Abstract
The E2F transcription factor family is known to play a key role in the timely expression of genes required for cell cycle progression and proliferation, but only a few E2F target genes have been identified. We explored the possibility that E2F regulators play a broader role by identifying additional genes bound by E2F in living human cells. A protocol was developed to identify genomic binding sites for DNA-binding factors in mammalian cells that combines immunoprecipitation of cross-linked protein–DNA complexes with DNA microarray analysis. Among ∼1200 genes expressed during cell cycle entry, we found that the promoters of 127 were bound by the E2F4 transcription factor in primary fibroblasts. A subset of these targets was also bound by E2F1. Most previously identified target genes known to have roles in DNA replication and cell cycle control and represented on the microarray were confirmed by this analysis. We also identified a remarkable cadre of genes with no previous connection to E2F regulation, including genes that encode components of the DNA damage checkpoint and repair pathways, as well as factors involved in chromatin assembly/condensation, chromosome segregation, and the mitotic spindle checkpoint. Our data indicate that E2F directly links cell cycle progression with the coordinate regulation of genes essential for both the synthesis of DNA as well as its surveillance.
Keywords: DNA microarray, E2F, cell cycle regulation, transcription, DNA damage
The E2F transcription factor family plays a crucial and well-established role in cell cycle progression (Dyson 1998). This family comprises six different polypeptides (E2F1–E2F6) that pair with a heterodimeric partner (DP1 or DP2). E2F is thought to function by activating a panel of genes involved in progression through the G1 phase as well as DNA replication. An important function of E2F is the recruitment of the retinoblastoma (pRB) tumor suppressor family of proteins (“pocket” proteins); this family includes the related proteins p107 and p130 (Harbour and Dean 2000). We and others have shown that E2F-mediated recruitment of pocket proteins in quiescent cells and the early G1 phase of the cell cycle results in repression of genes that are subsequently activated at the G1/S phase transition. The pRB family both inhibits transcriptional activation of E2F and globally represses promoters by recruiting histone deacetylases (HDACs) (Brehm et al. 1998; Luo et al. 1998; Magnaghi-Jaulin et al. 1998; Ross et al. 1999, 2001).
The E2F family can be functionally subdivided into repressors (E2F4 and E2F5) and activators (E2F1, E2F2, and E2F3) based on a number of findings. First, E2F target gene expression is reduced in E2F3-deficient mouse embryonic fibroblasts, and these cells show significant proliferative defects (Humbert et al. 2000). Chromatin immunoprecipitation (ChIP) experiments have shown that E2F1, E2F2, and E2F3 associate with the promoters of previously described human and mouse E2F-responsive genes coincident with their activation at the G1/S phase boundary (Takahashi et al. 2000; J. Rayman, Y. Takahashi, and B. Dynlacht, unpubl.). Unlike E2F1, E2F2, and E2F3, which are constitutively nuclear, entry of E2F4 and E2F5 into the nucleus is restricted to cells in G0 and early G1 and may depend on posttranslational modification, association with pRB family members, or heterodimerization with DP-2 (Magae et al. 1996; Lindeman et al. 1997; Muller et al. 1997; Verona et al. 1997). Binding of E2F4 to promoters in living quiescent cells coincides with recruitment of p130, diminished acetylation of histones at the promoter, and gene repression (Takahashi et al. 2000). However, it is formally possible that E2F4 and E2F5 could activate transcription under some circumstances, because E2F4 has been detected on certain mouse promoters in late G1/S phase (Wells et al. 2000).
Despite the considerable progress that has been made toward understanding how E2F and pRB regulate the cell cycle, it has not been possible until very recently to identify direct and physiologically relevant downstream targets of these important cell cycle regulators. Much work has been focused on elucidating targets of E2F and pRB by means of transient transfection assays and promoter analysis. Although these studies have identified a number of genes that are likely to be E2F regulated, they suffer from important caveats. First, transfected plasmids are not reconstituted into chromatin, a natural substrate for HDAC corepressors believed to be recruited by the pRB family. Although selective activation of certain promoters after ectopic expression of individual E2F family members has been observed in some settings (DeGregori et al. 1997), it is often difficult to recapitulate both target specificity and biologically relevant temporal regulation under these conditions. Furthermore, overexpression of E2F is known to drive cell proliferation, resulting in secondary transcriptional changes linked to cell cycle status. The recent adaptation of chromatin immunoprecipitation approaches to the study of E2F-regulated gene expression (Takahashi et al. 2000) permits an examination of direct targets of E2F and pRB family members in living cells at different stages of the cell cycle without the above caveats.
Identification of the complete set of physiologically important targets of E2F and other transcription factors in the human genome would provide a valuable foundation for further understanding of the roles of these factors in cellular regulation, but there are two important limitations to experiments that might produce this knowledge. First, a DNA microarray-based technique that identifies genomic sites bound directly by DNA-binding factors has been developed for living yeast cells (Ren et al. 2000; Iyer et al. 2001; Simon et al. 2001; Wyrick et al. 2001), but it is not clear that this method will produce significant signal-to-noise ratios with the more complex mammalian genome. Second, the sequence of the human genome is incomplete, making it impossible to produce a DNA microarray that represents all promoter sites. We describe here solutions to these two issues that allowed us to identify a large number of new E2F target genes that are cell cycle-regulated. We report modifications of the yeast factor location technique that allow robust identification of promoters in living human cells that are bound by specific transcription factors. We focused our studies on E2F and the proximal promoters of nearly 1200 human genes whose expression is cell cycle-dependent by producing a microarray with these promoter sequences. Our results indicate that E2F plays a heretofore unrecognized role in the regulation of genes required for DNA repair, chromatin assembly, condensation, and segregation, as well as multiple checkpoints that ensure genomic integrity. Therefore, E2F does not exclusively regulate genes important for the G1/S transition. Rather, E2F plays a role in the expression of genes involved in multiple cell cycle control points.
