Abstract
The mechanisms of RNA replication of plus-strand RNA viruses are still unclear. Here, we identified the first promoter element for RNA synthesis described in a flavivirus. Using dengue virus as a model, we found that the viral RdRp discriminates the viral RNA by specific recognition of a 5′ element named SLA. We demonstrated that RNA–RNA interactions between 5′ and 3′ end sequences of the viral genome enhance dengue virus RNA synthesis only in the presence of an intact SLA. We propose a novel mechanism for minus-strand RNA synthesis in which the viral polymerase binds SLA at the 5′ end of the genome and reaches the site of initiation at the 3′ end via long-range RNA–RNA interactions. These findings provide an explanation for the strict requirement of dengue virus genome cyclization during viral replication.
Keywords: Flavivirus, RNA-dependent RNA polymerase, RNA cyclization, viral RNA synthesis, AFM
Plus-strand RNA viruses cause numerous human, animal, and plant diseases, including hepatitis, encephalitis, and hemorrhagic fevers. Following viral infection, a rapid amplification of the viral genome from one single molecule to tens of thousands of copies occurs in only a few hours, which could lead to cell damage and disease. This process is catalyzed by viral-encoded RNA-dependent RNA polymerases (RdRps) in combination with other viral and cellular factors. Viral RdRps efficiently amplify viral RNAs but do not copy cellular mRNAs and, therefore, must discriminate these RNA molecules in the cytoplasm of the infected cell.
After viral entry, the genome of plus-strand RNA viruses directs the synthesis of viral proteins, and, once the viral polymerase and other essential proteins are synthesized, the viral RNA is copied, starting from the 3′ end, to generate a complementary minus-strand RNA that, in turn, is transcribed into new molecules of plus strand. RNA elements that regulate the level, polarity, and timing of RNA synthesis are present within both the minus-and plus-strand templates. These elements include promoters that bind the viral polymerase, as well as enhancers and repressors that modulate polymerase access to the sites of initiation (Song and Simon 1995; Dreher 1999; Sullivan and Ahlquist 1999; Olsthoorn and Bol 2002; Pogany et al. 2003; Ranjith-Kumar et al. 2003; Ray and White 2003; Grdzelishvili et al. 2005). Promoters are generally found at the 3′ termini in both the plus-and minus-strand RNAs. However, cis-acting RNA structures have been also found at the 5′ end or within the coding region of viral genomes. In the case of alphaviruses, a promoter element essential for minus-strand RNA synthesis was found at the viral 5′ UTR (Niesters and Strauss 1990; Frolov et al. 2001). Furthermore, a cloverleaf structure present at the 5′ end of the poliovirus genome was shown to be essential for both plus-and minus-strand RNA synthesis (Andino et al. 1990; Gamarnik and Andino 1998; Herold and Andino 2001), while a cis-acting replication element (CRE) located in the coding region of the viral RNA serves as a template for Vpg uridylation, which is a primer for genome replication (McKnight and Lemon 1998; Paul et al. 2000; Rieder et al. 2000). Therefore, even though plus-strand RNA viruses share fundamental similarities in the RNA replication process, these viruses have evolved an array of mechanisms to ensure efficient and specific genome amplification. How the viral polymerases discriminate viral from cellular RNAs and where the signals are that modulate RdRp activity are questions that remain largely unanswered for many medically important plus-strand RNA viruses. Here, we use dengue virus (DV), an important human pathogen, as a model to investigate the mechanism of specific viral RNA amplification.
DV is a member of the Flaviviridae family together with yellow fever (YFV), West Nile (WNV), Japanese encephalitis (JEV), and hepatitis C (HCV). DV infections can produce clinical illness ranging from dengue fever (DF), a nonspecific flu-like syndrome, to a severe and sometimes fatal dengue hemorrhagic fever (DHF) (Gubler 1998). The World Health Organization continues reporting outbreaks of severe forms of the disease in the Americas and Asia. Despite the wide morbidity and mortality associated with dengue infections, neither specific anti-viral therapies nor a licensed vaccine exist.
The genome of dengue and other flaviviruses is ∼11 kb long and encodes one open reading frame flanked by 5′ and 3′ UTRs (Rice 2001). The ∼100-nucleotide (nt)-long 5′ UTR of DV shows almost complete sequence conservation among different serotypes. Specific deletions within this region in the context of DV4 showed an important role for the 5′ UTR during viral replication (Ca-hour et al. 1995). The ∼450-nt DV 3′ UTR lacks a poly(A) tail but ends in a very conserved 3′ stem–loop (3′SL). A detailed structure–function analysis of the 3′SL in many flaviviruses revealed that it is absolutely required for viral replication (Brinton et al. 1986; Men et al. 1996; Proutski et al. 1997; Rauscher et al. 1997; Zeng et al. 1998; Elghonemy et al. 2005; Tilgner et al. 2005; Yu and Markoff 2005). Another essential RNA element for viral replication, the conserved sequence CS1, is present upstream of the 3′SL (Men et al. 1996). This element contains the cyclization sequence CS that is complementary to a sequence at the 5′ end present in all mosquito-borne flavivirus genomes (Hahn et al. 1987). 5′–3′ long-range RNA–RNA interactions have been proposed to be necessary for RNA replication of DV, WNV, Kunjin virus, and YFV (Khromykh et al. 2001; Corver et al. 2003; Lo et al. 2003; Alvarez et al. 2005b). Interestingly, de novo RNA synthesis with recombinant DV RdRp is greatly enhanced when the RNA templates contain the 5′ and 3′CS complementary sequences (You and Padmanabhan 1999; You et al. 2001). More recently (Alvarez et al. 2005b), we identified a second pair of complementary sequences named 5′–3′UAR that is required for DV genome cyclization and viral replication. Even though cis-acting elements have been identified to be essential for flavivirus RNA synthesis, the mechanisms by which these RNA structures participate in the viral process remain unclear.
Here, we discovered an RNA element that recruits the viral RdRp at the 5′ end of the DV genome and promotes RNA synthesis at the 3′ end through long-range RNA– RNA interactions. Using infectious DV RNAs and viral subgenomic replicons, we demonstrated an essential role of this RNA element during viral RNA amplification in infected cells. We propose the first mechanism for minus-strand RNA synthesis described in a flavivirus, in which the promoter element functions from the 5′ end of a circular viral genome. Because similar 5′–3′ long-range interactions were observed in many viral RNA genomes, the mechanism proposed for DV may represent a widespread strategy for viral RNA replication.
Results
An RNA element present at the 5′ end of DV genome promotes specific RNA synthesis
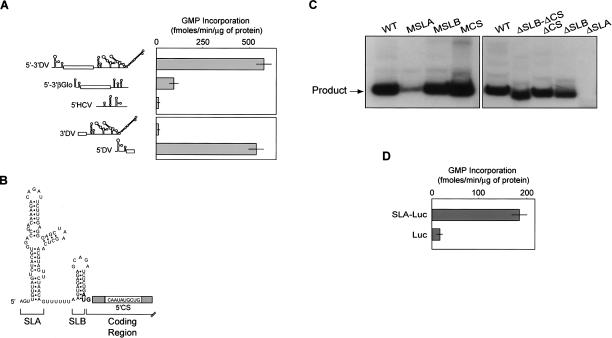
To gain insight into RNA recognition by DV RdRp, we first characterized the RNA synthesis activity of a recombinant protein corresponding to the polymerase domain of the viral protein NS5 (amino acids 270–901, NS5pol). Using a poly(C) RNA as template, the recombinant NS5pol showed high RNA synthesis activity, in agreement with previous reports (Selisko et al. 2006). RNA homopolymers, like poly(C), are efficient but non-specific templates for many viral RdRps. Thus, we analyzed whether NS5pol could discriminate secondary or tertiary structures present in natural RNAs. To this end, we compared RNA synthesis by the viral polymerase using different templates: (1) a subgenomic DV RNA, which encodes for the viral structural proteins flanked by the complete 5′ and 3′ viral UTRs (5′–3′DV RNA); (2) a cellular mRNA (βglobin mRNA); and (3) an unrelated viral RNA (5′HCV RNA). We observed that 5′–3′DV RNA was 10′ and 50′ fold more active as template for RNA synthesis than the cellular or the unrelated viral RNAs, respectively (Fig. 1A). To identify viral RNA elements recognized by NS5pol, we generated two RNA molecules carrying the 5′-terminal or the 3′-terminal sequences of the 5′–3′DV RNA molecule. We observed a remarkable preference (100′ fold) for the template carrying the 5′-terminal sequences (5′DV RNA) than for the RNA template containing the 3′ end sequences (Fig. 1A).
