Abstract
During early mammalian female development, one of the two X chromosomes becomes inactivated. Although X-chromosome coating by Xist RNA is essential for the initiation of X inactivation, little is known about how this signal is transformed into transcriptional silencing. Here we show that exclusion of RNA Polymerase II and transcription factors from the Xist RNA-coated X chromosome represents the earliest event following Xist RNA accumulation described so far in differentiating embryonic stem (ES) cells. Paradoxically, exclusion of the transcription machinery occurs before gene silencing is complete. However, examination of the three-dimensional organization of X-linked genes reveals that, when transcribed, they are always located at the periphery of, or outside, the Xist RNA domain, in contact with the transcription machinery. Upon silencing, genes shift to a more internal location, within the Xist RNA compartment devoid of transcription factors. Surprisingly, the appearance of this compartment is not dependent on the A-repeats of the Xist transcript, which are essential for gene silencing. However, the A-repeats are required for the relocation of genes into the Xist RNA silent domain. We propose that Xist RNA has multiple functions: A-repeat-independent creation of a transcriptionally silent nuclear compartment; and A-repeat-dependent induction of gene repression, which is associated with their translocation into this silent domain.
Keywords: X inactivation, Xist RNA, transcriptional silencing, 3D nuclear organization, histone modifications, differentiation
One of the most striking features of X inactivation is that it implies the differential treatment of two homologous chromosomes within the same nucleus. However, the nature of this differential treatment and the mechanisms that initiate, propagate, and maintain it remain poorly understood. Random X inactivation occurs in the embryonic lineage of female mouse embryos (Lyon 1961) and during the differentiation of female embryonic stem (ES) cells, the latter providing a powerful model system for dissection of the mechanisms underlying this process.
X inactivation is controlled by a complex locus on the X chromosome, known as the X-inactivation center (Xic). Within the Xic locus, the Xist gene produces a long untranslated RNA known to be essential for initiation and propagation of inactivation (for review, see Avner and Heard 2001; Plath et al. 2002). The accumulation of the Xist transcript on the X chromosome chosen to be inactivated (Xi) triggers the initiation of X inactivation, while expression of the other allele is progressively repressed. Xist RNA coating of the X chromosome induces inactivation rapidly, within one or two cell cycles (Penny et al. 1996; Marahrens et al. 1998; Wutz and Jaenisch 2000). Using inducible Xist cDNA transgenes in male ES cells, Wutz and Jaenisch (2000) have defined the critical time window during which Xist RNA is required for inactivation. Based on these studies, at least two phases have been defined in the inactivation process: an initiation phase (the first 48–72 h), which is dependent on Xist RNA coating, followed by an irreversible phase (>72 h), which is independent of Xist RNA (Wutz and Jaenisch 2000). Indeed, Xist RNA coating is not essential for the maintenance of transcriptional repression in somatic cells; rather, multiple epigenetic marks act in synergy to ensure the stability of the inactive state (Czankovszki et al. 2001). However, Xist RNA coating of the X chromosome does seem to be required for the recruitment of some kind of chromosomal memory, or epigenetic marking, during differentiation, although the exact nature of this chromosomal memory remains unclear (Kohlmaier et al. 2004). Some of the changes induced following Xist RNA coating include a shift to asynchronous replication timing (Takagi et al. 1982), incorporation of the histone variant macro H2A (Mermoud et al. 1999; Costanzi et al. 2000), DNA (CpG) methylation (Norris et al. 1991), and a variety of histone modifications (Chaumeil et al. 2002; Chadwick and Willard 2003; for review, see Heard 2004). Notably, changes in histone H3 and H4 modifications include hypoacetylation of H3K9 and of H4 at multiple lysines (Jeppesen and Turner 1993; Boggs et al. 1996; Keohane et al. 1996), as well as hypomethylation of H3K4, dimethylation of H3K9 (Heard et al. 2001; Boggs et al. 2002; Mermoud et al. 2002; Peters et al. 2002), and trimethylation of H3K27 (Plath et al. 2003; Silva et al. 2003; Rougeulle et al. 2004). The early timing of appearance of histone modifications during the X-inactivation process suggests that they could be involved in the initiation and/or early maintenance of the inactive state.
Inducible Xist cDNAs carrying different deletions have been used to define functional regions of this transcript (Wutz et al. 2002). A series of conserved repeats at the 5′ end of the transcript (known as the “A”-repeats) have thus been shown to be critical for its gene silencing function. Several regions of Xist are involved in its capacity to coat a chromosome in cis, as well as its ability to recruit potential epigenetic marks such as macro H2A and H3K27 trimethylation (Wutz et al. 2002; Plath et al. 2003; Kohlmaier et al. 2004). The latter is triggered by the Ezh2/Eed (PRC2) Polycomb group complex, which is recruited (directly or indirectly) by Xist RNA in an A-repeat-independent fashion (Plath et al. 2003; Silva et al. 2003; Kohlmaier et al. 2004). Analysis of Eed and Ezh2 mutant embryos suggests that PRC2 and associated H3K27 trimethylation are not required for initiation, but rather for maintenance of the inactive state of the X chromosome and that, furthermore, they are more critical for maintenance in extra-embryonic tissues than in the embryo-proper (Wang et al. 2001; Mak et al. 2002). The PRC1 Polycomb group complex is also recruited to the inactive X chromosome during early development, although its exact role in X inactivation remains to be defined (de Napoles et al. 2004; Plath et al. 2004; Hernandez-Nunoz et al. 2005). Xist RNA coating again seems to be the trigger for PRC1 recruitment, but as for the PRC2 complex, this can occur in the absence of Xist A-repeats and associated gene silencing. Thus, in addition to its silencing function, Xist RNA appears to be able to recruit various marks that may be involved in the early maintenance of the inactive state. However, the molecular mechanisms that underlie Xist RNA's capacity to induce transcriptional silencing and the nature of the changes induced on the X chromosome during development that result in the chromosomal memory of the inactivate state remain unclear.
Several hypotheses to explain Xist silencing function have proposed that Xist RNA could recruit a specific repressor to the X chromosome, although so far no such protein has been reported. Alternatively Xist RNA could have an architectural role by creating a repressive compartment or structure at the level of the X chromosome (Clemson et al. 1996). Xist RNA is intriguingly restricted in its nuclear localization to the vicinity of the chromosome territory from which it is expressed (Brown et al. 1992; Clemson et al. 1996). Cell biology studies have revealed that this restriction does not appear to be dependent on the DNA of the chromosome itself, as the Xist RNA compartment remains unperturbed after DNase treatment (Clemson et al. 1996). It has, therefore, been proposed that Xist RNA may show only an indirect association with the X chromosome and a closer association with the nuclear matrix. Potential support for this has come from the finding that scaffold attachment factor A (SAF-A) is enriched on the inactive X chromosome in an RNA-dependent manner (Helbing and Fackelmayer 2003). Xist RNA may thus form a stable structure with nuclear matrix or scaffold factors, which could be important for initiation and/or maintenance of the inactive state (Helbing and Fackelmayer 2003; Fackelmayer 2005).
In this context, we set out to determine whether Xist RNA might function at the level of nuclear organization. It is becoming increasingly clear that the spatial organization of chromatin in the nucleus is tightly correlated with the regulation of transcription. In particular, the position of a gene with respect to its chromosome territory may be linked to its transcriptional status (for reviews, see Cremer and Cremer 2001; Williams 2003). Although some genes can be transcribed from the interior of a territory, most active genes are found to be located outside the territory (Volpi et al. 2000; Mahy et al. 2002a, b; Chambeyron and Bickmore 2004b). Indeed, gene-rich regions often loop out of their chromosome territories when transcriptionally active (for review, see Chambeyron and Bickmore 2004a). In the case of the X chromosome, a recent study has shown that many X-linked genes tend to be located close to the periphery of the inactive X chromosome in human somatic cells, irrespective of their activity status (Clemson et al. 2006). However, a potential link between gene location and transcriptional status on human inactive X chromosome was revealed in another study, using high-resolution three-dimensional (3D) analysis of two X-linked genes (one is subject to inactivation [ANT2] whereas the other escapes inactivation [ANT3]) (Dietzel et al. 1999). The ANT2 gene on the active X and ANT3 on both active and inactive X chromosomes were found to be located close to the periphery of the chromosomal territory, whereas the silent ANT2 gene on the inactive X displayed a more internal location. Such studies have so far only been performed in somatic cells, where the X-inactivation process is complete and XIST RNA no longer plays any major role in silencing, as epigenetic marks have taken over.