Results
Mammalian factor location analysis
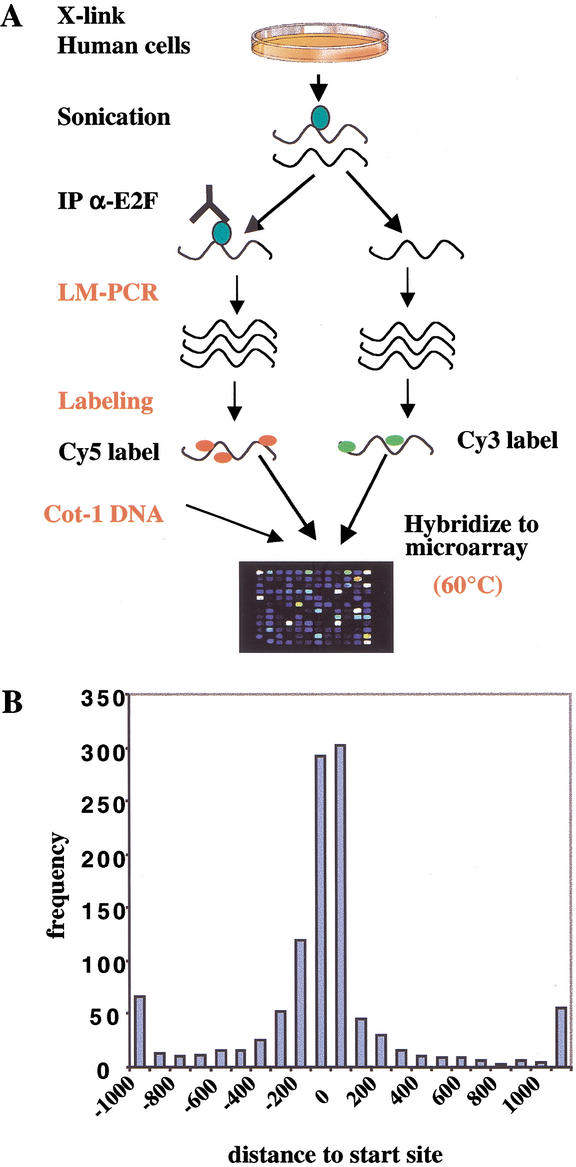
We first examined the occupancy of E2F4 on promoters during quiescence, since previous ChIP experiments have indicated that E2F4 directs recruitment of the pRB-related protein p130 to repress several genes during cell cycle exit (Takahashi et al. 2000). Primary human fibroblasts (WI-38) rendered quiescent through serum starvation were fixed with formaldehyde, harvested, and disrupted by sonication (Fig. 1A). To enrich for target genes bound by E2F, we immunoprecipitated the resulting chromatin fragments with antibodies that specifically recognize E2F4. At this point, we made several important modifications to protocols previously developed for yeast genome-wide location analysis (Ren et al. 2000) to compensate for the increased complexity of the human genome (see Materials and Methods). After reversal of cross links, the purified and enriched DNA was amplified by ligation-mediated polymerase chain reaction (LM-PCR) and subsequently labeled with the Cy5 fluorophore by random priming. For purposes of normalization, we also performed LM-PCR on DNA that was not enriched by immunoprecipitation and labeled the amplified sample with a second fluorescent dye, Cy3. ChIP-enriched and nonenriched (total input) pools of DNA were mixed with Cot-1 DNA to suppress annealing of repetitive sequences and hybridized under stringent conditions to a single DNA microarray composed of human promoters described below.
Figure 1.
Strategy for mammalian factor location analysis using the 1.5K DNA microarray. (A) An illustration of the experimental protocol. Modifications to previous yeast protocols (Ren et al. 2000) are indicated in red. Primary human cells (WI-38) were cross-linked and sonicated, and chromatin was immunoprecipitated with anti-E2F antibodies. The resulting ChIP-enriched DNA was purified, amplified by LM-PCR together with total genomic DNA, labeled with the Cy5 and Cy3 fluorophores using random priming, and hybridized to the 1.5K microarray in the presence of Cot-1 DNA to reduce nonspecific hybridization. (B) The frequency and distribution of transcription factors binding to human promoters. The distance of transcription factor-binding sites relative to the transcription start site is plotted versus the frequency of its occurrence. Most known transcription factor binding sites cluster between 200 nucleotides upstream and downstream of the start site (data source: http://transfac.gbf.de/TRANSFAC/).
It is not feasible to conduct a genome-wide microarray analysis of the location of DNA-binding proteins on human promoters because the human genome is not yet fully sequenced. Nevertheless, as an initial effort toward this ultimate goal, we developed a DNA microarray (henceforth referred to as the 1.5K array) that contained polymerase chain reaction (PCR) products spanning the proximal promoters of 1444 human genes, ∼1200 of which show regulated expression during cell cycle reentry (Iyer et al. 1999; Ishida et al. 2001). We included ∼200 genes with no apparent link to the cell cycle. Based on algorithms that predict DNA-binding motifs within promoters (Fig. 1B), we chose as end points of each PCR reaction regions ∼700 bp upstream and 200 bp downstream of putative transcription start sites. Known E2F sites are generally located within 200 nucleotides of the start site.