Figure 1.
RNA discrimination by NS5pol is mediated by an RNA structure present at the 5′ end of the DV genome. (A) In vitro RdRp activity of the viral NS5pol using cellular and viral RNA templates. Schematic representations of the RNAs used are indicated on the left: (5′–3′DV) Subgenomic RNA encoding DV structural proteins flanked by the viral 5′ and 3′ UTRs, (5′–3′βGlo) mRNA encoding βglobin, (5′HCV) 220′ nt RNA of HCV 5′ UTR, (3′DV) 454′ nt DV 3′ UTR, and (5′DV) 5′-end 160 nt of DV genome. RNA polymerase activity was carried out as described in Materials and Methods. (B) Schematic representation of the predicted secondary structure of an RNA encompassing the first 100 nt of the DV genome obtained by Mfold algorithm. Stem–loop A (SLA), stem–loop B (SLB), and the complementary sequence (5′CS) within the viral coding sequence are indicated. (C) Requirement of SLA for NS5pol activity in vitro. Analysis of radiolabeled RNA products in denaturing 5% polyacrylamide gels synthesized by NS5pol. 5′DV RNAs with mutations within SLA, SLB, and CS (MSLA, MSLB, and MCS) or deletions of the complete structures (ΔSLA, ΔSLB, ΔCS, and ΔSLB-CS) were used at 300 nM as templates. (D) In vitro RNA synthesis by DV NS5pol using unrelated RNA templates encoding firefly luciferase preceded or not by DV SLA (SLA-Luc and Luc, respectively). RNA polymerase activity was expressed as described in A.
To more precisely map the RNA elements responsible for the high 5′DV template activity, we made a set of deletions and mutations within this molecule. The predicted 5′DV RNA secondary structure, which is highly conserved among different flaviviruses, consists of a large stem–loop (SLA) that includes a side loop and a second short stem–loop (SLB) terminating in the translation initiator AUG (Fig. 1B). The 5′CS sequence, which is complementary to the 3′CS present at the 3′ end of the viral genome, is present within the coding sequence of the capsid protein following the AUG. We performed deletions and mutations of each of these 5′ elements (SLA, SLB, and 5′CS) and tested the template activity by quantifying the radiolabeled RNA products (Fig. 1C). We used a well-characterized assay in which template-length RNA products of negative polarity were demonstrated for DV RdRp activity (Ackermann and Padmanabhan 2001; Nomaguchi et al. 2004). We found that substitutions of specific nucleotides within SLB or 5′CS yielded RNAs that were very active as templates (MSLB and MCS RNAs). Similar results were obtained by complete deletion of SLB or 5′CS (ΔSLB and ΔCS RNAs) or deletion of both regions (ΔSLB-ΔCS RNA) (Fig. 1C). In contrast, mutations within SLA or deletion of the complete domain nearly abolished RNA synthesis (MSLA and ΔSLA RNAs). These results strongly suggest that the SLA present at the viral 5′ UTR contains signals that promote specific RNA synthesis by DV RdRp.
To further analyze the ability of SLA to promote RNA synthesis in the context of nonviral sequences, we generated RNAs containing the coding sequence of luciferase fused or not to the SLA structure at the 5′ end of the molecule. Using these RNAs as templates for in vitro NS5pol activity, we observed that RNA synthesis was >10′ fold higher when the RNA contained the SLA, strongly suggesting that the SLA structure is sufficient to promote RNA synthesis by the viral RdRp even in the context of nonviral sequences (Fig. 1D).
SLA is an essential element for DV RNA amplification in transfected cells
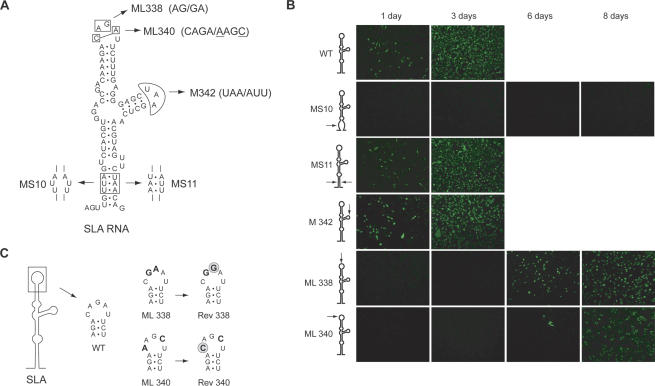
Viral replication in infected cells involves a complex machinery formed by several viral and cellular factors; therefore, it became crucial to investigate the role of SLA in the context of DV infections. To this end, we employed a full-length cDNA clone of DV type 2 (Kinney et al. 1997). We generated recombinant infectious RNAs with specific mutations that disrupted the predicted structure or changed the nucleotide sequence of SLA at the viral 5′ UTR. Five DV RNA mutants were designed (Fig. 2A): (1) a mutant with a 3′ nt substitution at the bottom of SLA that disrupted the predicted stem (MS10),(2) a mutant with a 6′ nt substitution at the bottom of SLA that maintained the stem structure (MS11), (3) a mutant with a 3′ nt substitution within the side loop of SLA (M342), and (4) two mutants each carrying two nucleotide substitutions at the top loop of SLA (ML338 and ML340). Equal amounts of in vitro transcribed RNAs corresponding to the wild type and mutants MS10, MS11, M342, ML338, and ML340 were transfected into BHK cells. Infectivity of the RNAs was assessed by immunofluorescence assays (IFA) using specific anti-DV 2 antibodies and by characterization of recovered infectious viruses. Cells transfected with the wild-type DV RNA showed positive IFA at day 1, and nearly 100% of the monolayer was antigen-positive at day 3 (Fig. 2B). At day 4, cells showed extensive cytopathic effect and death (data not shown). Cells transfected with MS10 RNA were IFA-negative up to day 8, and no infectious viruses were recovered during this time. In contrast, almost 100% of the cell monolayer transfected with MS11 RNA was positive at day 3 (Fig. 2B), suggesting that base-pairing at the bottom of SLA is required for viral replication regardless of the nucleotide sequence at these positions. The M342 RNA was infectious, and the isolated viruses replicated similarly to the wild type. In contrast, transfections with the two mutant RNAs with substitutions at the top of SLA (ML338 and ML340) yielded viruses with a significant delay in replication (Fig. 2B). To determine the nucleotide sequence of these replicating viruses, RNA was purified from isolated viral particles and used for RT-PCR and sequence analysis. Interestingly, the sequences of the viruses recovered from cells transfected with mutants ML338 and ML340 differed from the input RNA sequences. Reversion to the wild-type sequence was observed at position 32 in mutant ML338 and at position 30 in mutant ML340 RNA (Rev 338 and Rev 340; Fig. 2C). These sequences were found in three independent transfections, suggesting that the original mutations at the top of SLA ML338 and ML 340 yield nonreplicating RNAs, and only revertant viruses were rescued in cell culture. Altogether, these results indicate that mutations that disrupt the stem or the top loop of SLA impaired DV replication, while mutations within the stem that maintain the structure (at least in the positions tested) or mutations within the side loop did not alter significantly viral replication.