In this study we investigated the relationship between Xist RNA-mediated silencing and the 3D nuclear organization of the X chromosome in a developmental context. We provide evidence for an intimate link between Xist RNA function during X inactivation and nuclear compartmentalization. Immediately following Xist RNA accumulation on the X chromosome, it forms what appears to be a silent nuclear compartment that is depleted for RNA Polymerase II (RNA Pol II) and transcription factors and is associated with silencing of repetitive sequences on the X chromosome. We also make the striking finding that formation of this early compartment occurs independently of the Xist A-repeats. Gene repression occurs subsequently, requires Xist A-repeat action, and is accompanied by a shift of genes from outside to inside this silent nuclear compartment. We thus demonstrate, for the first time, that the onset of X inactivation is accompanied by a dynamic 3D reorganization of the X chromosome and we reveal a new function for the Xist transcript in generating a repressive nuclear compartment that is formed independently of its A-repeats.
Results
Early exclusion of RNA Pol II and transcription factors from the Xist RNA-coated X chromosome during female ES cell differentiation
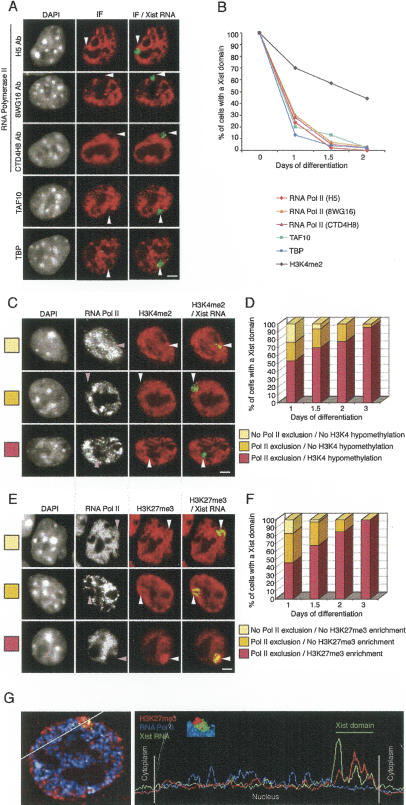
The mechanisms underlying the establishment of global transcriptional silencing of the X chromosome during early development and, more specifically, the manner in which Xist RNA accumulation is converted into a powerful repressive signal remain unknown. In order to explore this issue, we set out to analyze the relationship between Xist RNA accumulation and RNA Pol II. Immunofluorescence for RNA Pol II was coupled with Xist RNA FISH on differentiating female ES cells (Fig. 1A,B). An antibody that recognizes the C-terminal domain (CTD) of the largest subunit of RNA Pol II was first tested (8WG16). Xist RNA accumulation kinetics are described in Materials and Methods. Following 1 d of differentiation, the majority (72%), but not all, Xist RNA domains were already found to be associated with a clear exclusion in RNA Pol II (Fig. 1B; n = 60) (“exclusion” or “depletion” is equivalent to the fluorescence intensity of the extranuclear or nucleolar background; see blue signal in line scan in Fig. 1G). By day 3 of differentiation, the proportion of cells with Xist RNA domains showing an exclusion of RNA Pol II had increased to almost 100%. No exclusion of RNA Pol II could be seen across either of the two X chromosomes prior to inactivation in undifferentiated or early differentiating ES cells, based on line scans drawn around the Xist RNA primary transcript signals (see Supplementary Fig. 1). A second antibody that recognizes the CTD (CTD4H8) gave identical results. Phosphorylation of Ser 2 and Ser 5 of the CTD of the largest subunit of RNA Pol II characterizes the elongation and initiation forms of this enzyme, respectively. Immuno-Xist RNA FISH on differentiating female ES cells was performed using H5 (elongation-specific) and H14 (initiation-specific) anti-Pol II antibodies. Similar early kinetics of exclusion from the Xist RNA domain were found using these antibodies (Fig. 1A,B, H14 not shown). We also investigated the profiles of two partners of RNA Pol II, TAF10 and TBP (the latter is thought to be one of the first proteins to be recruited during transcription). Both proteins were excluded from the Xist RNA domain with similar kinetics to RNA Pol II (Fig. 1A,B). In order to control that exclusion of RNA Pol II and other factors was not simply due to antibody accessibility issues, we performed transient transfections with RNA Pol II fused to GFP. We found it to be similarly excluded from the Xist RNA-coated X chromosome in all cells (differentiating ES cells and MEFs [mouse embryonic fibroblast]) examined. Other GFP fusion proteins (e.g., H2A-GFP or H2B-GFP) showed no such exclusion (data not shown). Thus, exclusion of the transcription machinery represents a very early event in the time course of X inactivation.
Figure 1.
Exclusion of the transcription machinery from the Xist RNA domain during X inactivation of female ES cells undergoing differentiation. (A) Representative immunofluorescence (IF) using antibodies against several components of the transcription machinery (RNA Pol II, TAF10, and TBP; red) combined with Xist RNA FISH (green) on female ES cells after 2 d of differentiation. Three antibodies (H5, 8WG16, and CTD4H8) recognizing different phosphorylated forms of RNA Pol II were used. Arrowheads indicate the location of the Xist RNA domain. DNA is stained with DAPI (gray). Bar, 5 μm. (B) Kinetics of exclusion of the transcription machinery from the Xist RNA-coated X chromosome during female ES cell differentiation (day 1, n = 60; days 1.5 and 2, n = 100). (C,E) Dual IF for RNA Pol II (H5 Ab; gray) and histone H3K4me2 (C, red) or histone H3K27me3 (E, red) was combined with Xist RNA FISH (green) in female ES cells undergoing differentiation (day 1, top line; day 2, middle line; day 3, bottom line). The Xist RNA domain is shown with an arrowhead. DNA is stained with DAPI (gray). Bar, 5 μm. (D,F) Relative kinetics of the exclusion of the transcription machinery and the appearance of histone modifications during X inactivation on female ES cells undergoing differentiation (day 1, n = 60; days 1.5, 2, and 3, n = 100). (G) Representative example of 3D analysis, 3D reconstruction, and a scan line to demonstrate the relative exclusion of RNA Pol II (blue) and enrichment of H3K27me3 (red) with respect to the Xist RNA domain (green) in female ES cells differentiated for 3 d.
Exclusion of the transcription machinery is the earliest known event following Xist RNA accumulation
The above results suggest that the exclusion of transcription factors from the X chromosome might be an even earlier event than the establishment of specific histone modifications on the future inactive X chromosome. In order to define the relative order of these events more precisely, we performed dual immunofluorescence to detect transcription factors and histone modifications simultaneously, and combined this with Xist RNA FISH during female ES cell differentiation (Fig. 1C–F). Examples of RNA Pol II (H5 antibody) and histone H3 dimethylated on Lys 4 (H3K4me2) (Fig. 1C,D) or histone H3 trimethylated on Lys 27 (H3K27me3) (Fig. 1E,F) stainings are shown, alongside line scans (Fig. 1E). Three profiles were observed during early stages of differentiation. In the first category (Fig. 1D,F, white bars), the Xist RNA domain is neither depleted for RNA Pol II, nor is it associated with the Xi-specific histone modifications. In the second category (Fig. 1D,F, yellow bars), the Xist RNA domain is already depleted for RNA Pol II but is not yet associated with histone modifications. In the third category (Fig. 1D,F, purple bars), the Xist RNA domain is both depleted in RNA Pol II and associated with histone modifications (depletion of H3K4me2 and enrichment of H3K27me3). Nuclei where the Xist RNA domain is associated with Xi-specific histone modifications but is not depleted for RNA Pol II were never detected (n ≥ 50 at each differentiation stage). To determine the exact relative timing of these two events, we examined ES cells differentiated for 1, 1.5, 2, and 3 d. This showed that after 1 d of differentiation, 70%–80% of Xist RNA domains are depleted for RNA Pol II, whereas only 53% are additionally depleted of H3K4me2 and 46% are also enriched in H3K27me3 (n = 60). After 3 d of differentiation, these characteristic histone modifications are present on the Xi in almost 100% of cells with Xist RNA domains. Other histone modifications, including H3K9ac, H3K9me2, and H4ac, and other transcription factors such as TAF10, TBP, and RNA Pol II (8WG16 antibody) gave similar results (Supplementary Table 1). Taken together, these results demonstrate that exclusion of transcription factors from the Xist RNA-coated chromosome precedes the appearance of known histone modifications characteristic of the Xi. The exclusion of the transcription machinery is thus the earliest event following Xist accumulation described so far.
