∷kanMX/ SPC42∷RedStar∷kanMX SPC110∷CFP∷kanMX/SPC110∷CFP∷kanMX | Taxis et al (2005) |
| YMK828 | MATa/alpha SPC72/Δspc72∷natNT2 | This study |
| YMK834 | MATa/alpha SPC72/Δspc72∷natNT2 MPC54∷YFP∷hphNT1/MPC54∷YFP∷hphNT1 SPC110∷ CFP∷kanMX/SPC110-CFP∷kanMX SPC42∷RFP∷kanMX/SPC42∷RFP∷kanMX | This study |
| YCT716 | MATa/alpha MNN1/MNN1∷PYKL050c∷GFP∷kanMX | Taxis et al (2005) |
| YCT883-2 | MATa/alpha Δspc72∷hphNT1/Δspc72∷hphNT1 MNN1∷Pykl50c∷GFP∷kanMX/MNN1 pRS305-N-SPC72-ΔN-KAR1/pRS305-N-SPC72-ΔN-KAR1 | This study |
| YCT871 | MATa/alpha SPC72/Δspc72∷natNT2 MNN1/MNN1∷PYKL050c∷GFP∷kanMX | This study |
| YCT917 | MATa/alpha PTUB1∷GFP∷TUB1∷URA3/PTUB1∷GFP∷TUB1∷URA3 SPC42∷RFP∷kanMX/ SPC42∷RFP∷kanMX MPC54∷YFP∷hphNT1/MPC54∷YFP∷hphNT1 SPC72/Δspc72∷naNT2 | This study |
| YCT1106 | YCT716 containing natNT2∷CUP1pr∷SPC72/SPC72 | This study |
| YCT1107 | YCT716 containing natNT2∷CUP1pr∷ΔN-SPC72/SPC72 (codons 2–176 are deleted) | This study |
| Plasmids | ||
| pMR77 | KAR1, URA3, amp-r. | Rose and Fink (1987) |
| pSRN1 | RFP (RedStar) in pFA6a-natNT2 | This study |
| pAB2 | PYKL050c∷RFP(RedStar) in pFA6a-natNT2 | This study |
| pRS305 nud1-2 | Integrative LEU2 plasmid with nud1-2 allele | Gruneberg et al (2000) |
Sporulation, spore counting and microscopy
Sporulation in liquid cultures and spore counting were performed as previously described (Taxis et al, 2005). For the sporulation of temperature-sensitive strains, pre-growth regimens were performed at 23°C, and the duration of the growth period on presporulation medium was correspondingly extended (13.5 h at 30°C, 17 h at 23°C). Following induction of sporulation (in water containing the indicated potassium acetate (KAc) concentrations, see figure legends), cells were shifted to restrictive temperature after 90 min. Sporulation on plates was performed on SPO solid medium (1.5% KAc, 1.5% agar) and incubated for 2–7 days.
We used a microscope (IRBE; Leica) equipped with a × 63 NA 1.4 oil objective (Leica), a piezo stepper, a camera (CoolSNAP HQ; Photometrics) and DAPI/Cy3/Piston-GFP/YFP filter sets (Chroma Technology Corp.). The pictures were recorded using Metamorph software (Molecular Devices). For fluorescence images, always stacks encompassing the thickness of the cells were recorded (spacing 0.6 μm). Maximum projections of the fluorescence images were generated, colored and sometimes superimposed with the phase-contrast image using Metamorph software. Sporulation efficiency was calculated as a fraction of the maximal achievable amount of spores (all cells form tetrads=100%).