Figure 2.
Stem–loop A is required for DV replication in BHK cells. (A) Schematic representation of specific mutations within SLA are indicated with arrows, and the nucleotide changes are shown for each mutant MS10, MS11, ML338, ML340, and M342. (B) Expression of DV proteins in cells transfected with wild-type and mutated viral RNAs was monitored by IFA. (C) Sequence of revertant viruses obtained after transfecting BHK cells with full-length DV RNA carrying the mutations ML338 and ML340 at the top loop of SLA. (Bold) Nucleotide substitutions of ML338 and ML340 RNAs, (gray circles) sequences of revertant viruses, Rev 338 and Rev 340.
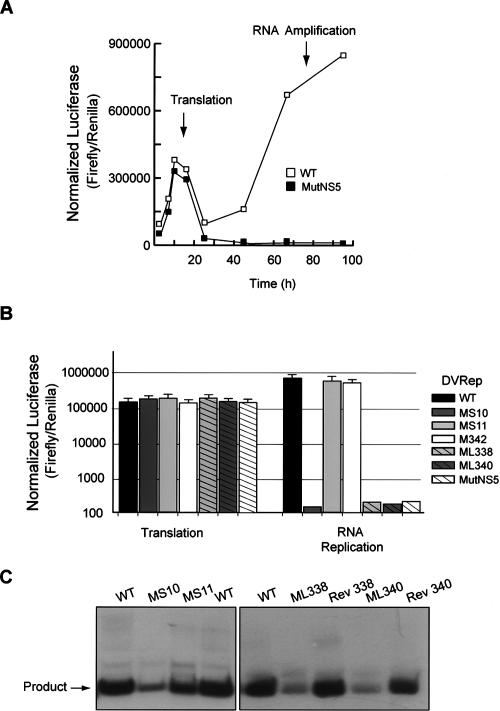
The DV genome is translated in the infected cell by a cap-dependent mechanism. In this process, the small subunit of the ribosome scans through SLA and SLB to reach the initiator AUG codon. Therefore, the finding that SLA plays an important role during viral replication could be attributed to a function of this RNA element in translational regulation and/or in further steps of viral replication. In order to discriminate the role of SLA in translation or RNA synthesis, we used a recently developed DV replicon system (Alvarez et al. 2005a). This RNA replicon includes a firefly luciferase reporter gene (Luc) replacing the viral structural proteins. After transfection of replicon RNA, the levels of Luc activity peaked between 8 and 10 h, reflecting translation of the input RNA (Fig. 3A). RNA synthesis can be detected by an exponential increase of Luc after 20 h of transfection. As a control to ensure that replicon RNA amplification was mediated by the viral replicase, a replication-defective RNA was designed by mutating the essential GDD motif of NS5 (RepMutNS5). This control RNA was efficiently translated but was not replicated (Fig. 3A).
Figure 3.
Stem–loop A is required for DV replicon RNA amplification in BHK cells. (A) Time course of luciferase activity showing the replication of the DV replicon in BHK cells. The times used to assess translation of input RNA or RNA amplification are shown by arrows. (B) Translation and RNA replication of wild-type and mutant DV replicons carrying mutations within SLA described in Fig. 2A (MS10, MS11, M342, ML338, and ML340) and the replication-incompetent MutNS5 were evaluated in BHK cells. Normalized luciferase levels are shown in logarithmic scale for each replicon RNA (DVRep) as indicated on the right. (C) In vitro NS5pol activity using 5′DVRNA templates corresponding to the first 160 nt of the DV genome with specific mutations around SLA. 5′DV RNAs with mutations within the stem of SLA (MS10 and MS11), the top loop of SLA (ML338 and ML340), and the 5′DV RNAs carrying the sequences from the revertant viruses (Rev 338 and Rev 340) as indicated on the top were used at 300 nM concentration as templates for the in vitro activity of viral NS5pol.
In the context of the replicon system, we generated RNAs carrying the mutations described above for the infectious clone (RepMS10, RepMS11, RepM342, RepML338, and RepML340; Fig. 2A). Quantified amounts of all in vitro transcribed RNAs, as well as a Renilla Luc mRNA serving as a transfection control, were transfected into BHK cells, and both Luc activities were monitored. The normalized Luc signal at 10 h was used to evaluate translation efficiency, while the Luc signal measured at 72 h was used to assess RNA amplification. The levels of Luc at 10 h were not significantly different between cells transfected with RepWT or mutant RNAs (Fig. 3B). In contrast, RNA amplification of replicons carrying modifications within the stem or the top loop of SLA (RepMS10, RepML338, and RepML340) was undetectable (Fig. 3B). In addition, mutations that changed the sequence but not the structure of the stem and mutations within the side loop of SLA (RepMS11 and RepM342) yielded replicons that amplified their RNAs as efficiently as the RepWT. These results are consistent with the observations made with the infectious clones and indicate that alterations in the sequence or structure of SLA directly compromised viral RNA amplification without affecting protein synthesis.
To further investigate the role of SLA during viral RNA synthesis, we analyzed the in vitro template activity of RNA molecules corresponding to the first 160 nt of the genome of recombinant viruses MS10, MS11, ML338, ML340, and M342 along with RNAs with the sequences of the isolated viruses with reversions within the top loop of SLA, Rev 338 and Rev 340. RNA synthesis by NS5pol was determined by quantifying radiolabeled RNA products. Template activity of MS10 RNA was eightfold lower than that observed with the wild-type sequences, while MS11 displayed ∼80% of wild-type activity (Fig. 3C). RNA synthesis promoted by ML338 or ML340 RNAs was very inefficient, about 20′ and 30′ fold lower, respectively, than that measured for the wild-type template. In contrast, Rev 338 and Rev 340 RNAs were excellent templates, indicating a striking dependence of the NS5pol activity on the SLA sequence and a direct correlation with the in vivo observations (Fig. 3C). These results suggest that the in vivo nonreplicating phenotypes of ML338 and ML340 RNAs can be explained by a lack of NS5 activity and that single nucleotide reversions within the SLA are sufficient to restore promoter activity.
DV NS5pol is an RNA-binding protein that interacts with the 5′end of the viral genome
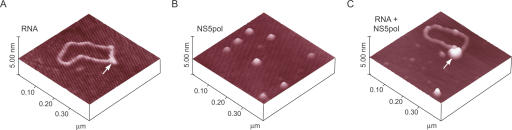
We have previously demonstrated using AFM that a model RNA molecule carrying the 5′ and 3′ end sequences of the DV genome is capable of acquiring a circular conformation mediated by specific RNA–RNA interactions (Alvarez et al. 2005b). Here, in an attempt to analyze binding of NS5pol to the RNA, we visualized the model RNA molecule in the presence or absence of NS5pol. We prepared samples containing RNA or NS5pol, or both together and examined individual molecules using tapping mode AFM in air. The apparent volume of the viral 5′ and 3′ UTR region (shown by an arrow in Fig. 4A) was determined by analysis of a large number of molecules. In agreement with previous measurements (Alvarez et al. 2005b), the apparent volume in the absence of NS5pol was 150 ± 30 nm3 while in the presence of NS5pol it was 300 ± 50 nm3, consistent with binding of NS5 to the RNA (Fig. 4). In addition, under conditions in which NS5pol and the RNA were in a 5:1 molar ratio, ∼70% of the RNA molecules were decorated with the viral protein, providing evidence for DV NS5pol–RNA interaction.