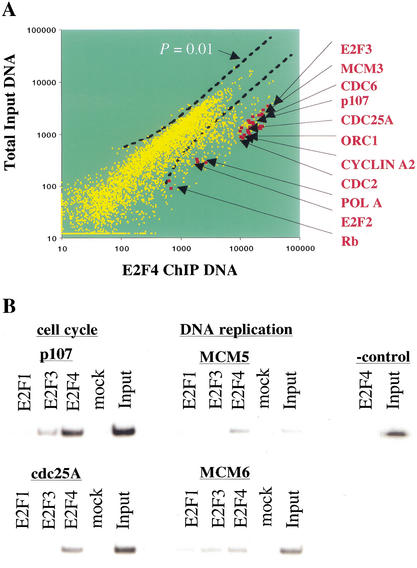
The results of two independent E2F4 location analysis experiments are shown in Figure 2A. The analysis indicates that E2F4 is associated with the promoters of 127 genes based on a P-value ≤ 0.002 in the error model (see Materials and Methods). Several lines of evidence indicate that this is a robust and accurate method for identifying factor-binding sites in mammalian cells. First, the fold-enrichment was at least as good as that observed in analogous experiments with yeast, where conventional approaches to identify direct factor-binding sites confirmed essentially all of the microarray results (Ren et al. 2000). Second, we observed striking enrichment of a majority of previously reported E2F-responsive promoters, including a number involved in cell cycle control (p107, pRB, cdc2, cdc25A, cyclin A, E2F2, and E2F3) and DNA replication (DNA polymerase α, cdc6, PCNA, and thymidine kinase). Indeed, we identified 17 of 20 previously established E2F targets that were printed on the microarray (see Discussion). Third, as described below, all of the target promoters selected for further study were confirmed to be bound by E2F using conventional ChIP.
Figure 2.
Location analysis of E2F4 on human promoters and identification of previously known E2F target genes. (A) Scatter plot analysis of Cy3-labeled total genomic DNA versus Cy5-labeled, E2F4 ChIP-enriched DNA. A P-value cutoff of 0.01 is shown. (B) Confirmation of promoter occupancy by E2Fs in quiescent WI-38 cells using a standard ChIPs protocol. E2Fs were enriched at several promoters involved in cell cycle control and DNA replication. In contrast, we did not detect enrichment of a negative control β-interferon fragment with anti-E2F4 antibodies. Nor did we observe significant promoter enrichment with an irrelevant antibody control (mock) lanes. Input lanes correspond to PCR reactions with 0.5% of total chromatin in immunoprecipitation reactions.
The strong enrichment of known E2F-responsive genes, coupled with the observation that only a small percentage of the genes regulated during cell cycle progression were enriched, attests to the highly selective nature of our ChIP and microarray protocols. Nevertheless, we used standard ChIPs to confirm our findings regarding previously identified E2F target genes. As expected from previous studies (Takahashi et al. 2000), we observed significant binding by E2F to the p107 and cdc25A promoters (Fig. 2B). We extended these findings by showing that two genes involved in replication, MCM5 and MCM6, were also occupied by E2F. In contrast, we observed no enrichment of unrelated control genes β-interferon and actin (Fig. 2B; data not shown).
E2F4 target genes cluster into seven functional categories
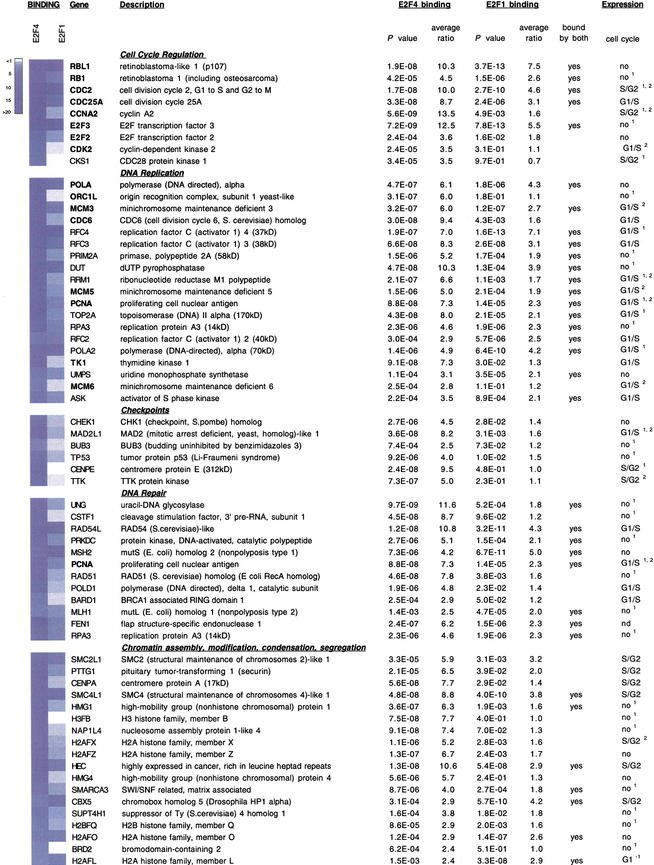
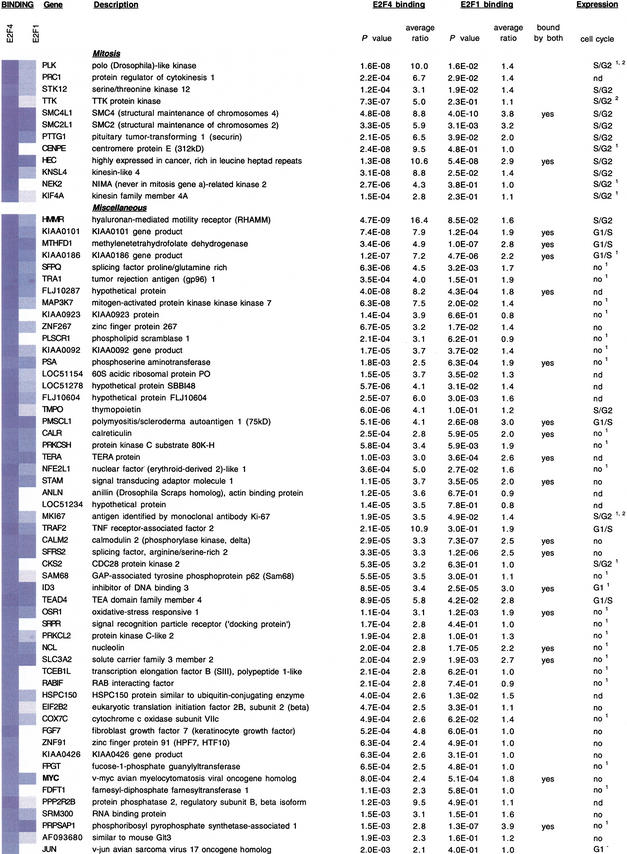
In addition to previously identified DNA replication and cell cycle regulatory genes, the genomic location analysis revealed a substantial number of novel E2F targets (Fig. 3). The E2F4 gene targets that we have identified clustered into seven functional groups related to cell cycle regulation, DNA replication, DNA repair, DNA damage and G2/M checkpoints, chromosome transactions, and mitotic regulation (Fig. 3). Several genes could be clustered into multiple categories as indicated. Each of the clusters showed, on average, similar levels of enrichment (Fig. 3). Interestingly, several genes identified in the screen encode proteins that are not only functionally related but also interact with one another in multimeric complexes, suggesting that components of the DNA damage repair and checkpoint responses are transcriptionally coregulated (see Discussion). Therefore, we conclude that E2F regulates an array of genes that function during, and subsequent to, the G1-to-S phase transition.