Exclusion of the transcription machinery from the Xist RNA-coated X precedes gene silencing during the course of X inactivation
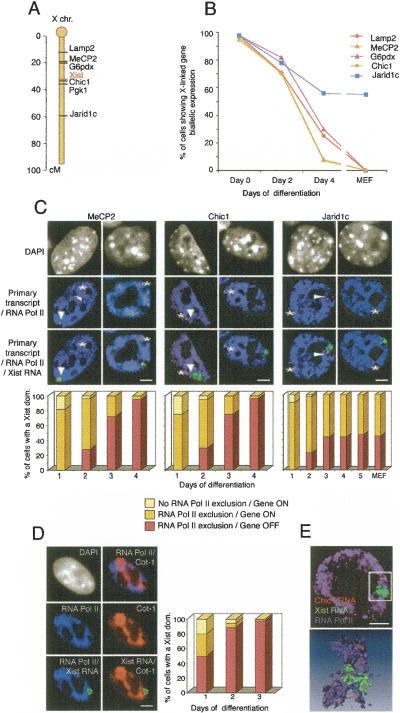
This early exclusion of the transcription machinery, within 1–2 d, would appear to be in contradiction with previous studies, which showed that X-linked gene repression only begins after 2 d of differentiation (Keohane et al. 1996; Heard et al. 2001). In order to resolve this paradox, we examined the kinetics of RNA Pol II exclusion and X-linked gene activity simultaneously, using immunofluorescence (RNA Pol II H5 antibody) combined with dual RNA FISH (Xist and a gene-specific probe for primary transcript detection) during the course of X inactivation (Fig. 2C). In this assay, a transcriptionally active gene is detectable as a punctate RNA signal at its locus corresponding to its nascent transcript (Lawrence and Singer 1985). The kinetics of transcriptional silencing of several X-linked genes was analyzed (Fig. 2A). Two of these, MeCP2 and G6pdx, are separated by ~300 kb and are located 30 Mb proximal to Xist; Lamp2 is located 66 Mb proximal to Xist; Chic1 is located just 70 kb 3′ to Xist; Pgk1 and Jarid1c are located 3 and 47 Mb distal to Xist, respectively. Transcription on the active X could be detected with high efficiency (>70%) for some of these genes, whereas for others, such as MeCP2, a primary transcript was detectable in only 10% of cells. The primary transcript of Pgk1 was barely detectable and only in a low percent of cells, thus precluding analysis of its kinetics, although previous RT– PCR studies have established that it is silenced around day 2 of differentiation (Keohane et al. 1996; Panning et al. 1997). All of the above genes have been found to be subject to X inactivation during ES cell differentiation (Fig. 2B; Keohane et al. 1996; Simmler et al. 1997; Heard et al. 2001; Wutz et al. 2002). In addition to these genes, we also examined Jarid1c, which has been reported to be subject to X inactivation initially and then to escape later on in development (Tsuchiya et al. 2004).
Figure 2.
Exclusion of the transcription machinery occurs earlier than the transcriptional repression of three X-linked genes during X inactivation. (A) Locations of the Lamp2, MeCP2, G6pdx, Xist, Chic1, Pgk1, and Jarid1c genes on the X chromosome. (B) Kinetics of repression of Lamp2, MeCP2, G6pdx, Chic1, and Jarid1c during X inactivation (n > 50 at every time point). The percentage of cells with Xist RNA domains (except at day 0) showing biallelic expression of different X-linked genes is shown in ES cells differentiated for 0, 2, or 4 d, as well as in MEFs where X inactivation has been completed. (C) Representative IF of RNA Pol II (H5 Ab; blue) combined with Xist RNA FISH (green) and RNA FISH showing the primary transcript of three X-linked genes (MeCP2, Chic1, or Jarid1c; red) on female ES cells at day 2 (left column) and day 4 (right column) of differentiation. Arrowheads show primary transcripts on the inactive X chromosome, whereas asterisks indicate primary transcripts on the active X chromosome. DNA is stained with DAPI (gray). Bar, 5 μm. Relative kinetics of RNA Pol II exclusion and gene repression during X inactivation on female ES cell undergoing differentiation (day 1, n = 60; following days, n = 100). (D) Representative IF for RNA Pol II (H5 Ab; blue) combined with Cot-1 (red) and Xist (green) RNA FISH on female ES cells at day 2 of differentiation. Arrowheads show the exclusion of RNA Pol II and Cot-1 RNA on the Xist RNA domain. DNA is stained with DAPI (gray). Bar, 5 μm. Relative kinetics of RNA Pol II exclusion and Cot-1 repression during X inactivation on female ES cell undergoing differentiation (day 1, n = 60; days 2 and 3, n = 100). (E) Example of 3D analysis of RNA Pol II IF together with Chic1 and Xist RNA FISH in early differentiated ES cells, using Metamorph software (top) and after Amira reconstruction (bottom).
Biallelic expression of these genes could be detected in both undifferentiated and day 1 differentiated ES cells with equivalent frequencies (data not shown), despite the fact that >70% of Xist RNA domains at day 1 already show an exclusion of RNA Pol II (n = 60) (Fig. 2C). By day 2 of differentiation, MeCP2 is repressed on the Xist RNA-coated X chromosome in 27%, Chic1 in 30%, G6pdx and Lamp2 in 18%, and Jarid1c in 23% of cells (n ≥ 100) (Fig. 2B,C). The kinetics of gene silencing found for all of the above genes confirm previous studies and demonstrate that the transcription machinery is indeed excluded from the Xist RNA-coated X chromosome prior to the initiation of gene silencing.
In the case of Jarid1c, we were not able to detect >50% of cells in which this gene was silenced throughout ES cell differentiation and even in somatic cells (MEFs) where inactivation is known to be complete (Fig. 2C, right panel). Although this result is in agreement with previous studies showing that Jarid1c is one of the few genes escaping X inactivation in mice, in differentiating ES cells we did not find that Jarid1c was initially submitted to inactivation and then subsequently escaped at later stages (Tsuchiya et al. 2004). This could reflect differences between embryos and differentiating ES cells. As X inactivation during ES cell differentiation is not totally synchronous, we may have missed transient full inactivation followed by reactivation of Jarid1c.
As another assay for the onset of gene repression, we used Cot-1 RNA FISH combined with the immunostaining of RNA Pol II (Fig. 2D). The Cot-1 fraction of the genome represents middle repetitive DNA which is present in transcribed regions such as introns, 5′ and 3′ UTRs, as well as in intergenic regions. When Cot-1 DNA is used as a probe for RNA FISH, this allows visualization of the overall transcriptional status of the genome (Hall et al. 2002). After 1 d of differentiation, 63% of the Xist RNA domains that were depleted of RNA Pol II also showed a loss of Cot-1 RNA signal within the domain (Fig. 2D; Supplementary Fig. 2), and by day 2 this proportion had increased to almost 100%. Thus, depletion of the Cot-1 RNA signal from the Xist RNA domain was detected following the exclusion of RNA Pol II during the time course of X inactivation. This result confirmed that exclusion of the transcription machinery from the Xist RNA-coated X chromosome occurs prior to gene repression. Intriguingly, the onset of X-chromosome silencing detected by Cot-1 RNA FISH appears to occur slightly earlier, at day 1 (Fig. 2), than the silencing of any of the genes analyzed, which only begins at day 2 (see above).