Figure 4.
Visualization of RNA-NS5pol interaction by AFM. Twenty microliters of samples containing RNA (1ng/μL) and/or NS5pol(0.4ng/μL) was deposited on freshly cleaved mica for imaging analysis. (A) A representative RNA molecule in a circular conformation is observed (Alvarez et al. 2005b). This 2.3′kb RNA contains the 5′ and 3′ end sequences of DV flanking the sequence of the luciferase, which is annealed to a 1633′nt antisense RNA. In this RNA, the viral sequences at the 5′ and 3′ ends of the molecule remain as single-stranded overhangs and mediate RNA cyclization by long-distance RNA–RNA contact (arrow). (B) A representative image showing individual molecules of purified NS5pol. (C) A representative image obtained with a sample containing both RNA and the viral NS5pol. The arrow indicates the polymerase bound to the RNA. Image sizes are 400 nm × 400 nm.
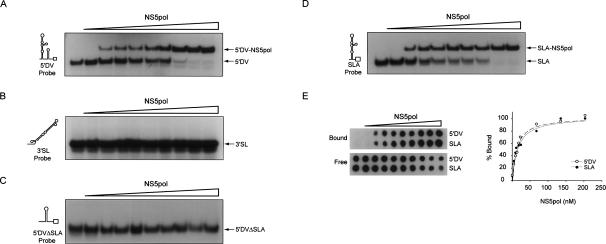
To further investigate the RNA-binding ability of DV NS5pol, we performed electrophoretic mobility shift (EMSA) and filter-binding assays using different radiolabeled RNAs and purified NS5pol protein. We synthesized two RNA probes corresponding to the 5′ end 160 nt (5′DV) or the last 106 nt (3′SL) of the viral genome. These RNAs were incubated with increasing concentrations of NS5pol, and the interactions were analyzed on native polyacrylamide gels. A well-defined and stable heparin-resistant complex was observed when the viral protein was incubated with the 5′DV probe (Fig. 5A), while no complex formation was detected with the 3′SL probe (Fig. 5B). To better define the RNA element recognized by the polymerase at the viral 5′ end, we designed two additional RNA probes: one containing a deletion of SLA (5′DVΔSLA probe) and a second one carrying a deletion of SLB and CS (SLA probe of 130 nt) containing the complete SLA structure. Binding of NS5pol to the 5′DVΔSLA was undetectable, while binding to SLA probe was readily observed (Fig. 5C,D), indicating specific recognition of SLA by the viral polymerase. In addition, to compare the binding affinity of NS5pol to the 5′DV and the SLA, we used EMSA and filter-binding assays. The dissociation constants (K ds) obtained for the two complexes were similar: 17 nM for the NS5pol-5′DV and 16 nM for the NS5pol-SLA using the filter-binding assay (Fig. 5E), and 10 and 8 nM, respectively, using the EMSA assay, showing high affinity of NS5pol for the viral RNA. These results indicate that the viral NS5pol is an RNA-binding protein that interacts with the 5′ SLA but not with the 3′ end of DV RNA.
Figure 5.
DV NS5pol binds to 5′ end sequences of the viral genome. (A) EMSA showing the interaction between the 5′DV RNA probe, corresponding to the first 160 nt of the DV genome (schematically represented on the left), and the purified NS5pol. Uniformly 32P-labeled 5′DV RNA (0.1 nM, 30,000 cpm) was titrated with increasing concentrations of NS5pol from 0 to 240 nM. (B) EMSA showing the lack of interaction between the 3′SL RNA probe corresponding to the last 106 nt of the DV genome and the purified NS5pol. (C) EMSA showing the lack of interaction between the 5′DV RNA probe with a deletion of SLA (5′DVΔSLA probe, left) and the purified NS5pol. (D) EMSA showing the interaction between SLA probe and purified NS5pol. The binding reactions were as described in A. (E) Interaction of NS5pol with the 5′DV and the SLA probes monitored by a filter-binding assay. Uniformly 32P-labeled RNA (0.1 nM) was incubated with increasing concentrations of NS5pol. (Left) RNA probes bound to each membrane as a function of NS5pol concentration visualized by PhosphorImaging. “Bound” indicates RNA–protein complexes retained in the nitrocellulose membrane, and “free” denotes the unbound probes retained in the nylon membrane. (Right) Quantification of the percentage of RNA probe bound was plotted as a function of NS5pol concentration, and Equation 1 was fitted.
Dissecting the role of long-range RNA–RNA interactions in the DV genome during RNA synthesis
It has been previously proposed that interactions between the 5′ and 3′ ends of the flavivirus genome could constitute the structure recognized by the polymerase during minus-strand RNA synthesis (Ackermann and Padmanabhan 2001; You et al. 2001; Nomaguchi et al. 2004; Alvarez et al. 2005b). In addition, it has been reported that RNA elements at the 5′ end of DV genome, besides the CS sequence, enhance in vitro polymerase activity (You et al. 2001). However, molecular details of how the 5′–3′ communication promotes flavivirus RNA synthesis remain unclear. Taking into account (1) that SLA is an essential element for DV NS5pol specificity and activity, (2) that SLA is located at the 5′ end of the genome, and (3) that the viral polymerase must initiate RNA synthesis using the 3′ end of the viral RNA as template, it is reasonable to propose that long-range RNA–RNA interactions could play a role in positioning the 3′ end of the template near the SLA promoter at the 5′ end of the viral genome.
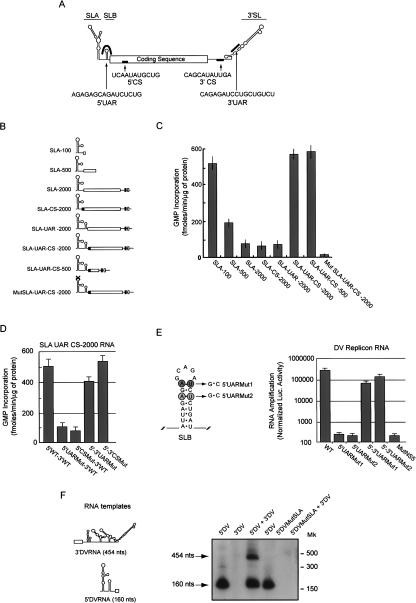
Two pairs of complementary sequences, 5′–3′UAR and 5′–3′CS, are necessary for DV genome cyclization (Fig. 6A). To investigate how the long-range RNA–RNA interactions participate during RNA synthesis by NS5pol, we first analyzed the ability of SLA RNA, without or with the cyclization sequences present at varying distances, to promote RNA synthesis. To this end, we designed RNAs carrying the SLA followed by 100-, 500-, or 2000-nt-long unrelated sequences (SLA-100, SLA-500, and SLA-2000 RNAs; Fig. 6B). The highest template activity was observed with the shortest RNA used (SLA-100 RNA; Fig. 6C). Increasing the length of the RNA template from 100 to 500 or to 2000 nt resulted in three-and sixfold reduction in RNA synthesis, respectively (Fig. 6C). Incorporation of one pair of cyclization sequences, 5′–3′UAR or 5′–3′CS, did not increase template activity (SLA-UAR-2000 and SLA-CS-2000 RNAs). In contrast, incorporation of both 5′–3′CS and 5′–3′UAR cyclization sequences greatly increased SLA-500 or the SLA-2000 RNA template activity (Fig. 6C). In these cases, the template efficiency was similar to the efficiency observed with the SLA-100 RNA. Therefore, while shorter RNAs were better templates in the absence of the cyclization sequences, the level of RNA synthesis was independent on RNA template length in the presence of the cyclization sequences. Furthermore, the template activity of an RNA molecule competent for cyclization but with specific mutations at the top of SLA (Mut SLA-UAR-CS-2000) was almost abolished, indicating that the cyclization sequences per se do not promote RNA synthesis (Fig. 6C).