Simulation of spore number control in the nud1-2 kar1Δ15 mutant
A mathematical model of spore number control (Taxis et al, 2005) was used to analyze the nud1-2 kar1Δ15 mutant. The simulation describes the self-organizing system underlying MP assembly at the four different SPBs. It requires four input parameters that characterize the individual SPBs' fitness with respect to promoting MP assembly. The model furthermore considers population statistics to model entire populations. For this purpose, it implements two types of variations. One distribution function describes the quantitative variation in acetate uptake for individual cells, for example, owing to variations in meiotic progression. The other distribution function considers the individual regulation of the abundance of MP components with available acetate. The sporulation profile of the wild-type strain was fitted with the mathematical model, as described for the wild-type strain by Taxis et al (2005). As we used different pre-growth conditions (23°C instead of 30°C) in order to account for the experimental needs of the temperature-sensitive nud1-2 kar1Δ15 mutant, the associated experimental wild-type sporulation profiles had to be measured anew. Subsequently, the model was reapplied to fit the sporulation profile of the nud1-2 kar1Δ15 mutant.
Immunofluorescence and electron microscopy
Immunofluorescence microscopy, antibodies used for immunolabelling, as well as the electron microscopy protocol (permanganese staining) of sporulating yeast cells have been described previously (Riedel et al, 2005).
Acknowledgments
We thank E Schiebel and M Rose for providing plasmids. This work was supported by the German Israeli Foundation (GIF Grant G-664-71.13/2000 to M Knop, E Stelzer and G Simchen).
References
- Adams IR, Kilmartin JV (1999) Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol 145: 809–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E, Cao L, Kleckner N (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116: 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajgier BK, Malzone M, Nickas M, Neiman AM (2001) SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol Biol Cell 12: 1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Amon A (2001) Men and sin: what's the difference? Nat Rev Mol Cell Biol 2: 815–826 [DOI] [PubMed] [Google Scholar]
- Brachat A, Kilmartin JV, Wach A, Philippsen P (1998) Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol Biol Cell 9: 977–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullitt E, Rout MP, Kilmartin JV, Akey CW (1997) The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell 89: 1077–1086 [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L (1975) Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol 124: 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L (1982) Reversible pachytene arrest of Saccharomyces cerevisiae at elevated temperature. Mol Gen Genet 187: 47–53 [DOI] [PubMed] [Google Scholar]
- Coluccio A, Bogengruber E, Conrad MN, Dresser ME, Briza P, Neiman AM (2004) Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot Cell 3: 1464–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidow LS, Goetsch L, Byers B (1980) Preferential occurrence of nonsister spores in two-spored asci of Saccharomyces cerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics 94: 581–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Kilmartin JV (1996) Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J Cell Biol 132: 887–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S, Knop M, Schlenstedt G, Schiebel E (1999) Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc Natl Acad Sci USA 96: 6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Krapp A, Bagley S, Simanis V, Hagan IM (2004) Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev 18: 1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Jurczyk A, Sillibourne J, Halilovic E, Mogensen M, Groisman I, Blomberg M, Doxsey S (2003) A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J Cell Biol 161: 535–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ (2005) Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 123: 75–87 [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E (2000) Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J 19: 6475–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR (1991) Guide to yeast genetics and molecular biology. Meth Enzymol 194: 429, 663. [PubMed] [Google Scholar]
- Hawthorne DC, Mortimer RK (1960) Chromosome mapping in Saccharomyces: centromere-linked genes. Genetics 45: 1085–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962 [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Huneycutt BJ, Giddings TH Jr, Resing KA, Ahn NG, Winey M (2004) Cdc28/Cdk1 regulates spindle pole body duplication through phosphorylation of Spc42 and Mps1. Dev Cell 7: 263–274 [DOI] [PubMed] [Google Scholar]
- Johnson LJ, Antonovics J, Hood ME (2005) The evolution of intratetrad mating rates. Evolution 59: 2525–2532 [PubMed] [Google Scholar]