Figure 3.
Identification of E2F1- and E2F4-binding sites in human promoters and functional clustering of target genes. Each of the clusters is indicated, and previously identified E2F-responsive genes are indicated with bold type. In some cases, genes clustered into two or more functional categories. For each gene, P-values and average enrichment ratios for E2F4 and E2F1 binding are shown. Binding ratios are also shown using a blue-and-white color scheme. Those genes bound by both factors are indicated. The final column illustrates the cell cycle phase during which the indicated genes are induced. Data were obtained using Affymetrix microarray analysis (see Materials and Methods). In most cases, these expression profiles were also confirmed either by Iyer et al. 1999 (superscript 1), Ishida et al. 2001 (superscript 2), or both. (nd) Not determined; (G1−) repression in G1.
E2F1 and E2F4 bind an overlapping set of target genes
We have thus far shown that E2F4 binds a set of target genes with a high degree of specificity in quiescent cells. Given that E2F1 appears to associate in vivo with active promoters during the G1-to-S transition (Takahashi et al. 2000), we examined the binding of E2F1 at promoters of cells entering S phase. Quiescent WI-38 cells were stimulated by serum addition, and we confirmed by flow cytometry (FACS) that a substantial fraction of cells had entered S phase 26 h after serum addition (data not shown). G1/S phase samples were cross-linked, and chromatin immunoprecipitations were performed with anti-E2F1 antibodies. ChIP-enriched DNA was then hybridized to the 1.5K microarray. Interestingly, we observed significant overlap (50 genes) between the set of genes bound by E2F4 in quiescent cells and the targets bound by E2F1 in G1/S phase (Fig. 3). Although E2F1 was found to be associated with genes involved in mitosis, the vast majority of targets clustered in the DNA replication and repair categories.
In order to link our findings on E2F location to gene regulation, we performed a gene expression profiling experiment to determine when each of the E2F target genes we identified was induced during cell cycle reentry. Quiescent WI-38 cells were stimulated with serum, and RNA was prepared from nonstimulated cells and those collected 12, 16, and 26 h poststimulation, corresponding to G1, G1/S, and S/G2 phase populations. Target cRNA was prepared and hybridized to Affymetrix high-density DNA microarrays that contained most of the genes represented on our 1.5K array. The results are presented in Figure 3 and are largely consistent with data obtained from murine and human fibroblasts (Iyer et al. 1999; Ishida et al. 2001). As expected, most of the DNA replication genes were induced in late G1/S phase. Interestingly, many of the DNA repair, checkpoint, and mitotic genes were also induced in late G1 and S phase, suggesting that E2F could link expression of DNA replication and repair genes and play a role in the activation of genes required not only during S phase but at subsequent stages of the cell cycle (see Discussion).
These findings are consistent with our previous data indicating that many E2F-responsive genes are collaboratively regulated by E2F1 and E2F4 at different stages of the cell cycle (Takahashi et al. 2000). Furthermore, the widespread overlap in targets enriched by both factors again points to the high degree of specificity achieved in our ChIP and microarray experiments.
Confirmation of novel target genes
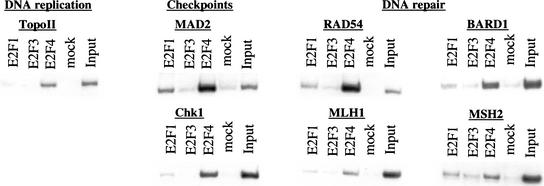
The genome location experiment revealed a large number of previously unidentified E2F target genes. It was therefore important to confirm promoter occupancy by E2F family members on a subset of genes identified in our microarray analysis using conventional ChIP analysis (Fig. 4). We observed striking enrichment of E2F4 on each of the genes we examined in quiescent WI-38 cells. Interestingly, several DNA repair genes (rad54, bard1), as well as G2 and mitotic checkpoint genes (chk1, mad2) were highly enriched with E2F4 antibodies; the enrichment was comparable to, or exceeded, that observed with the previously characterized cell cycle regulators, p107 and cdc25A (Fig. 2B; Takahashi et al. 2000). We did not observe significant promoter enrichment in ChIPs with irrelevant antibodies (“mock” lanes), nor was enrichment observed after amplification of unrelated promoter fragments immunoprecipitated with anti-E2F antibodies (data not shown). As for other mammalian cell cycle synchronization techniques, it is not feasible to obtain a homogeneous population of quiescent WI-38 cells, and the presence of residual cycling cells results in the detection of E2F1 (and E2F3) at this stage.
Figure 4.
Conventional ChIP analysis confirms the enrichment of genes identified in the 1.5K microarray screen. ChIP was performed with chromatin from quiescent WI-38 cells using the indicated antibodies, and enriched DNA was amplified with primers corresponding to representative genes from several different clusters.