Location of transcribed genes at the periphery of or outside the Xist RNA domain
The above findings demonstrate that exclusion of the transcription machinery from the Xist RNA-coated X chromosome occurs while some X-linked genes are still being transcribed and thus raises the paradox of the time-lag between these two events. In the course of our analysis, we noted that the primary transcripts of all of the X-linked genes analyzed were detected at the periphery of, or outside, the Xist RNA domain, in 100% of cases (Fig. 2C). To assess this more carefully, we performed 3D microscopy and reconstruction using dedicated softwares (Metamorph and Amira; see Materials and Methods) (Fig. 2E). This confirmed that the primary transcripts of genes still being transcribed on the X chromosome undergoing inactivation are still within nucleoplasm containing RNA Pol II and transcription factors. This external location, although rarely >0.8 μm away from the Xist RNA domain (see Materials and Methods), explains how genes can remain expressed during early stages of differentiation even though the transcription machinery has already become excluded from the Xist RNA domain.
Relocalization of X-linked genes within the Xist RNA domain during X inactivation
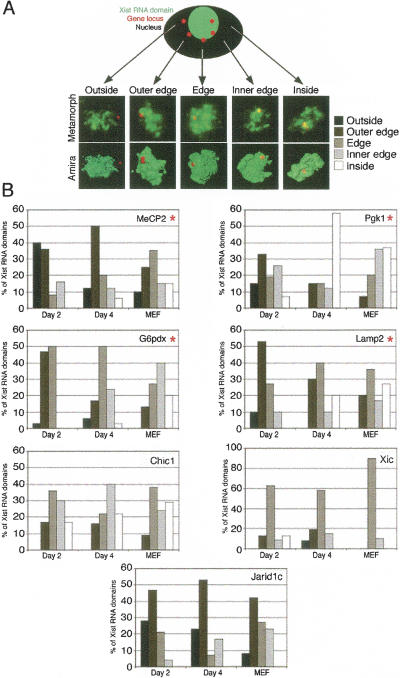
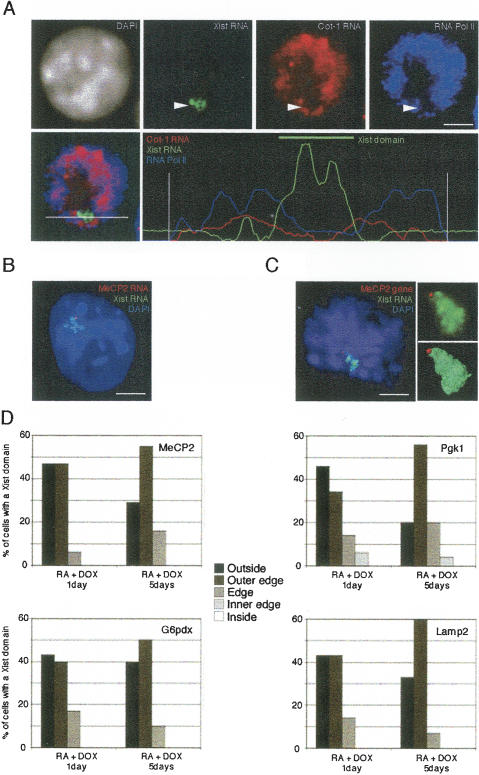
Given that, prior to their silencing, genes are detected by RNA FISH at the periphery or outside of the Xist RNA domain, this raises the question of the location of the genes after they become inactivated. In order to address this, we performed DNA FISH to assess how positions of X-linked genes might change with respect to the Xist RNA domain following their inactivation. The locations of the genes tested above were examined at different stages of ES cell differentiation (Fig. 3). Cells were analyzed at day 2 of differentiation, when Xist RNA has already accumulated in a substantial proportion of cells (~30%) but X inactivation has only just begun and X-linked genes are still expressed biallelically in the majority of these cells; at day 4 of differentiation, when inactivation is virtually complete and monoallelic expression is seen in the majority of cells containing a Xist RNA domain for all of the X-linked genes examined (except Jarid1c), and in MEFs as a control for a fully differentiated and inactivated state. Following 3D microscopy analysis, Amira reconstruction, and measurements (see Materials and Methods), profiles of gene location with respect to the Xist RNA domain were classified into five distinct categories (Fig. 3A; see Materials and Methods for classification), and the proportions of cells showing different profiles for each gene are presented in Figure 3B.
Figure 3.
Location of three X-linked genes with respect to the Xist RNA domain during X inactivation. (A) 3D analysis of Xist RNA FISH (green) combined with DNA FISH to detect the location of X-linked genes (Lamp2, MeCP2, G6pdx, Pgk1, Chic1, Jarid1c, and the Xic locus; red) on female ES cells undergoing differentiation (days 2 and 4) and MEFs. Different profiles of X-linked gene location outside, at the outer edge, at the edge, at the inner edge, or inside the Xist RNA domain are shown following Amira reconstruction. (B) Distribution of the different locations of X-linked genes with respect to the Xist RNA domain before, during, and after X inactivation in differentiated ES cells at days 2 and 4, and in MEFs (n > 30). Genes showing a statistical difference in position distributions are indicated with red asterisks.
In the cases of the MeCP2, G6pd, Lamp2, and Pgk-1 genes, following 2 d of differentiation (genes still active in >70% of cells, see above), they are located outside or at the outer edge of (i.e., no overlap of green and red signals) the Xist RNA domain in most cells (Fig. 3B) and are thus in contact with the transcription machinery, as predicted by the RNA FISH analysis (Fig. 2C). However, after4dof differentiation, when these genes have mostly undergone X inactivation, they all show a shift in position toward the interior of the Xist RNA domain (i.e., the red signal appears yellow due to overlap with green). This internalization is even more pronounced in MEFs where X inactivation is fully established. The most dramatic change was found for the Pgk1 gene, which was located outside or at the outer edge of the Xist RNA domain in almost 50% of cells at day 2 and shifted to the inside or inner edge in >70% of day 4 differentiated cells or MEFs. For MeCP2, Pgk1, G6pdx, and Lamp2, gene locations relative to the Xist RNA domain were also found to differ significantly (p < 0.05) between days 2 and 4 of differentiation and MEFs, using a χ2 test (see Materials and Methods).
In the case of Chic1, although a shift to a more internal location within the Xist RNA domain was observed, the change was not found to be statistically significant. Indeed, this gene was always found to be located rather close to the edge of the Xist RNA domain, and the shift in location was only from outer to inner edge, never to the inside (Fig. 3B). The position of the Chic1 gene is probably restricted to this peripheral location because of its tight linkage to the Xist gene and the fact that Xist has to remain transcriptionally active on the otherwise silent X chromosome following X inactivation. To verify that this is the case, we analyzed the position of the Xic region with respect to the Xist RNA domain, using a 460-kb-length probe (including Xist and Chic1) (Fig. 3B). The Xic locus was found to be located at or close to the periphery of the Xist RNA domain in the majority of cells and showed no significant shift in position through differentiation (2 d;4din MEFs). Thus at least part of the region remains in contact with the transcription machinery. This specific location of the Xic region straddling the periphery of the Xist RNA domain thus reflects the fact that, on the one hand, it carries the Xist gene which remains active and located outside of the domain, while on the other hand it also contains other genes, such as Chic1, that are subject to inactivation and located at the inner edge of the Xist RNA domain.
Finally, the Jarid1c gene, which escapes inactivation in a proportion of cells, showed a rather unique distribution of positions relative to the Xist RNA domain compared with all of the other loci examined. This was the only gene to show an external location in a strikingly high proportion of cells (>70% outside or outer edge) at day 4 of differentiation. In MEFs, where Jarid1c is active (on the otherwise inactive X chromosome, see Fig. 2C) in ~50% of cells, it is located on the outside or the outer edge of the Xist RNA domain (Fig. 3B). Moreover, even in those MEFs where it is silent, it remains situated at the periphery of the Xist RNA domain and never shows a fully internal location, contrary to all other X-linked genes examined, except the Xic (Fig. 3B). This external/ peripheral location of the Jarid1c gene correlates well with, and might even explain, its tendency to escape from X inactivation (Tsuchiya et al. 2004).