Figure 6.
Role of long-range 5′–3′ RNA–RNA interactions during DV RNA synthesis. (A) Schematic representation of the DV genome showing the location and nucleotide sequence of the complementary regions 5′UAR, 5′CS, 3′CS, and 3′UAR. The locations of stem–loop A (SLA), stem–loop B (SLB), and 3′ stem–loop (3′SL) are also indicated. (B) Schematic representation of RNA molecules used as templates for the viral NS5pol RdRp activity. RNAs carrying the viral SLA followed by unrelated sequences of 100, 500, or 2000 nt are labeled SLA-100, SLA-500, and SLA-1000, respectively. Incorporation of the 5′CS, 5′UAR, or 5′CS-5′UAR in the SLA-2000 yielded the SLA-CS 2000, SLA-UAR 2000, or SLA-UAR-CS 2000 RNAs, respectively. The MutSLA UAR-CS 2000 corresponds to the respective RNA with a 4-nt substitution at the top of SLA (CAGA was replaced by AGAC). The white and black boxes shown at the 3′ of the molecules represent the 3′UAR and 3′CS regions, respectively. (C) In vitro RNA synthesis by the viral NS5pol using RNA templates described in B. The RdRp assay was carried out as described in Materials and Methods. (D) In vitro RNA synthesis by the viral NS5pol using the RNA template SLA-UAR CS 2000 described above, carrying specific mutations at 5′–3′UAR or 5′–3′CS regions that disrupted or reconstituted base-pairings. RNA mutants within 5′UAR or 5′CS (5′UARMut and 5′CSMut), which generated three and four mismatches when hybridized with the 3′UAR and 3′CS, respectively, and double mutants at the 5′ and 3′ regions (5′– 3′UARMut and 5′–3′CSMut RNAs), which compensated complementarity, were used as templates for polymerase activity. RdRp activity for the viral NS5pol was determined for each template and expressed as described in A. (E) DV replicon RNA amplification in BHK cells requires 5′–3′UAR complementarity. RNA synthesis of DV replicons with specific mutations within 5′UAR (5′UARMut1 and5′UARMut2, left) or double mutations at 5′ and 3′UAR (5′–3′UAR-Mut1 and 5′–3′UARMut2) that restored complementarity was evaluated by reporter activity 72 h after RNA transfections into BHK cells and compared with wild-type and replication-incompetent (MutNS5) replicons. Normalized luciferase levels were obtained using a control RNA encoding Renilla luciferase that was cotransfected with each replicon RNA. The normalized luciferase activity is shown in logarithmic scale. (F) SLA promotes RdRp activity of the viral NS5pol in trans. (Left) Schematic representations of the RNA templates used; (right) a denatured polyacrylamide gel showing the radiolabeled products from in vitro RdRp activity using NS5pol (6 ng/μL) and 300 nM of the templates described at the top of the gel. Arrows indicate the mobility of the full-size products of 454 and 160 nt. Mobilities of RNA markers (Mk) are also indicated on the right.
To further analyze the requirement for sequence complementarity within both CS and UAR in the in vitro system, we designed SLA-UAR-CS-2000 RNA molecules with specific mutations in UAR or CS. We synthesized RNAs carrying substitutions in 5′UAR or 5′CS, and in both 5′–3′UAR or 5′–3′CS, which were predicted to disrupt or reconstitute the 5′–3′ complementarity, respectively. The 5′UAR and the 5′CS mutants contained 3-and 4-nt substitutions, respectively (80-ACAGAGCA CAUCUGUG and 134-UCCAAAUGAUA). These mutations were shown previously to abolish RNA–RNA complex formation in RNA-binding assays (Alvarez et al. 2005b). Mutations within 3′UAR and 3′CS were designed to restore complementarity (3′UAR 10634-CACAGAUCGUGCUGUGU and 3′CS 10618-UAUCAUUUGGA). As shown in Figure 6D, mutations in 5′UAR or 5′CS decreased template activity four-and fivefold, respectively, while reconstitution of sequence complementarity restored RNA synthesis to near wild-type template levels, confirming that sequence complementarity and not the nucleotide sequence per se is important to enhance RNA synthesis.
We have previously reported that CS complementarity is necessary for in vivo DV RNA amplification (Alvarez et al. 2005a); however, the requirement for 5′–3′ UAR interaction during RNA synthesis has not been examined. Therefore, we introduced mutations within 5′ and 3′ UAR to disrupt or reconstitute the RNA–RNA interactions in the context of the replicon system, and tested the ability of these RNAs to replicate in BHK cells. Because the 5′UAR is located within the very conserved SLB, mutations were incorporated at both sides of the stem to avoid altering the predicted stem–loop structure (Fig. 6E). In addition, compensatory mutations were introduced within the 3′SL to restore base-pairings. We designed two RNA mutants with substitutions within the 5′UAR (5′UARMut1 and 5′UARMut2 RNAs) to disrupt complementarity, and double mutants (5′–3′UARMut1 and 5′–3′UARMut2 RNAs) to restore RNA–RNA interaction. Replicon RNAs corresponding to the wild type and the 5′UARMut1, 5′UARMut2, 5′–3′UARMut1, 5′– 3′UARMut2, and NS5Mut were transfected into BHK cells together with the mRNA encoding Renilla luciferase. Translation of the input RNA corresponding to the Rep wild type and mutants was very efficient (data not shown). In contrast, viral RNA synthesis showed a strict requirement for base-pairing around UAR (Fig. 6E). No RNA amplification was detected with the 5′UARMut1, 5′UARMut2, or the MutNS5 RNAs; however, restitution of UAR complementarity restored RNA synthesis. The RNA amplification of the double mutants was efficient, although it did not reach wild-type Rep levels, suggesting an additional role of the wild-type nucleotide sequences for an efficient process in vivo.
Altogether, the in vitro assays for DV RdRp showed that template activity becomes cyclization-sequences-dependent as the length of the RNA increases and that both pairs of complementary sequences are required to enhance RNA synthesis, which is strictly dependent on an intact SLA. In addition, RNA amplification becomes absolutely dependent on the complementary regions in the context of a self-replicating 10-kb-long RNA.
SLA promotes trans initiation by NS5pol at the viral 3′ UTR
5′–3′ RNA–RNA interactions were observed between two RNA molecules carrying the end sequences of the DV genome in the absence of proteins (You and Padmanabhan 1999; Alvarez et al. 2005b), and these interactions were able to support RNA synthesis from two separate RNA molecules (Ackermann and Padmanabhan 2001; You et al. 2001). Therefore, we wondered whether the SLA could function in trans to promote RNA synthesis of an RNA molecule containing only the viral 3′ end sequences. To that end, we determined NS5pol activity using the following RNA templates: (1) a 160-nt RNA containing the 5′-terminal DV sequences corresponding to the SLA-UAR-CS (5′DV RNA); (2) an RNA bearing the 454-nt viral 3′ UTR (3′DV RNA); (3) the two RNA molecules together, 5′DV and the 3′DV; and (4) two RNA molecules as in 3, with the 5′DV containing a mutation within SLA (MSLA; 30-CAGA was changed to AGAC). Analysis of the radiolabeled RNA products indicated that the 5′DV was an efficient template yielding a 160-nt-long product, while the 3′DV was inactive (Fig. 6F). However, when the two templates were present, NS5pol was able to synthesize two RNA products, one of 160 nt and the other of 454 nt (Fig. 6F). In addition, no NS5pol activity was detected when the 5′DV template with a mutation in SLA was used together with the 3′DV molecule. These results indicate that the 5′DV with an intact SLA could promote RNA synthesis in trans from the inactive 3′DV template.