The observed connection between E2F and a significant number of DNA repair genes as well as genes involved in the DNA damage and spindle checkpoints was unexpected. We focused on these promoters and examined the nucleotide sequence of the amplified products printed on our microarray for consensus E2F-binding sites (TTTC/GC/GCGC). Interestingly, inspection of these promoters revealed the existence of multiple E2F-binding sites in many of the target genes (data not shown). However, several promoters lacked obvious E2F recognition motifs, suggesting either that E2F may recognize noncanonical binding sites or that in some settings, it may be recruited to the promoter through association with other factors. Indeed, it is possible that specific temporal expression profiles of E2F-responsive genes could result from combinatorial interactions with other proximal transcription factors in a manner similar to the control of certain developmental programs in yeast (Madhani and Fink 1997). A mutational analysis of individual promoters will ultimately be required to determine the contribution of E2F-binding sites and proximal promoter elements to the expression of each gene.
Multiple E2F4 targets are deregulated in the absence of p107 and p130
Our studies identified novel genes that are bound by E2F4. Because E2F4 functions as a transcriptional repressor in quiescent and early G1 cells when bound to the pRB-related proteins p107 and p130, we predict that loss of these proteins would result in derepression of some or all of our newly identified E2F4 target genes. We tested this idea by synchronizing wild-type and p107−/−; p130−/− mouse embryonic fibroblasts (MEFs) by serum starvation and restimulation and then harvesting cells at various points after restimulation. We isolated RNA and assessed the level of each transcript by reverse transcription coupled with the polymerase chain reaction (RT-PCR) (Fig. 5). The results confirmed that one well-established target of this repressor, cyclin A2, was indeed derepressed in p107−/−; p130−/− MEFs relative to wild-type cells (Hurford et al. 1997). In contrast, cyclin E expression was not altered in the mutant cells, again confirming previous findings (data not shown). This result, when combined with our FACS analysis, also confirms that these cells were synchronized effectively (data not shown). Importantly, transcription of a control gene, actin, was not affected by loss of p107 and p130.
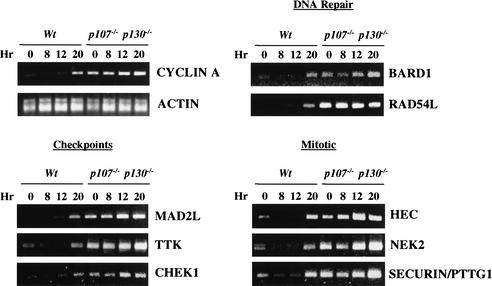
Figure 5.
Novel E2F targets identified in this study are derepressed in mouse embryonic fibroblasts (MEFs) lacking p107 and p130. Wild-type and p107−/−; p130−/− MEFs made quiescent by serum deprivation (t = 0 h) were stimulated to reenter the cell cycle. Cells were harvested at various times after restimulation, as indicated, and RNA was isolated. Cells entered S phase at the 20-h time point. RT-PCR analysis was performed using primers corresponding to selected DNA repair, checkpoint control, and mitotic genes as shown. Cyclin A and actin serve as positive and negative controls for cell cycle synchronization and RNA normalization, respectively.
We then compared transcript levels in both wild-type and mutant MEFs for a subset of the genes identified in our location analysis. We examined genes involved in DNA repair, mitotic and DNA damage checkpoints, and mitosis. Remarkably, we found that loss of p107 and p130 led to dramatic derepression in G0/G1 of each of the E2F4 targets that we identified, although the degree of derepression varied from one gene to another. The DNA repair gene RAD54L, mitotic checkpoint genes MAD2L and TTK, and the HEC and NEK2 mitotic genes were among the most significantly derepressed. These genes were deregulated to at least the same degree as cyclin A2, a known target of the E2F4 transcription factor. We conclude that these genes, first identified by our location analysis and surveyed in this assay, are bona fide targets of the E2F4–p107/p130 transcriptional repressor.
Discussion
Our understanding of the functions of sequence-specific transcriptional regulators is limited in part by our knowledge of their target genes. We developed a robust protocol to identify genomic binding sites for DNA-binding factors in mammalian cells that combines immunoprecipitation of cross-linked protein–DNA complexes with DNA microarray analysis of promoters for genes that are cell cycle-regulated. This method was used to identify novel E2F target promoters, which we verified using two independent assays. Our results indicate that E2Fs are involved in regulating genes that encode components of the DNA damage checkpoint and repair pathways, as well as factors involved in chromatin assembly/condensation, chromosome segregation, and the mitotic spindle checkpoint.
Previous experiments have identified E2F targets by profiling genes activated and repressed in response to ectopic expression of E2F1, E2F2, and E2F3 (Ishida et al. 2001; Kalma et al. 2001; Muller et al. 2001). However, two potential limitations of such experiments are introduced by this approach. First, there is the potential for loss of target specificity resulting from overexpression. Second, this type of experiment does not permit a straightforward or unequivocal identification of direct targets, as secondary changes in gene expression resulting from progression through the cell cycle are possible (Ishida et al. 2001). In contrast, the direct binding experiments described here circumvent both complications and allow for the identification of promoters that are bound directly by E2F in primary cells under physiological conditions.
The binding of a transcriptional activator to the promoter region of a gene suggests that the activator has a regulatory effect on the gene, but it is also possible that the activator does not fully or even partially control the gene. For this reason, we have identified the set of genes where factor binding correlates with gene expression, an approach that produced highly accurate information on transcription factor function in previous studies with other factors (Ren et al. 2000). We have also confirmed the importance of E2F binding to several of the novel targets identified here by examining gene expression in cells that lack the p107 and p130 repressors recruited by E2F4. We found that expression of each of these targets was significantly deregulated in cells lacking p107 and p130. These findings confirm the functional relevance of E2F binding in vivo to targets identified in our location analysis.
Classification of E2F targets
Using a novel DNA microarray that allows us to monitor promoter binding for a majority of human genes whose expression is known to oscillate with the cell cycle, we have identified a large number of putative E2F-responsive genes. A substantial number of these E2F target genes have not been previously linked to E2F function. Several important conclusions can be drawn from this work based on the functional clustering of the E2F targets we have identified.