- Knop M, Barr F, Riedel CG, Heckel T, Reichel C (2002) Improved version of the red fluorescent protein (drFP583/DsRed/RFP). Biotechniques 33, 592, 594, 596–598 passim [DOI] [PubMed] [Google Scholar]
- Knop M, Miller KJ, Mazza M, Feng D, Weber M, Keranen S, Jantti J (2005) Molecular interactions position Mso1p, a novel PTB domain homologue, in the interface of the exocyst complex and the exocytic SNARE machinery in yeast. Mol Biol Cell 16: 4543–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Schiebel E (1999) Microtubule organization by the budding yeast spindle pole body. Biol Cell 91: 291–304 [PubMed] [Google Scholar]
- Knop M, Schiebel E (1998) Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J 17: 3952–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Strasser K (2000) Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J 19: 3657–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Cano E, Simanis V (2004) Analysis of the S. pombe signalling scaffold protein Cdc11p reveals an essential role for the N-terminal domain in SIN signalling. FEBS Lett 565: 176–180 [DOI] [PubMed] [Google Scholar]
- Kusch J, Liakopoulos D, Barral Y (2003) Spindle asymmetry: a compass for the cell. Trends Cell Biol 13: 562–569 [DOI] [PubMed] [Google Scholar]
- Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M (2001) Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol Cell Biol 21: 6972–6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H, Usui T, Knop M, Schiebel E (2003) Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J 22: 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Rose MD (1998) Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol 140: 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Rapport E (1971) Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J Cell Biol 50: 344–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM (1998) Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol 140: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas ME, Diamond AE, Yang MJ, Neiman AM (2004) Regulation of spindle pole function by an intermediary metabolite. Mol Biol Cell 15: 2606–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas ME, Schwartz C, Neiman AM (2003) Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot Cell 2: 431–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Iino T (1981) Selective abortion of two nonsister nuclei in a developing ascus of the hfd1-1 mutant in Saccharomyces cerevisiae. Genetics 99: 197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Iino T (1982) Genetic block of outer plaque morphogenesis at the second meiotic division in an hfd1-1 mutant of Saccharomyces cerevisiae. J Gen Microbiol 128: 1309–1317 [DOI] [PubMed] [Google Scholar]
- Pereira G, Grueneberg U, Knop M, Schiebel E (1999) Interaction of the yeast gamma-tubulin complex-binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J 18: 4180–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth K, Schiebel E (2001) Modes of spindle pole body inheritance and segregation of the Bfa1p–Bub2p checkpoint protein complex. EMBO J 20: 6359–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M (2001) Centrosome-dependent exit of cytokinesis in animal cells. Science 291: 1550–1553 [DOI] [PubMed] [Google Scholar]
- Riedel CG, Mazza M, Maier P, Korner R, Knop M (2005) Differential requirement for phospholipase D/SPO14 and its novel interactor SMA1 for regulation of exocytotic vesicle fusion in yeast meiosis. J Biol Chem 280: 37846–37852 [DOI] [PubMed] [Google Scholar]
- Rose MD, Fink GR (1987) KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell 48: 1047–1060 [DOI] [PubMed] [Google Scholar]
- Schaerer F, Morgan G, Winey M, Philippsen P (2001) Cnm67p is a spacer protein of the Saccharomyces cerevisiae spindle pole body outer plaque. Mol Biol Cell 12: 2519–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M, Bloom K (2001) Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol 11: 160–166 [DOI] [PubMed] [Google Scholar]
- Smeets MF, Segal M (2002) Spindle polarity in S. cerevisiae: MEN can tell. Cell Cycle 1: 308–311 [DOI] [PubMed] [Google Scholar]
- Taxis C, Keller P, Kavagiou Z, Jensen LJ, Colombelli J, Bork P, Stelzer EH, Knop M (2005) Spore number control and breeding in Saccharomyces cerevisiae: a key role for a self-organizing system. J Cell Biol 171: 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Kato KH, Washitani-Nemoto S, Nemoto S (2002) Nonequivalence of maternal centrosomes/centrioles in starfish oocytes: selective casting-off of reproductive centrioles into polar bodies. Dev Biol 247: 149–164 [DOI] [PubMed] [Google Scholar]
- Vallen EA, Hiller MA, Scherson TY, Rose MD (1992) Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J Cell Biol 117: 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharov IA (2005) [Intratetrad mating and its genetic and evolutionary consequences]. Genetika 41: 508–519 [PubMed] [Google Scholar]