Constant Xist RNA coating surface of the X-chromosome territory during the course of X inactivation
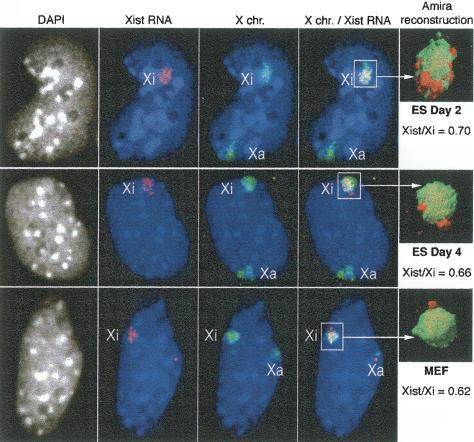
The relocation of genes into the Xist RNA domain during X inactivation could be explained by several hypotheses: an increase in the extent of the X chromosome coated by Xist RNA, an increase in the compaction of chromatin of the X chromosome within the Xist RNA domain, or a spatial reorganization of genes. In order to test the first two possibilities, we measured the relative volumes of the Xist RNA domain and the X-chromosome territory, before and after inactivation. We combined Xist RNA FISH with DNA FISH using an X-chromosome paint probe in female ES cells at days 2 and 4 of differentiation, as well as in MEFs (Fig. 4). After 3D analysis and reconstruction, volumes were measured using Amira software. At all stages, Xist RNA covers ~70% of the X-chromosome territory. Thus, we find no evidence for an increase in the degree of X-chromosome coating by the Xist transcript. Furthermore, although we cannot exclude that some local compaction of chromatin occurs during differentiation that would be undetectable at the level of DNA FISH, we find no evidence for any global compaction of X-linked chromatin within the Xist RNA domain during differentiation.
Figure 4.
Comparative analysis of the relative volumes of Xist RNA domain and X-chromosome territory during X inactivation. Xist RNA FISH (red) combined with an X-chromosome paint DNA FISH (green) on female ES cells undergoing differentiation (days 2 and 4) and MEFs. 3D analysis using Amira software reconstruction (n = 30). DNA is stained with DAPI (gray or blue). Bar, 5 μm. Relative volumes were calculated using Amira software and are shown on the right of each panel.
Gene relocation into the X-chromosome territory is dependent on the silencing A-repeat domain of the Xist transcript
The above results suggested that relocation of X-linked genes into the Xist compartment might be linked to their silencing. In order to test this hypothesis and to determine whether the silencing activity of Xist RNA is linked to its capacity to exclude the transcription machinery and/or to cause internalization of genes, we analyzed a male ES cell line (30–24–46) in which the endogenous Xist gene carries a deletion of its A-repeats (XistΔA) and is under the control of an inducible tetracycline-responsive promoter (Wutz et al. 2002). The XistΔA RNA mutant cannot induce gene silencing but can induce correct coating in cis, and histone modifications such as H3K27me3, as well as macroH2A, are correctly recruited (Wutz et al. 2002; Kohlmaier et al. 2004). This thus provides a useful means of distinguishing between changes due to Xist RNA silencing and those due to chromatin modulation.
We first analyzed the profile of RNA Pol II and Cot-1 RNA on the Xist RNA-coated chromosome in these cells following induction of differentiation and expression of XistΔA for 2 d. Although it is not known if the XistΔA RNA associates with exactly the same chromosomal regions as wild-type Xist RNA, the accumulation of XistΔA RNA in interphase nuclei was indistinguishable from wild-type XX ES cells (Fig. 5D). We were, however, very surprised to find that RNA Pol II becomes excluded from the XistΔA RNA-coated chromosome early on after induction of XistΔA expression and that this is accompanied by silencing of repeat-containing transcription detected by Cot-1 RNA FISH (Fig. 5A). Indeed, at day 1, >90% of cells with a XistΔA RNA-coated X chromosome showed exclusion of RNA Pol II and 70% showed repression of transcripts detected by Cot-1 DNA within the XistΔA RNA domain. The nuclear patterns of RNA Pol II and Cot-1 RNA depletion at the XistΔA RNA domain were indistinguishable from those found in wild-type female ES cells (see line scan in Fig. 5D). The more rapid kinetics of RNA Pol II exclusion and Cot-1 repression found in XistΔA cells following 1 d of induction, compared with female ES cells differentiated for the same time, is due to the rapid expression of high levels of XistΔA RNA owing to the strong inducible promoter (similar kinetics were found with other inducible Xist transgene lines) (data not shown). Indeed other changes, including histone modifications, appear more rapidly in such inducible Xist lines (ΔA or wild type) (Kohlmaier et al. 2004; data not shown). Our findings show that the absence of the A-repeats does not appear to impair Xist RNA's capacity to form a silent nuclear compartment and to induce silencing of at least some X-linked transcription (the Cot-1 fraction), even though X-linked genes are not subject to silencing by XistΔA RNA in this cell line (Wutz et al. 2002).
Figure 5.
Role of Xist RNA A-repeats using an inducible Xist gene. (A) RNA Pol II immunostaining (blue) combined with Cot-1 RNA FISH (red) and Xist RNA FISH (green) in 30–24–46 male ES cells after 2 d of differentiation and Doxicyclin-induction of XistΔA RNA transcription. DNA is stained with DAPI (gray). Bar, 5 μm. A line scan shows the relative intensities of the three signals across the nucleus. The large and the small peaks of Xist RNA likely correspond to the bulk of the X chromosome and the transcribed Xist locus itself, respectively. (B) Dual RNA FISH for MeCP2 RNA (red) and Xist RNA (green) following 1 d of differentiation and XistΔA expression. DNA is stained with DAPI (gray). Bar, 5 μm. (C) Representative 3D analysis of MeCP2 DNA FISH (red) combined with a Xist RNA FISH (green) using Metamorph software and Amira reconstruction, after 5 d of differentiation and XistΔA expression. DNA is stained with DAPI (gray). Bar, 5 μm. (D) Distribution of the different profiles of MeCP2, G6pdx, Lamp2, and Pgk1 locations with respect to the Xist RNA domain (based on the categories described in Fig. 3) after 1 or 5 d of differentiation and XistΔA expression (n = 30). No statistical differences in position distributions between days 1 and 5 are found.
We went on to analyze X-linked gene expression and localization with respect to the XistΔA RNA-coated X chromosome, in this cell line. Cells were analyzed at days 1 and 5 of differentiation and XistΔA induction (Fig. 5). Even after5dof XistΔA induction and differentiation, MeCP2, G6pdx, and Lamp2 all remained active (Fig. 5B), as previously shown for Pgk1 (Wutz et al. 2002). We next examined the positions of X-linked genes in this cell line. In the case of Pgk1, this locus remained outside the XistΔA RNA domain in ~75% of nuclei after both 1 and 5 d of induction (with no statistically significant change in distribution), in striking contrast to wild-type female cells where it becomes very internally located in the majority (85%) of nuclei even at day 4 of differentiation. Similar results were found for the Mecp2, G6pdx, and Lamp2 genes (Fig. 5C).
In summary, we can conclude that the A-repeats of the Xist transcript are indeed necessary for silencing of X-linked genes (Wutz et al. 2002). We have further demonstrated that the Xist A-repeats, or their silencing function, are required for the relocation of X-linked genes into the Xist RNA compartment during X inactivation. Finally, we have also made the surprising and novel finding that the silent compartment associated with the Xist RNA domain is formed independently of the Xist A-repeats. This raises the possibility of further silencing roles mediated by different regions of the Xist transcript.
Discussion
Xist RNA forms a silent nuclear compartment
In this study we have examined the events underlying the onset of X inactivation in the context of the 3D organization of the X chromosome. We have shown that Xist RNA chromosome coating leads to the rapid exclusion of RNA Pol II and associated transcription factors and that this represents the earliest event following Xist RNA accumulation described so far. Only subsequently do genes cease to be transcribed, as detected at the primary transcript level by RNA FISH. The Xist gene is essential for silencing of chromatin in cis and this repressive function was thought to be solely dependent on the A-repeat region of the Xist transcript (Wutz et al. 2002). However, we have made the surprising finding that Xist RNA is able to trigger the exclusion of RNA Pol II and the repression of repeat sequence transcription within the Xist RNA domain, even in the absence of the A-repeat silencing region of Xist. Nevertheless, this transcript is deficient in its capacity to trigger X inactivation at the level of all the X-linked genes so far examined. Our study thus provides evidence for a new and early step in the X-inactivation process and for a novel role for the Xist transcript. This new function is independent of the A-repeats and results in the formation of a silent nuclear compartment with exclusion of the transcription machinery from the X chromosome chosen to be inactivated.