Cell cycle regulation
We identified eight genes with a well-established role in cell cycle control. This cluster includes cyclin A, Cdc2, Cdc25A, CDK2, two members of the E2F family (E2F2 and E2F3), and two members of the RB family (RB and p107). Each of these genes had been regarded as E2F-responsive, and our work confirms that they are, indeed, direct, physiological targets of E2F.
DNA replication
A role for E2F in the activation of several DNA replication genes has been well established. Known E2F targets include genes encoding proteins involved in the initiation of replication (Orc1, Mcm proteins, Cdc6), nucleotide metabolism (thymidine kinase), and the enzymatic synthesis of DNA (DNA polymerase α). Our studies have expanded this list to include additional nucleotide synthesis and replication factor genes. These results are consistent with the work of Nevins and colleagues and others (Ishida et al. 2001; Kalma et al. 2001), who recently identified by expression-profiling several of the genes we have isolated as direct targets of E2F.
Mitosis and mitotic spindle checkpoint
Previous work over the past decade had established a primary role for E2F in regulating the G1/S phase transition, and it is now clear that cyclins important for this transition and a cohort of replication factors and enzymes are physiological targets of E2F. However, an emerging idea from recent studies suggests that E2F may be required to regulate cyclin gene expression beyond S phase (Lukas et al. 1999), and expression-profiling identified several potential downstream targets involved in mitosis (Ishida et al. 2001). However, given the caveats associated with this type of experiment described above, it was not possible to distinguish direct from indirect targets. We identified a significant number of genes that function in mitosis. Among this list were genes known to play a role in cytokinesis (Plk, PRC1), chromosome condensation (SMC4, SMC2), and chromosome segregation (securin or PTTG1, CENP-E, HEC1). Our experiments strongly suggest that E2F plays a direct role in regulating several genes involved in mitosis. It is interesting to note that we have also identified several components of the mitotic spindle checkpoint, including CENP-E, Bub3, and Mad2, proteins that interact with Bub1, which was identified as an E2F target by Nevins and colleagues (Ishida et al. 2001). Thus, our work establishes that E2F directly binds the promoters of genes linked to post-S-phase events and suggests that it plays an important role in their expression.
DNA repair
Interestingly, we identified a large cluster of 12 genes required for DNA repair. These genes are involved in the full spectrum of repair processes, including mismatch repair (MSH2, MLH1), base excision repair (UNG), nucleotide excision repair (RPA3), homologous recombination (RAD51 and RAD54), and nonhomologous end-joining (DNA-dependent protein kinase). These experiments may point to the conservation of a phenomenon first noted in Saccharomyces cerevisiae, namely, that genes involved in repair of DNA damage may be required during replication to ensure genomic integrity (Myung et al. 2001). Many of the genes isolated in this screen are members of multimeric complexes, and our data suggest that they may be coordinately regulated. For example, FEN1–PCNA, Rad51–Rad54, Msh2–Mlh1, and Bard1–Cstf1 interactions have been reported, and several pairs of interacting proteins were found among the other functional categories as well. Many of these genes appear to be bound by E2F4 in quiescent cells when each is inactive and by the E2F1 activator in G1/S phase cells when each is expressed (Fig. 3). Furthermore, our analysis of wild-type and p107−/−; p130−/− MEFs indicates that at least two of these genes (RAD54L and BARD1) are repressed in vivo by these pRB-related proteins during G0 and early G1 (Fig. 5). Consistent with our findings, murine Rad51 was induced severalfold in cells ectopically expressing E2F1 or E2F2 (Ishida et al. 2001). Therefore, our findings link the processes of DNA replication and repair in mammalian cells and suggest that their expression could be regulated through a common factor, E2F.
Checkpoint controls
One of our most surprising findings was the identification of a cluster of genes involved in several different checkpoints. We identified two genes involved in the DNA damage checkpoint, p53 and Chk1. p53 is induced in response to DNA damage and acts to enforce a cell cycle block in G1 phase (Zhou and Elledge 2000). The identification of p53 as an E2F target was unanticipated, because the p53 promoter lacks a recognizable E2F consensus site (data not shown). This finding may be explained by the indirect recruitment of E2F by additional promoter-bound factors. E2F has been shown previously to indirectly increase levels of p53 through activation of the p14ARF gene, a component of the p14ARF–Mdm2 stabilization pathway (DeGregori et al. 1997; Bates et al. 1998; Robertson and Jones 1998). Our results suggest that E2F may also directly control p53 expression levels. This finding is also intriguing in light of previous reports implicating an essential role for both p53 and the pRB family in the G1 DNA damage arrest checkpoint (Harrington et al. 1998; Dannenberg et al. 2000; Sage et al. 2000). The mechanisms underlying the pRB requirement for this G1 block are not known, although a role for E2F-responsive genes has been postulated (Harrington et al. 1998). A second checkpoint gene, Chk1, was also identified in our E2F location analysis. CHK1 is required for the G2 DNA damage (and perhaps an S phase) checkpoint (Zhou and Elledge 2000). Interestingly, pRB was required for Chk1 down-regulation and resumption of G2 after DNA damage, suggesting that E2F could be involved in Chk1 gene expression (Gottifredi et al. 2001).
We also identified several components of the mitotic spindle checkpoint, including CENP-E, Bub3, Mad2, and the TTK kinase (a homolog of yeast Mps1p). These proteins have been shown to interact, again suggesting that this checkpoint may be controlled coordinately by E2F. Interestingly, we found that two of these genes, MAD2L and TTK, as well as several mitotic genes (HEC, NEK2, and PTTG1), were significantly derepressed in quiescent and early G1 cells deficient for the E2F4–p107/p130 repressor.