Discussion
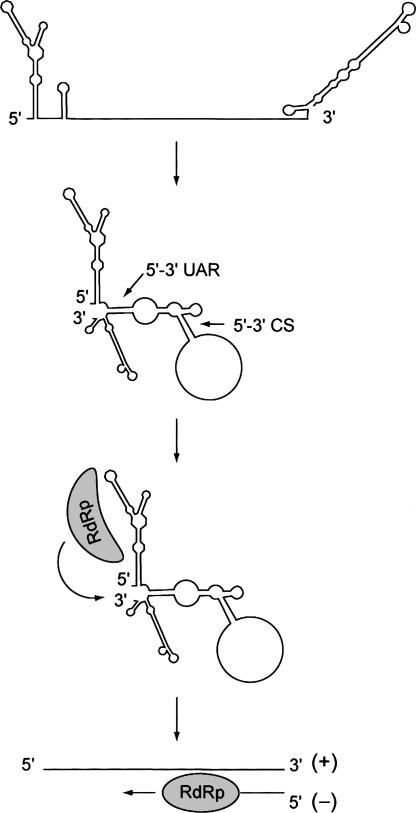
We found that DV RdRp discriminates viral RNAs by specific recognition of a novel RNA element at the 5′ end of the viral genome. Based on our findings and previous reports, we propose a new model for DV minus strand RNA synthesis in which the viral polymerase recognizes the 5′ end of the RNA, and through long-range RNA–RNA interactions the 5′ promoter and the 3′ end of the genome are brought together to facilitate RNA synthesis (Fig. 7).
Figure 7.
Model for DV minus strand RNA synthesis. The viral genome circularizes in the absence of proteins mediated by 5′– 3′ UAR and 5′–3′ CS hybridization. The viral RdRp binds to a 5′ stem–loop (SLA), and by long-range RNA–RNA interactions the polymerase is transferred to the site of initiation at the 3′ end of the genome.
It has been previously proposed that the 5′–3′ hybridized complementary sequences present in the DV genome constituted the promoter for RNA synthesis (Ackermann and Padmanabhan 2001; You et al. 2001; Nomaguchi et al. 2004; Alvarez et al. 2005b). Here, we demonstrated that these sequences do not promote viral RNA synthesis per se and that the RNA element responsible for this activity is the SLA structure. Importantly, the 5′ location of the promoter explains the need for long-range 5′–3′ communication found in dengue and other mosquito-borne flaviviruses (Khromykh et al. 2001; Corver et al. 2003; Lo et al. 2003; Alvarez et al. 2005b). The folding prediction of the 5′–3′ interacting DV RNA shows that the bottom half of the highly conserved 3′SL opens while the upper half is retained (Supplemental Fig. S1A). Therefore, the 5′–3′ UAR interaction could have a role in modulating the 3′SL structure during the initiation of RNA synthesis. In this case, besides bringing the 3′ end of the RNA near the 5′ promoter, the long-range interaction could make the 3′-terminal nucleotides of the viral genome available for the viral RdRp. Interestingly, a recent report identified novel complementary sequences that were essential for replication of tick-borne encephalitis virus (Kofler et al. 2006). These sequences located upstream of the initiator AUG and at the bottom of the 3′SL resembled the 5′–3′ UAR sequences of DV, providing evidence of structural changes around the 3′SL during RNA replication of both mosquito-and tick-borne flaviviruses.
Using RNA-binding assays, we found that the viral polymerase does not interact with sequences corresponding to the 3′ terminus of the viral genome; instead, the binding was specific to the SLA RNA (Fig. 5). In addition, we observed interaction of NS5pol with an RNA–RNA complex formed between the 5′ and 3′ ends of the viral genome, suggesting that NS5pol recognizes SLA even in the context of interacting 5′ and 3′ end viral sequences (Fig. 4; Supplemental Fig. S1B). The apparent K d of the complex NS5pol-RNA when the 5′DV RNA was interacting with the 3′SL was 12 nM (Supplementary Fig. S1C,D). This affinity was similar to that observed for the complexes NS5pol-SLA and NS5pol-5′DV (Fig. 5), suggesting that 5′–3′ interactions do not modify the binding affinity of NS5pol-RNA in these experimental conditions.
While it is not surprising to find a core promoter for RNA synthesis at the 3′ end of a viral genome, it is intriguing why certain plus-strand RNA viruses such as DV developed mechanisms in which essential elements for RNA replication are located at the 5′ end of the RNA. Genome cyclization may provide several advantages for viral replication, including (1) a control mechanism to amplify only full-length templates, (2) coordination of translation and RNA synthesis by overlapping signals at the 5′ and 3′ ends of the genome, (3) increasing RNA stability, (4) locating the viral polymerase or accessory proteins of the replication complex at the appropriate start site, and (5) controlling the levels of minus strand RNA synthesis. Regarding this last possibility, hybridization of the plus-strand with the nascent minus-strand RNA would not allow cyclization to occur, providing a mechanism to limit the levels of minus-strand RNA made. As with other plus-strand RNA viruses, the amount of minus-strand in infected cells is ∼10′ to 100′ fold lower than plus-strand, and it is likely that its synthesis is regulated during RNA replication. Moreover, if the minus-strand RNA is not free in the flavivirus-infected cell, as previously reported (for review, see West-away et al. 2003), we expect that NS5 should be able to use the double-stranded RNA (RF) as template for plus-strand amplification, presumably using a viral NS3 heli-case activity. Based on our observations that SLA was able to function in trans to promote RNA synthesis (Fig. 6F), and due to the fact that the 3′ end of the minus strand is in contact with the 5′ end of the plus strand in the RF, we speculate that SLA could also function as a promoter in trans for plus strand RNA synthesis. Interestingly, the crystal structure of NS5 identified the RNA cap methyl transferase (MT) and a novel GTP-binding site at the N terminus of the protein (Egloff et al. 2002). Therefore, interaction of the NS5 MT subdomain with the cap structure, which is right next to SLA, and binding of the RdRp subdomain to SLA could be a coordinated event with functional significance during flavivirus RNA replication.
Taken together, our data support a novel mechanism for DV minus-strand RNA synthesis that involves a promoter element present at the 5′ end of the genome. The identified promoter binds the viral RdRp and facilitates template recognition at the 3′ end of the genome via long-range RNA–RNA interactions. Since similar interactions have been found in other viral RNA genomes, the model proposed here may shed light on general strategies for RNA virus replication.
Materials and methods
RNA template preparation
RNAs were obtained by in vitro transcription using T7 RNA polymerase (90 min, 37°C) and treated with RNase-free DNase I to remove templates. The RNAs were purified using the RNeasy Mini Kit (Qiagen Inc.) to remove free nucleotides and quantified spectrophotometrically, and their integrity was verified by electrophoresis on agarose gels. All numbers given below in parentheses refer to nucleotide positions of DV2 16681 infectious cDNA clone (GenBank accession no. U87411; Kinney et al. 1997). Nucleotide sequences of primers used for PCRs are listed in Supplemental Table 1.