Previous studies have suggested that Xist RNA may associate with components of the nuclear scaffold or matrix, such as SAF-A, in order to form a stable architectural structure (Clemson et al. 1996; Helbing and Fackelmayer 2003) which could be, as shown here, responsible for the early exclusion of the transcription machinery and, thus, for the early transcriptional repression of sequences at the interior of the X-chromosome territory. It will be interesting now to assess to what extent the Xist RNA domain might represent a true physical compartment, using accessibility assays such as the injection of dextran beads of different sizes (Verschure et al. 2003). Other candidates that might participate in the formation of a repressive compartment at the level of the inactive X chromosome are the Polycomb group proteins. Polycomb group complexes are known to be involved in silencing and in the creation of repressive environments (Bantignies et al. 2003), thus the recruitment of PRC2 and PRC1 complexes by Xist RNA might play a similar role. However, the exclusion of the transcription machinery from the Xist RNA domain occurs well prior to the enrichment of H3K27 trimethylation triggered by PRC2 (Plath et al. 2003; Silva et al. 2003). This is consistent with our previous study in embryos (Okamoto et al. 2004). Even if recruitment of PRC2 and PRC1 is not directly involved in creating such a silent compartment, they may nevertheless play a role in stabilizing a repressive environment which facilitates the maintenance of silencing. Indeed, PRC2 recruitment is also Xist A-repeat-independent and H3K27me3 is not critical for gene silencing, but rather participates in the maintenance of the inactive state (Wang et al.2001; Plath et al. 2003; Silva et al. 2003; Kohlmaier et al. 2004).
Genes become relocated into the Xist RNA domain during X inactivation
Several studies have shown that gene-rich regions tend to be located at the periphery of chromosome territories while repetitive regions tend to be located more internally (Mahy et al. 2002b). Indeed, this also seems to be the case for the human X chromosome (Clemson et al. 2006). Consistent with this, we find that the silent compartment formed after Xist RNA accumulation initially contains more repeat-rich regions, as detected by Cot-1. X-linked genes, on the other hand, tend to be located outside or at the periphery of this Xist RNA domain when still active. It should be noted that all of the X-linked genes we have examined so far tend to be fairly peripherally located on the X-chromosome territory, whatever their activity status, as in the study by Clemson et al. (2006). Indeed, the shift from the outside toward the inside of the Xist RNA domain that we describe during X inactivation, although statistically significant, is only in the order of 0.1–0.8 μm (data not shown). It will be interesting to identify genes that are more internally located on the X-chromosome territory, in order to assess their kinetics of inactivation. We predict that such genes, if they exist, might be subject to silencing in a Xist A-repeat-independent fashion.
Our work also provides new insights into the function of the Xist A-repeats, as we show that the shift of genes to a more internal location within the RNA Pol II-depleted Xist RNA domain is linked to Xist A-repeat-mediated gene silencing, as it does not occur in the XistΔA mutant. We have demonstrated that this shift in position of genes does not appear to be due to an increase in the size of the Xist RNA domain, nor to a global condensation of the X chromosome. Rather, genes become relocated into the RNA Pol II-depleted Xist RNA compartment when they become silenced, thus demonstrating that there is an intimate link between gene silencing and location within a chromosomal territory during X inactivation. Several studies have previously reported dynamic changes in the location of genes with respect to their chromosome territory, with looping out of genes being correlated with the activation of expression (Volpi et al. 2000; Chambeyron and Bickmore 2004a,b). Our study provides the first evidence in a developmental context for dynamic changes in gene position that are associated with the X-inactivation process.
The gene relocation that accompanies X inactivation is dependent on the A-repeat silencing function of the Xist transcript. How could this occur? Given that the movement of genes toward the interior of the transcriptionally inert Xist RNA domain does not precede gene silencing, but rather accompanies or follows it, a likely explanation is that the transcriptional repression induced by the Xist A-repeat containing RNA results in the capacity of genes to become internalized. Indeed, actively transcribed genes may be located or even sequestered in putative “transcription factories” (see Osborne et al. 2004). When Xist A-repeat-induced silencing occurs, a gene may become unleashed from such a transcription compartment and thus become more internally located by default. An alternative but not mutually exclusive explanation for gene relocation could be that the Xist transcript and the ribonucleoprotein structure it forms participate in “reeling” genes into the silent domain, either through local chromatin condensation or through active translocation. As PRC2-associated H3K27 trimethylation, in the absence of silencing (in the XistΔA mutant), does not induce internalization, this chromatin change alone is not sufficient for gene internalization. However, this does not exclude that it might be necessary for relocation to occur. Examination of PRC2 mutant female ES cells will be required to address this issue.
Finally, our findings with the Jarid1c gene also provide potential insights into the process of escape from X inactivation. The fact that the Jarid1c gene remains on the outside or at the edge of the Xist RNA domain at all differentiation stages may reflect a resistance of this locus to be internalized, despite the silencing action of the Xist A-repeats. This would be consistent with the recent finding that Jarid1c is flanked by CTCF boundary elements, which could render it more resistant to relocation compared with other genes (Filippova et al. 2005).
Conclusion: multiple roles for the Xist transcript?
In conclusion, we provide novel and unexpected insights into the multiple roles of Xist (Fig. 6). Previous studies have suggested that Xist RNA might be multivalent, not just in the initiation of transcriptional silencing but also in the early maintenance of the silent state, through the action of histone modifying enzymes and PRC2/PRC1 complexes, which are important for the memory and the maintenance of inactivation and could lead to the Xist-independent locking-in of the inactive state (Wang et al. 2001; Mak et al. 2002; Plath et al. 2003, 2004; Silva et al. 2003; de Napoles et al. 2004; Kohlmaier et al. 2004; Hernandez-Nunoz et al. 2005). In the present study we have shown that the accumulation of the Xist transcript appears to be capable of triggering the rapid formation of a silent nuclear compartment. However, we have also shown that this is not in itself sufficient to induce silencing of all X-linked genes, as it can occur independently of the A-repeats. The exact nature of the sequences present within this compartment is currently under investigation. On the other hand, Xist A-repeat RNA can induce the silencing of genes, and we demonstrate that these genes then become relocated within the silent nuclear compartment delineated by Xist RNA.
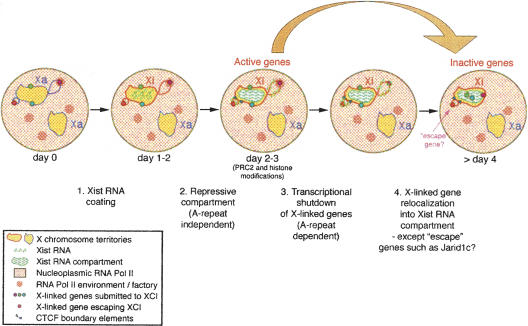
Figure 6.
Model for Xist RNA-mediated X-chromosome inactivation. In undifferentiated female ES cells, both X chromosomes are active (yellow). Only genes on the X chromosome undergoing inactivation (Xi) are depicted. (1) During early differentiation, Xist RNA starts to accumulate on the X chromosome chosen for inactivation. (2) The formation of this silent compartment is associated with the exclusion of the transcription machinery and the repression of internal repeat-rich regions. This step is independent of Xist A-repeats. (3) X-linked gene silencing is triggered via a Xist A-repeat-dependent mechanism, and histone modifications as well as PRC2/PRC1 accumulation (Xist A-repeat-independent) are observed. (4) Genes then become relocated into the Xist RNA compartment by default because they are no longer constrained by their requirement to be transcribed in an RNA Pol II-enriched environment (or potential transcription factory, see Osborne et al. 2004), and/or by an active mechanism of relocation, which may be dependent on the Xist A-repeats. This locus internalization may help maintain the silent state. Genes escaping inactivation, such as Jarid1c, have special elements (such as CTCF boundaries) that may prevent efficient internalization. Their location outside of the Xist repressive compartment could participate in, or facilitate, their reactivation.