Taken together with expression-profiling experiments and our analysis of wild-type and mutant MEFs, the above clustering suggests that the DNA damage-response and checkpoint controls may be entrained to the cell cycle through E2F regulation (Fig. 6). That is, each of these repair and checkpoint genes is inactive in cells that have exited the cell cycle and have no immediate need for repair. This inactive state correlates with binding by E2F4. This idea appears to be consistent with recent studies in mouse cells in which it was shown that proliferation was required for repair of mutations (Bielas and Heddle 2000). However, as cells reenter the cell cycle and approach S phase, it is necessary to activate genes needed for repair of the DNA damage that occurs during replication. This period coincides with the time at which E2F is expressed and bound in vivo to genes involved in the DNA replication and repair pathways.
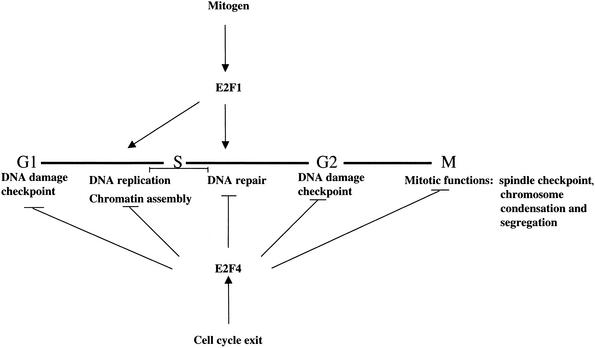
Figure 6.
Model illustrating the pervasive role of E2F in several phases of the cell cycle and in multiple checkpoints. The bold horizontal line denotes the cell cycle. In addition to regulating several cell cycle regulatory genes, including those encoding cyclins, CDKs, other E2Fs, and the pRB family, E2F4 is bound to genes involved in the G1 and G2 DNA damage checkpoints, DNA replication, and DNA repair. E2F4 also binds to genes that function to promote chromosome condensation and segregation as well as the spindle checkpoint. Although E2F1 bound several genes involved in mitosis, most genes bound by this transcription factor in our location analysis clustered in the DNA replication and repair pathways.
Recent gene expression-profiling experiments had also identified a number of genes involved in apoptosis, signaling, and differentiation (Muller et al. 2001). Despite the fact that promoter fragments for a subset of these genes were represented on our cell cycle microarray, it is worth noting that these genes were not identified in our microarray screen. The basis for this discrepancy is unknown at present, although divergent experimental protocols, including the use of different cell lines and ectopic versus endogenous expression of E2Fs, might account for the variance.
Conclusions
We and others showed previously that several genes are regulated by multiple E2F family members (Takahashi et al. 2000; Wells et al. 2000). These experiments allowed us to discriminate between opposing transcriptional signals: E2F4 is recruited to promoters repressed in quiescent cells, whereas E2F1 and E2F3 are enriched at promoters activated during the G1-to-S phase transition. This strongly suggests that the E2F family can be grouped into activator (E2F1, E2F2, and E2F3) and repressor (E2F4 and E2F5) subclasses. The microarray approach described here, in combination with gene expression-profiling experiments, will allow us to extend these findings on a much larger scale. Furthermore, the robust method we have developed to identify genomic binding sites for DNA-binding factors in mammalian cells will be a powerful tool for identifying the genomic loci that are directly regulated by most DNA-associated proteins, including those involved in gene expression and chromosomal maintenance. Such studies in yeast have begun to reveal the transcriptional networks that regulate the cell cycle and the response to nutrients (Ren et al. 2000; Iyer et al. 2001; Simon et al. 2001) and have identified origins of DNA replication throughout the genome (Wyrick et al. 2001). Similar studies can now be envisioned in mammalian cells.
Cancer cells are frequently unable to arrest in G1 in response to DNA-damaging agents (for review, see Zhou and Elledge 2000). Knowledge of the location of key transcription factors on their immediate downstream targets induced by DNA damage may suggest new therapeutic strategies. Indeed, our screen has revealed the direct binding by E2F to several tumor suppressors and other genes whose mutation results in genomic instability and cancer, including securin, mitotic checkpoint genes, and mismatch repair genes (Fishel et al. 1993; Cahill et al. 1998; Jallepalli et al. 2001). A more complete understanding of the targets of cell cycle regulators throughout the human genome will be an important goal for future studies.
Materials and methods
Cell culture
Primary human WI-38 cells were obtained from ATCC. Cells were grown in DMEM containing 10% fetal bovine serum (FBS) and were made quiescent by removal of serum for 3 d. Where indicated, cells were stimulated to enter the cell cycle by adding 10% FBS to the medium. Cell cycle position was monitored by propidium iodide staining and flow cytometry.
1.5K and Affymetrix DNA microarrays
Approximately 900 nucleotide fragments were amplified by PCR from human genomic DNA based on previous cell cycle gene expression-profiling data (Iyer et al. 1999; Ishida et al. 2001). Primer sequences are available upon request, and the identity and sequence for all genes on the 1.5K array are available as Supplemental Data (http://www.genesdev.org). Target preparation for Affymetrix DNA microarrays and hybridization to U95A microarrays were carried out according to the manufacturer's instructions. For the Affymetrix experiments, we identified an induced gene as one that was enhanced ≥2.5-fold relative to 0 h for two consecutive time points.
Amplification, labeling, and hybridization of DNA to 1.5K DNA microarray
The immunoprecipitated (ChIP) DNA and 1/200 of total input DNA were amplified in parallel according to a ligation-mediated PCR protocol (Ren et al. 2000), with the following modifications: (1) After ligation to universal linkers, the DNA was amplified in 25 cycles of PCR, without the addition of fluorescence-labeled dUTP. (2) After PCR, the DNA was first purified with the use of QIAQuick PCR purification kit (QIAGEN).