RNA template 5′–3′DV corresponding to the viral 5′ and 3′ UTRs flanking the coding sequence of the structural proteins was obtained from the full-length DV cDNA clone by digestion with HpaI. The 6305-bp fragment was ligated to generate a new plasmid, pDVΔHpa. The PCR fragment obtained using pDVΔHpa as template and primers AVG 1 and AVG 5 was then used as template for in vitro transcription to obtain 5′–3′DV RNA. The 5′DV and 3′DV RNAs were also obtained using PCR fragments from pDVΔHpa using primers AVG 1–AVG 130 and AVG 2–AVG 5, respectively. The 5′HCV RNA (nucleotides 1–220 of HCV 1b) was obtained from a PCR using primers AVG 242–AVG 243. Mutations and deletions within 5′DV RNA (ΔSLA, ΔSLB, ΔCS, ΔSLB-ΔCS, MSLA, MCS, MSLB, MS10, MS11, ML338, and ML340) were obtained by PCR and overlapping PCRs as described in Supplemental Table 2. To obtain the 5′DVRNAs with the sequences of the revertant viruses Rev 338 and Rev 340, we performed RT-PCR using as templates the TRIZOL-extracted RNA from the supernatant of transfected cells with ML338 and ML340 RNAs, respectively, using primers AVG 1 and AVG 130 in both cases. Luc and SLA-Luc DNA templates to transcribe the respective RNAs were obtained from plasmid pGL5′3′DV described elsewhere (Alvarez et al. 2005b), using primers AVG 107–AVG 426 and AVG 1–AVG 426, respectively. To obtain RNAs SLA 100, SLA 500, and SLA UARCS 2000, the plasmid pGL5′3′DV was used to generate PCR products using the primers AVG 1–AVG 130, AVG 1–AVG 426, and AVG 1–AVG 5, respectively. To obtain RNAs SLA 2000, SLA CS 2000, and SLA UAR 2000, three new plasmids derived from pGL5′3′DV were constructed: pGL5′3′DVΔSLBΔCS, pGL5′3′DVΔCS, and pGL5′3′DVΔSLB. These plasmids were generated digesting the pGL5′3′DV with SacI and AatII and introducing a PCR fragment corresponding to SLA, SLA-SLB, or SLA-CS, respectively. Finally, SLAMut-UAR-CS 2000 DNA was generated by overlapping PCR with primers AVG 485 and AVG 486 to obtain a DNA fragment that was cloned between SacI and SphI sites in the plasmid pGL5′3′DV.
AFM sample preparation and imaging
The 2.3-kb model RNA molecule that encodes the luciferase gene flanked by the 5′ and 3′ UTRs of DV hybridized with an antisense RNA of 1.6 kb corresponding to the luciferase coding sequence was obtained as previously described (Alvarez et al. 2005b). Samples were prepared in buffer A containing 20 mM Hepes (pH 7.9) and 4 mM MgCl2. Buffer A and MilliQ water were filtered before use, employing a 0.02-μm pore size (Anotop10, Whatman). RNA and purified NS5pol samples were diluted in buffer A to 1 ng/μL and 0.4 ng/μL, respectively. In all cases, 20 μL of the samples was deposited onto freshly cleaved muscovite mica. After 5 min, samples were gently washed with.
0.5 mL of milliQ water to remove molecules that were not firmly attached to the mica, and then blown-dried with nitrogen. Tapping mode AFM was performed using a Nanoscope III Multimode-AFM (Digital Instruments, Veeco Metrology) as previously described (Alvarez et al. 2005b).
RdRp in vitro assay
To amplify the sequence of DV NS5pol protein (nucleotides 8377–10270), we used a PCR product obtained from the infectious cDNA clone of DV2 16681. The PCR fragment was cloned into a pQE30 expression vector, which carries the N-terminal six-histidine tag, and purified from Escherichia coli soluble fraction using affinity chromatography on a His-trap Niquel-agarose column (Amersham). Purified NS5pol was stored until use in 40% glycerol-containing buffer at −80°C. For AFM analysis, a fraction of the protein was stored in 10% glycerol.
The in vitro standard RdRp assay was performed in a total volume of 25 μL in buffer containing 50 mM Hepes (pH 8.0); 10 mM KCl; 5 mM MgCl2; 2 mM MnCl2; 10 mM dithiothreitol; 4 U RNase inhibitor; 500 μM (each) ATP, CTP, and UTP; 10 μM [α-32P] GTP; 0.5 μg of template RNA; and 0.15 μg of recombinant purified NS5pol. The reaction was carried out for 30 min at 30°C and was stopped by adding a denaturing solution to a final 7% (w/v) trichloroacetic acid (TCA) and 50 mM H3PO4 at 0°C. The TCA-precipitated RNA was then collected by vacuum filtration using a V-24 apparatus carefully adding the mixture onto the center of a Millipore filter (type HAWP, 0.45-μm pore size). The filters were washed eight times with 5 mL each of cold 7% (w/v) TCA–50 mM H3PO4 and dried, and then the radioactivity was measured. For PAGE analysis of the RNA products, the standard mix was the same as the one described above except that the reaction was ended by phenol extraction followed by ethanol precipitation. The RNA products were resuspended in TBE containing formamide (80%) and heated for 5 min at 65°C. The samples were then analyzed by electrophoresis on a 5% denaturing polyacrylamide gel–6 M urea and visualized by autoradiography or PhosphorImaging analysis.
RNA-binding assays
RNA-NS5 interactions were analyzed by electrophoretic mobility shift assays (EMSA) and filter-binding assays (FBA). Uniformly 32P-labeled RNA probes were obtained by in vitro transcription using T7 RNA polymerase and purified on 5% polyacrylamide gels–6 M urea. For this analysis four different probes were designed. The 5′DV probe (nucleotides 1–160), 3′SL probe (nucleotides 10617–10723), SLA probe (nucleotides 1–130 from template pGL5′3’DVΔSLBΔCS), and 5′DVΔSLA probe (nucleo-tides 72–160) were obtained using PCR fragments with the primers described in Supplemental Table 2 and in vitro transcription.
The binding reactions contained 5 mM Hepes (pH 7.9), 25 mM KCl, 2 mM MgCl2, 3.8% glycerol, 0.12 mg/mL heparin, 0.1 nM 32P-labeled probe, and increasing concentrations of NS5pol (0, 1, 5, 10, 15, 20, 25, 80, 160, and 240 nM). RNA-NS5 complexes were analyzed by electrophoresis through native 5% polyacrylamide gels supplemented with 5% glycerol.
For FBA, Nitrocellulose (Protran BA 85, Whatman-Schleider & Schuell) and Hybond N+ nylon (Amersham Bioscience) membranes were presoaked in 5 mM Hepes (pH 7.9) and assembled in a dot-blot apparatus. A 20-μL aliquot of each protein–RNA mixture was applied to the filters and rinsed with 100 μL of 5 mM Hepes (pH 7.9). Membranes were air-dried and visualized by autoradiography. The macroscopic binding constants were estimated by nonlinear regression (Sigma Plot), fitting Equation 1: Bound % = Boundmax · [Prot] / (K d + [Prot]), where Bound % is the percentage of bound RNA, Boundmax is the maximal percentage of RNA competent for binding, [Prot] is the concentration of purified NS5pol, and K d is the apparent dissociation constant.
Construction of recombinant DV and replicons
The plasmid pDVRep containing the firefly luciferase in place of DV structural proteins was previously described (Alvarez et al. 2005a). Plasmids pDVRepMS10, pDVRepMS11, pDVRepML338, pDVRepML340, and pDVRepML342 were generated by replacing a SacI–SphI fragment from pDVRepWT with the respective fragment containing the mutation obtained by overlapping PCR, using a similar strategy as the one described in Supplemental Table 2. To generate full-length DV cDNAs with the same mutations as the ones described for the replicon (pDVMS10, pDVMS11, pDVML338, pDVML340, and pDVML342), the same strategy as the one described for the replicons was used.