In the future it will be important to define the exact mechanism of action of the Xist A-repeat sequence that triggers gene silencing and relocation. It will also be important to define which region of the Xist transcript is involved in the formation of the RNA Pol II-depleted nuclear compartment and to analyze the exact nature of this domain. Finally, we would like to determine whether only X-linked genes become silenced within this Xist RNA compartment, or whether it could be the host to other stably silenced sequences in the genome.
Materials and methods
Cell culture
Female MEFs, the LF2 female ES cell line (a gift from Dr. Austin Smith), and the 30–24–46 male ES cell line (Wutz et al. 2002) were prepared and cultured as described previously (Chaumeil et al. 2002, 2004). In the LF2 cell line, ~2%–5% of cells show a Xist RNA domain after 1 d of differentiation, 10% after 2 d, and 60% after 4 d. In the 30–24–46 cell line, Xist expression was induced by adding 1 μg/mL doxycycline in the culture medium. Around 30%–40% of cells show a Xist RNA domain after 1 d of induction, and 80% after 5 d. All cells were grown at 37°Cin8% CO2, and medium was changed daily.
Immunofluorescence and RNA FISH analysis
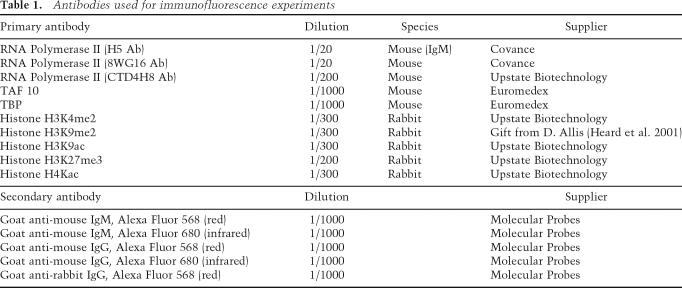
The antibodies and dilutions used are listed in Table 1.
Table 1.
Antibodies used for immunofluorescence experiments
Fixation and permeabilization conditions for immunofluorescence combined with RNA FISH were performed as described previously (Chaumeil et al. 2002, 2004). Note that in these experiments, cells were fixed and then permeabilized for 5 min. DNA probes for RNA and DNA FISH were a genomic λ clone (510) to detect Xist, overlapping λ clones (IIIC2 and IG10) to detect Chic1, and a plasmid probe to detect MeCP2 (Heard et al. 2001). Jarid1c, G6pdx, and Lamp2 were detected using genomic BAC probes (RP24-148H21 [Okamoto et al. 2005]; RP2313D21; RP24173A8). Xic DNA FISH was performed using YAC PA-2 (Heard et al. 1999). Pgk1 was detected using a cosmid probe. Mouse Cot-1 DNA (Invitrogen) was used for the Cot-1 RNA FISH assay. The Xist probe was labeled with SpectrumGreendUTP (Vysis), while X-linked genes (Chic1, MeCP2, and Jarid1c) and Cot-1 probes were labeled with SpectrumRed-dUTP. Histone modifications were detected using Alexa Fluor 568 highly cross-adsorbed secondary antibody (red; Molecular Probes), while RNA Pol II and transcription factors were detected using highly cross-adsorbed secondary antibodies conjugated to Alexa Fluor 568 or 680 (red or infrared; Molecular Probes), depending on the experiment.
RNA FISH and RNA/DNA FISH analysis
ES cells and MEFs cultured on coverslips were fixed in 3% paraformaldehyde for 10 min, permeabilized in 1× PBS/0.5% Triton X-100/2 mM Vanadyl Ribonucleoside Complex (Biolabs) on ice for 7 min, and progressively dehydrated in ethanol. For dual RNA FISH experiments, cells were then hybridized with probes overnight at 37°C in a dark and humid chamber. DNA probes detecting Xist RNA and X-linked genes primary transcripts were prepared as mentioned above. After three washes in 50% formamide/2× SSC, three washes in 2× SSC at 42°C, and DAPI counterstaining, coverslips were mounted on slides and visualized under fluorescent microscope. For RNA/DNA FISH experiments, cells were denaturated in 50% formamide/2× SSC at 80°C for 32–40 min (depending on cells) and rinsed several times in ice-cold 2× SSC prior to overnight hybridization with probes at 42°C. X-linked gene DNA FISH probes were denatured, competed with Cot-1 DNA (5 μg/coverslip) for 1 h at 37°C, and combined with Xist RNA FISH probe just prior to hybridization. Preparation of the X-chromosome paint probe was performed according to the supplier's instructions (Cambio). All of these protocols are detailed on the Epigenome Network of Excellence Web site (http://www.epigenome-noe.net/ researchtools/protocol.php?protid=3).
Microscopy and image analysis
Two-dimensional acquisitions were made with an upright DMR microscope (Leica) equipped with a CoolSNAP FX camera (Roper Scientific) and a 63× objective (NA 1.32). Images were analyzed using Metamorph software (Universal Imaging). For each time point, 100 nuclei were analyzed, except for day 1 (n = 60). 3D images were acquired with a DMRA2 microscope (Leica) equiped with a piezzo motor and a CoolSNAP HQ camera (Roper) and piloted by Metamorph software. Using a 100× objective (NA 1.4), images of ~50 optical sections separated by
0.2 μm were acquired at different λs (DAPI [360/40, 470/40], SpectrumGreen and Alexa 488 [470/40, 525/50], spectrumRed and Alexa 568 [545/30, 610/75], and Alexa 680 [620/60, 700/75]). For each time point, 30 nuclei were analyzed. After deconvolution of 3D data (iterative-constrained algorithm implemented in Metamorph software, Universal Imaging), line scans drawn across the nucleus (SoftWorx software, Applied Precision) were used to measure the relative intensities of fluorescence signals obtained by Cot-1 RNA FISH, histone modifications, or RNA Pol II (immunofluorescence) relative to the Xist RNA domain (RNA FISH). Positions of genes (red signal) relative to the Xist RNA domain (green signal) were determined through the analysis of 3D data sets using Amira software (Mercury-TGS). For each combination of Xist RNA FISH and gene RNA or DNA FISH, a segmentation threshold was determined and applied to the 30 nuclei analyzed at each stage of differentiation. Positions were classified in five categories: outside (no juxtaposition or overlap of red and green voxels; d ≥ 0.2 μm), outer edge (juxta-position but no overlap of red and green voxels; d ≤ 0.2 μm), edge (some overlap of red and green voxels), inner edge (juxta-position and complete overlap of red and green voxels; d ≤ 0.2 μm), and inside (complete overlap but no juxtaposition of red and green voxels; d ≥ 0.2 μm).
The statistical significance of the changes in gene location during differentiation was assessed using a χ2 test where p values < 0.05 were taken to be significant.
The volumes of X-chromosome territories and Xist RNA domains were measured in voxels using Amira software. A segmentation threshold was determined for both. Relative volumes (Xist RNA domain/X territory) were calculated on 30 cells for each time point.
Acknowledgments
We thank C. Disteche for providing the Jarid1c BAC; C.D. Allis, D. Reinberg, and D. Spector for helpful discussions and for antibodies; C. Bacher and R. Eils for helpful discussions; C. Patrat for providing gene-specific BAC probes; S. Huart for informatics assistance; G. Filion for help with statistical analysis; and members of the Heard team and the UMR 218 laboratory for stimulating discussions. J.C. was supported by the French Ministry of Research and by La Ligue Nationale contre le Cancer. E.H. and A.W. were funded by the Epigenome Network of Excellence; funding to E.H. was also provided by the HFSP, the Schlumberger Foundation, the EU HEROIC IP, and the Canceropole (IDF).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.380906.