After amplification, one-half of the resulting DNA was used as template for random priming reactions in the presence of 3 μL of either Cy5-dUTP (Amersham Pharmacia) or Cy3-dUTP (Amersham Pharmacia) with the use of a BioPrime DNA labeling kit (Invitrogen). The Cy5-labeled DNA that was prepared from the immunoprecipitated chromatin sample was combined with Cy3-labeled DNA corresponding to the total input DNA and was then hybridized to a 1.5K DNA microarray chip according to Ren et al. (2000), with modification of the hybridization condition. Each hybridization reaction included the Cy5- and Cy3-labeled DNA, 20 μg of human Cot-1 DNA enriched for repetitive DNA sequences such as the Alu and Kpn family members (Invitrogen), 40 μg of yeast tRNA (Sigma), 2.2× SSC, 0.1% SDS, 0.12% BSA (Sigma), 28% formamide (Sigma), and 5.7% dextran sulfate (Sigma), in a total of 50 μL. The hybridization was carried out in microarrray hybridization chambers (Corning) and incubated at 60°C for 16 h. The microarrays were then washed according to Ren et al. (2000) and scanned with the use of ScanArray5000 scanner (Packard). Detailed protocols are available upon request.
Microarray data analysis
Analysis of microarray scanning images was performed according to Ren et al. (2000). An error model was used to quantify the enrichment of each DNA species by immunoprecipitation. A probability value (P-value) was calculated to indicate the significance of enrichment for each DNA species on the array. Data from independent replicate experiments were combined according to Ren et al. (2000). An average enrichment ratio was calculated for each DNA species on the array from 4–6 replicate data sets. The binding of a factor to DNA is deemed significant if the averaged P-value is <0.002. Using these criteria, no DNA was judged significant in control experiments where input DNA was compared to the same DNA.
Standard chromatin immunoprecipitation
Chromatin immunoprecipitations were performed essentially as described previously (Takahashi et al. 2000), except that the cesium chloride purification step was omitted. Antibodies against E2F1 (sc-193) and E2F4 (sc-1082) were obtained from Santa Cruz. The sequence of primers used to amplify ChIP-enriched DNA is available upon request.
RT-PCR
Mouse embryonic fibroblasts (kind gift of J. Dannenberg and H. te Riele, Netherlands Cancer Institute) were rendered quiescent by starvation for 3 d with DMEM-lacking serum. Cells were stimulated to reenter the cell cycle with the addition of 20% FBS, harvested at various times, and RNA was extracted using Trizol reagent. RT-PCR was performed using Invitrogen RT-PCR Superscript One Step kit as described by the manufacturer. Primer sequences are available upon request. We ensured linear amplification in all cases, and reactions were performed a minimum of two, but most often three, times.
Acknowledgments
We thank E. Balciunaite and J. Rayman for the RNA used in our gene expression-profiling and RT-PCR experiments. We are grateful to J. Dannenberg and H. te Riele for providing wild-type and p107−/−; p130−/− MEFs. B.D.D. is grateful for the support of the American Cancer Society (RPG-0034201CCG) and the Pew Scholars Program in the Biomedical Sciences. B.R is supported by a postdoctoral fellowship from the Helen Hay Whitney Foundation. R.A.Y is supported by funds from NIH and Corning, Inc.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dynlacht@biosun.harvard.edu; FAX (617) 496-1391.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.949802.
References
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- Bielas JH, Heddle JA. Proliferation is necessary for both repair and mutation in transgenic mouse cells. Proc Natl Acad Sci. 2000;97:11391–11396. doi: 10.1073/pnas.190330997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G1 control causing immortalization and increased cell turnover under growth-restricting conditions. Genes & Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol. 2001;21:1066–1076. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes & Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- Harrington EA, Bruce JL, Harlow E, Dyson N. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc Natl Acad Sci. 1998;95:11945–11950. doi: 10.1073/pnas.95.20.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. E2f3 is critical for normal cellular proliferation. Genes & Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- Hurford RK, Jr, Cobrinik D, Lee M-H, Dyson N. PRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes & Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Boguski MS, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:17–18. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, Kinzler KW, Vogelstein B, Lengauer C. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- Kalma Y, Marash L, Lamed Y, Ginsberg D. Expression analysis using DNA microarrays demonstrates that E2F-1 up-regulates expression of DNA replication genes including replication protein A2. Oncogene. 2001;20:1379–1387. doi: 10.1038/sj.onc.1204230. [DOI] [PubMed] [Google Scholar]
- Lindeman GJ, Gaubatz S, Livingston DM, Ginsberg D. The subcellular localization of E2F-4 is cell-cycle dependent. Proc Natl Acad Sci. 1997;94:5095–5100. doi: 10.1073/pnas.94.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Sorensen CS, Kramer E, Santoni-Rugiu E, Lindeneg C, Peters JM, Bartek J, Lukas J. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 1999;401:815–818. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Magae J, Wu CL, Illenye S, Harlow E, Heintz NH. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J Cell Sci. 1996;109:1717–1726. doi: 10.1242/jcs.109.7.1717. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Muller H, Moroni MC, Vigo E, Petersen BO, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes & Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JF, Liu X, Dynlacht BD. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol Cell. 1999;3:195–205. doi: 10.1016/s1097-2765(00)80310-x. [DOI] [PubMed] [Google Scholar]
- Ross JF, Naar A, Cam H, Gregory R, Dynlacht BD. Active repression and E2F inhibition by pRB are biochemically distinguishable. Genes & Dev. 2001;15:392–397. doi: 10.1101/gad.858501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes & Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes & Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Verona R, Moberg K, Estes S, Starz M, Vernon JP, Lees JA. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J, Boyd KE, Fry CJ, Bartley SM, Farnham PJ. Target gene specificity of E2F and pocket protein family members in living cells. Mol Cell Biol. 2000;20:5797–5807. doi: 10.1128/mcb.20.16.5797-5807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick, J.J., Aparicio, J.G., Chen, T., Barnett, J.D., Jennings, E.G., Young, R.A., Bell, S.P., and Aparicio, O.M. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science 294: 2357–2360. [DOI] [PubMed]
- Zhou B-BS, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]