To generate pDVRep 5′UAR Mut1 and 5′UAR Mut2, a PCR product carrying the desired mutations was obtained by overlapping PCR using primers AVG194–AVG351 and AVG350– AVG84 for 5′UAR Mut1 and primers AVG194–AVG347 and AVG346–AVG84 for 5′UAR Mut2. Recombinant plasmids were obtained by replacing a SacI–SphI fragment from pDVRepWT with the fragment generated by PCR. Compensatory mutations in the 3′ UTR were obtained by overlapping PCR using primers AVG90–AVG353 and AVG352–AVG263 for 5′–3′UAR Mut1 and primers AVG90–AVG349 and AVG348–AVG263 for 5′–3′UAR Mut2. pDVRep 5′–3′UARMut1 and pDVRep 5′– 3′UARMut2 were generated by cassette substitution between AflII and XbaI sites in pDVRep 5′UARMut 1 and pDVRep 5′UARMut 2, respectively.
To obtain infectious and replicon DV RNAs in vitro transcription by T7 RNA polymerase in the presence of m7GpppA, a cap analog was used as previously described (Alvarez et al. 2005b). RNA transcripts (3 μg) were transfected with Lipofectacmine 2000 (Invitrogen) into BHK-21 cells. For sequencing analysis, TRIZOL-extracted RNAs from 200 μL of supernatants were used for reverse transcription, PCR amplification, and sequencing using an ABI 377 (Applied Biosystems).
Acknowledgments
We thank Bruno Canard and Barbara Selisko for technical advice and expression plasmids for NS5pol. We are grateful to Richard Kinney for dengue virus cDNA clone. We also thank Lucia Rothman-Denes, Raul Andino, and Alejandra Tortorici for helpful discussions concerning the manuscript. This work was funded by Howard Hughes Medical Institute, Fundación Bunge & Born, Agencia de Promoción Científica y Tecnológica de Argentina, and ICGEB-OPS-RELAB.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1444206.
References
- Ackermann M., Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymer-ase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Alvarez D.E., De Lella Ezcurra A.L., Fucito S., Gamarnik A.V. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005a;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Alvarez D.E., Lodeiro M.F., Ludueña S.J., Pietrasanta L.I., Gamarnik A.V. Long-range RNA–RNA interactions circularize the dengue virus genome. J. Virol. 2005b;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Rieckhof G.E., Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Brinton M.A., Fernandez A.V., Dispoto J.H. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- Cahour A., Pletnev A., Vazielle-Falcoz M., Rosen L., Lai C.J. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology. 1995;207:68–76. doi: 10.1006/viro.1995.1052. [DOI] [PubMed] [Google Scholar]
- Corver J., Lenches E., Smith K., Robison R.A., Sando T., Strauss E.G., Strauss J.H. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 2003;77:2265–2270. doi: 10.1128/JVI.77.3.2265-2270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T.W. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Benarroch D., Selisko B., Romette J.L., Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: Crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elghonemy S., Davis W.G., Brinton M.A. The majority of the nucleotides in the top loop of the genomic 3′ terminal stem loop structure are cis-acting in a West Nile virus infectious clone. Virology. 2005;331:238–246. doi: 10.1016/j.virol.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Frolov I., Hardy R., Rice C.M. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus-and plus-strand RNA synthesis. RNA. 2001;7:1638–1651. doi: 10.1017/s135583820101010x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A.V., Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes & Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grdzelishvili V.Z., Garcia-Ruiz H., Watanabe T., Ahlquist P. Mutual interference between genomic RNA replication and subgenomic mRNA transcription in brome mosaic virus. J. Virol. 2005;79:1438–1451. doi: 10.1128/JVI.79.3.1438-1451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler D.J. Dengue and dengue hemorrhagic fever. Clin.Microbiol. Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C.S., Hahn Y.S., Rice C.M., Lee E., Dalgarno L., Strauss E.G., Strauss J.H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Herold J., Andino R. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Meka H., Guyatt K.J., Westaway E.G. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001;75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney R.M., Butrapet S., Chang G.J., Tsuchiya K.R., Roehrig J.T., Bhamarapravati N., Gubler D.J. Construction of infectious cDNA clones for dengue 2 virus: Strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230:300–308. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- Kofler R.M., Hoenninger V.M., Thurner C., Mandl C.W. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J. Virol. 2006;80:4099–4113. doi: 10.1128/JVI.80.8.4099-4113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M.K., Tilgner M., Bernard K.A., Shi P.Y. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003;77:10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight K.L., Lemon S.M. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men R., Bray M., Clark D., Chanock R.M., Lai C.J. Dengue type 4 virus mutants containing deletions in the 3' noncoding region of the RNA genome: Analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 1996;70:3930–3937. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesters H.G., Strauss J.H. Defined mutations in the 5′ nontranslated sequence of Sindbis virus RNA. J. Virol. 1990;64:4162–4168. doi: 10.1128/jvi.64.9.4162-4168.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomaguchi M., Teramoto T., Yu L., Markoff L., Padmanabhan R. Requirements for West Nile virus (−)-and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli . J. Biol. Chem. 2004;279:12141–12151. doi: 10.1074/jbc.M310839200. [DOI] [PubMed] [Google Scholar]
- Olsthoorn R.C., Bol J.F. Role of an essential triloop hairpin and flanking structures in the 3′ untranslated region of Alfalfa mosaic virus RNA in in vitro transcription. J. Virol. 2002;76:8747–8756. doi: 10.1128/JVI.76.17.8747-8756.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A.V., Rieder E., Kim D.W., van Boom J.H., Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 2000;74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., Fabian M.R., White K.A., Nagy P.D. A replication silencer element in a plus-strand RNA virus. EMBO J. 2003;22:5602–5611. doi: 10.1093/emboj/cdg523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proutski V., Gould E.A., Holmes E.C. Secondary structure of the 3′ untranslated region of flaviviruses: Similarities and differences. Nucleic Acids Res. 1997;25:1194–1202. doi: 10.1093/nar/25.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar C.T., Zhang X., Kao C.C. Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol. 2003;77:1830–1839. doi: 10.1128/JVI.77.3.1830-1839.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher S., Flamm C., Mandl C.W., Heinz F.X., Stadler P.F. Secondary structure of the 3′-noncoding region of flavivirus genomes: Comparative analysis of base pairing probabilities. RNA. 1997;3:779–791. [PMC free article] [PubMed] [Google Scholar]
- Ray D., White K.A. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol. 2003;77:245–257. doi: 10.1128/JVI.77.1.245-257.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. Flaviviridae: The viruses and their replication. In: Knipe D., Howley P., editors. Fields virology. Vol. 1. Lippincott-Raven; Philadelphia: 2001. pp. 991–1041. [Google Scholar]
- Rieder E., Paul A.V., Kim D.W., van Boom J.H., Wimmer E. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 2000;74:10371–10380. doi: 10.1128/jvi.74.22.10371-10380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selisko B., Dutartre H., Guillemot J.C., Debarnot C., Benarroch D., Khromykh A., Despres P., Egloff M.P., Canard B. Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymer-ases. Virology. 2006 doi: 10.1016/j.virol.2006.03.026. (in press) [DOI] [PubMed] [Google Scholar]
- Song C., Simon A.E. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- Sullivan M.L., Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner M., Deas T.S., Shi P.Y. The flavivirus-con-served penta-nucleotide in the 3′ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology. 2005;331:375–386. doi: 10.1016/j.virol.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Westaway E.G., Mackenzie J.M., Khromykh A.A. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 2003;59:99–140. doi: 10.1016/s0065-3527(03)59004-2. [DOI] [PubMed] [Google Scholar]
- You S., Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′-and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 1999;274:33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- You S., Falgout B., Markoff L., Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′-and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 2001;276:15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- Yu L., Markoff L. The topology of bulges in the long stem of the flavivirus 3′ stem-loop is a major determinant of RNA replication competence. J. Virol. 2005;79:2309–2324. doi: 10.1128/JVI.79.4.2309-2324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Falgout B., Markoff L. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 1998;72:7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]