References
- Avner P., Heard E. X-chromosome inactivation: Counting, choice and initiation. Nat. Rev. Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- Bantignies F., Grimaud C., Lavrov S., Gabut M., Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes & Dev. 2003;17:2406–2420. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs B.A., Connors B., Sobel R.E., Chinault A.C., Allis C.D. Reduced levels of histone H3 acetylation on the inactive X chromosome in human females. Chromosoma. 1996;105:303–309. doi: 10.1007/BF02524648. [DOI] [PubMed] [Google Scholar]
- Boggs B.A., Cheung P., Heard E., Spector D.L., Chinault A.C., Allis C.D. Differentially methylated forms of his-tone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Hendrich B.D., Rupert J.L., Lafreniere R.G., Xing Y., Lawrence J., Willard H.F. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Chadwick B.P., Willard H.F. Chromatin of the Barr body: Histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum. Mol. Genet. 2003;12:2167–2178. doi: 10.1093/hmg/ddg229. [DOI] [PubMed] [Google Scholar]
- Chambeyron S., Bickmore W.A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes & Dev. 2004a;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———2004bDoes looping and clustering in the nucleus regulate gene expression? Curr. Opin. Cell Biol. 16256–262. [DOI] [PubMed] [Google Scholar]
- Chaumeil J., Okamoto I., Guggiari M., Heard E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet. Genome Res. 2002;99:75–84. doi: 10.1159/000071577. [DOI] [PubMed] [Google Scholar]
- Chaumeil J., Okamoto I., Heard E. X-chromosome inactivation in mouse embryonic stem cells: Analysis of his-tone modifications and transcriptional activity using immunofluorescence and FISH. Methods Enzymol. 2004;376:405–419. doi: 10.1016/S0076-6879(03)76027-3. [DOI] [PubMed] [Google Scholar]
- Clemson C.M., McNeil J.A., Willard H.F., Lawrence J.B. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/ chromosome structure. J. Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C.M., Hall L.L., Byron M., McNeil J., Lawrence J.B. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl. Acad. Sci. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C., Stein P., Worrad D.M., Schultz R.M., Pehrson J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development. 2000;127:2283–2289. doi: 10.1242/dev.127.11.2283. [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Czankovszki G., Nagy A., Jaenisch R. Synergism of Xist RNA, DNA methylation and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 2001;153:773–783. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M., Mermoud J.E., Wakao R., Tang Y.A., Endoh M., Appanah R., Nesterova T.B., Silva J., Otte A.P., Vidal M., et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dietzel S., Schiebel K., Little G., Edelmann P., Rappold G.A., Eils R., Cremer C., Cremer T. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity. Exp. Cell Res. 1999;252:363–375. doi: 10.1006/excr.1999.4635. [DOI] [PubMed] [Google Scholar]
- Fackelmayer F.O. A stable proteinaceous structure in the territory of inactive X chromosomes. J. Biol. Chem. 2005;280:1720–1723. doi: 10.1074/jbc.C400531200. [DOI] [PubMed] [Google Scholar]
- Filippova G.N., Cheng M.K., Moore J.M., Truong J.-P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Hall L.L., Byron M., Sakai K., Carrel L., Willard H.F., Lawrence J.B. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc. Natl. Acad. Sci. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 2004;16:247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Heard E., Mongelard F., Arnaud D., Avner P. Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol. Cell Biol. 1999;19:3156–3166. doi: 10.1128/mcb.19.4.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Rougeulle C., Arnaud D., Avner P., Allis C.D., Spector D.L. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:1–20. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Helbing R., Fackelmayer F.O. Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma. 2003;112:173–182. doi: 10.1007/s00412-003-0258-0. [DOI] [PubMed] [Google Scholar]
- Hernandez-Nunoz I., Lund A.H., van der Stoop P., Boutsma E., Muijrers I., Verhoeven E., Nusimow D.A., Panning B., Marahrens Y., van Lohuizen M. Stable X chromosome inactivation involves the PRC1 polycomb complex and requires histone MACROH2A1 and the CULLIN3/ SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P., Turner B.M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- Keohane A.M., O'Neill L.P., Belyaev N.D., Lavender J.S., Turner B.M. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev. Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Kohlmaier A., Savarese F., Lachner M., Martens J., Jenuwein T., Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:991–1003. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.B., Singer R.H. Quantitative analysis of in situ hybridization methods for the detection of actin gene expression. Nucleic Acids Res. 1985;13:1777–1799. doi: 10.1093/nar/13.5.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Mahy N.L., Perry P.E., Gilchrist S., Baldock R.A., Bick-more W.A. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 2002a;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy N.L., Perry P.E., Bickmore W.A. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 2002b;159:753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak W., Baxter J., Silva J., Newall A.E., Otte A.P., Brock-dorff N. Mitotically stable association of Polycomb group proteins Eed and Enx1 with the inactive X chromosome in trophoblast stem cells. Curr. Biol. 2002;12:1016–1020. doi: 10.1016/s0960-9822(02)00892-8. [DOI] [PubMed] [Google Scholar]
- Marahrens Y., Loring J., Jaenisch R. Role of the Xist gene in X chromosome choosing. Cell. 1998;92:657–664. doi: 10.1016/s0092-8674(00)81133-2. [DOI] [PubMed] [Google Scholar]
- Mermoud J.E., Costanzi C., Pehrson J.R., Brockdorff N. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J. Cell Biol. 1999;147:1399–1408. doi: 10.1083/jcb.147.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud J.E., Popova B., Peters A.H.F.M., Jenuwein T., Brockdorff N. Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr. Biol. 2002;12:247–251. doi: 10.1016/s0960-9822(02)00660-7. [DOI] [PubMed] [Google Scholar]
- Norris D.P., Brockdorff N., Rastan S. Methylation status of CpG-rich islands on active and inactive mouse X chomosomes. Mamm. Genome. 1991;1:78–83. doi: 10.1007/BF02443782. [DOI] [PubMed] [Google Scholar]
- Okamoto I., Otte A.P., Allis C.D., Reinberg D., Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Okamoto I., Arnaud D., Le Baccon P., Otte A.P., Disteche C.M., Avner P., Heard E. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature. 2005;438:369–373. doi: 10.1038/nature04155. [DOI] [PubMed] [Google Scholar]
- Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Panning B., Dausman J., Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brock-dorff N. The Xist gene is required in cis for X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Peters A.H.F.M., Mermoud J.E., O'Carroll D., Pagani M., Schweizer D., Brockdorff N., Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- Plath K., Mlynarczyk-Evans S., Nusinow D.A., Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., de la Cruz C.C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Plath K., Talbot D., Hamer K.M., Otte A.P., Yang T.P., Jaenisch R., Panning B. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J. Cell Biol. 2004;167:1025–1035. doi: 10.1083/jcb.200409026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C., Chaumeil J., Sarma K., Allis C.D., Reinberg D., Avner P., Heard E. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol. Cell. Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Mak W., Zvetkova I., Appanah R., Nesterova T.B., Webster Z., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed–Enx1 polycomb group complexes. Dev. Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Simmler M.C., Heard E., Rougeulle C., Cruaud C., Weissen-bach J., Avner P. Localization and expression analysis of a novel conserved brain expressed transcript, Brx/ BRX, lying within the Xic/XIC candidate region. Mamm. Genome. 1997;8:760–766. doi: 10.1007/s003359900561. [DOI] [PubMed] [Google Scholar]
- Takagi N., Sugawara O., Sasaki M. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- Tsuchiya K.D., Greally J.M., Yi Y., Noel K.P., Truong J.P., Disteche C.M. Comparative sequence and X-in-activation analyses of a domain of escape in human xp11.2 and the conserved segment in mouse. Genome Res. 2004;14:1275–1284. doi: 10.1101/gr.2575904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschure P.J., van der Kraan I., Manders E.M., Hoogstraten D., Houtsmuller A.B., van Driel R. Condensed chromatin domains in the mammalian nucleus are accessible to large macromolecules. EMBO Rep. 2003;4:861–866. doi: 10.1038/sj.embor.embor922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi E.V., Chevet E., Jones T., Vatcheva R., Williamson J., Beck S., Campbell R.D., Goldsworthy M., Powis S.H., Ragoussis J., et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000;113:1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Wang J., Mager J., Chen Y., Schneider E., Cross J.C., Nagy A., Magnusson T. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- Williams R. Transcription and territory: The ins and outs of gene positioning. Trends Genet. 2003;19:298–302. doi: 10.1016/S0168-9525(03)00109-4. [DOI] [PubMed] [Google Scholar]
- Wutz A., Jaenisch R.A. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Wutz A., Rasmussen T.P., Jaenisch R.A. Chromosomal silencing and chromosomal localization are mediated by different domains of Xist RNA. Nat. Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]