Abstract
Cross-regulatory cascades between hepatic transcription factors have been implicated in the determination of the hepatic phenotype. Analysis of recruitments to regulatory regions and the temporal and spatial expression pattern of the main hepatic regulators during liver development revealed a gradual increase in complexity of autoregulatory and cross-regulatory circuits. Within these circuits we identified a core group of six transcription factors, which regulate the expression of each other and the expression of other downstream hepatic regulators. Changes in the promoter occupancy patterns during development included new recruitments, release, and exchange of specific factors. We also identified promoter and developmental stage-specific dual regulatory functions of certain factors as an important feature of the network. Inactivation of HNF-4α in embryonic, but not in adult, liver resulted in the diminished expression of most hepatic factors, demonstrating that the stability of the network correlates with its complexity. The results illustrate the remarkable flexibility of a self-sustaining transcription factor network, built up by complex dominant and redundant regulatory motifs in developing hepatocytes.
Keywords: Hepatocyte development, transcription factor, regulatory network
The expression of a given array of genes determining the phenotype of each individual cell type is primarily regulated at the transcriptional level. Studies on the regulation of liver-specific genes have identified a set of transcription factor families that govern their tissue-restricted expression. These families, characterized by their structurally related DNA-binding domains, include the variant homeodomain-containing proteins (HNF-1α, HNF-1β); the winged helix family proteins HNF-3α, β, and γ (also called FoxA1, 2, and 3); members of the nuclear hormone receptor family (HNF-4, COUP-TFII, LRH-1, FXRα, and PXR); the basic leucine zipper-containing factor C/EBPα; and the onecut homeodomain protein HNF-6 (Cereghini 1996; Costa et al. 2003). Although none of these factors are expressed exclusively in hepatocytes and none of them alone can induce the hepatic program in nonhepatic cells, transcriptional regulation of most of the liver-specific genes requires a combinatorial action of the above activators. The requirement for such multifactor regulatory schemes was first suggested by the identification of multiple functionally important cis-acting elements on the promoter regions of many liver-specific genes (Cereghini 1996; Costa et al. 2003). A recent genome-wide promoter occupancy study performed in adult human liver further substantiated the combinatorial mode of action of hepatic regulators. >40% of the promoters of active genes were bound by HNF-4α, and most of the promoters bound by HNF-1α or HNF-6 were also occupied by HNF-4α (Odom et al. 2004).
The crucial role of the hepatocyte transcription factors in determining the hepatic phenotype suggests that mechanisms need to be in place for the liver-specific activation and the maintenance of transcription of their own genes. The identification of functional binding sites for the members of the hepatic transcription factor families on their own promoters put forward the idea that these factors establish a complex cross-regulatory network that ensures their high level of expression in hepatic cells. Growing in vivo evidence suggests that such a network is operating in differentiated hepatocytes. For example, reciprocal regulation of the HNF-4α and the HNF-1α genes in hepatic cell lines was demonstrated to be important for their strict coexpression in the same cell (Kuo et al. 1992; Bulla 1997; Bailly et al. 1998). Some in vivo evidence coming from genome-scale studies using a human 13K promoter array also supports the network model. In adult human liver, HNF-1α and HNF-4α were found to occupy one another's promoter, HNF-4α was detected on the LRH-1 and PXR promoters, and HNF-6 and HNF-1α were found on the HNF-4α7 promoter (Odom et al. 2004). In several cases the simultaneous recruitment of two or more factors to the promoters is necessary for high levels of expression, as exemplified by the synergism between C/EBPα and HNF-6 on the mouse HNF-3β gene (Yoshida et al. 2006).
The above results provide compelling initial evidence for the operation of a complex regulatory circuitry in differentiated hepatocytes. In order to elucidate the biological significance of the hepatic transcription factor network, especially its potential role in conferring redundant functions for certain factors, a more complete map of the interactions combined with expression and genetic data is required. Moreover, very little is known about when and how this network is established during hepatic development. This latter question is a fundamental one, since the same set of transcription factors has been shown to play crucial roles at distinct phases of liver development. Specification of hepatoblasts within the endoderm during the initial phases of liver development requires the concerted action of HNF-3α and HNF-3β (Duncan et al. 1998; Zaret 2002; Lemaigre and Zaret 2004; Lee et al. 2005). Gata-6 and HNF-4α1 are dispensable for hepatic specification, but are essential for liver bud expansion and hepatocyte differentiation (Li et al. 2000; Zhao et al. 2005). Control of the lineage split of hepatoblasts into hepatocytes and cholangiocytes involves HNF-6 and HNF-1β (Clotman et al. 2002; Coffinier et al. 2002).
The distinct developmental roles of the above hepatic regulators raise the idea that hierarchical networks may operate during hepatocyte differentiation. This prompted us to map the cross-regulatory interactions between the hepatic regulators during embryonic and postnatal liver development. We analyzed the expression levels of the 12 main hepatic regulators and the occupancy of their regulatory regions by each other after embryonic day (E12.5), when the specification phase is complete and hepatoblast differentiation is taking place. We also performed a similar analysis in HNF-4α-deficient embryonic and adult mouse livers to assess the role of this central activator in the composition and stability of the network. The comprehensive map of interactions, combined with expression studies, reveal that, during the studied developmental periods, cross-regulatory motifs are built up progressively to generate a highly flexible, self-sustaining transcription factor network.
Results
mRNA levels of the hepatic regulators during liver development
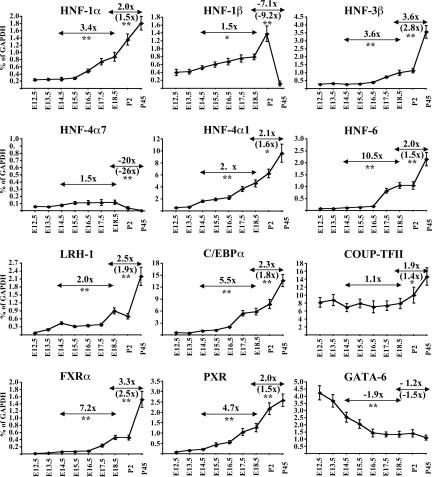
To determine the mRNA levels of the main hepatic transcription factors in developing livers, we performed quantitative RT-PCR assays with RNA samples prepared from pools of embryonic and postnatal mouse livers. In order to obtain a comparative assessment of factor levels, the absolute amounts of mRNAs were measured and expressed relative to the amounts of GAPDH mRNA. At E12.5, transcripts of all factors were already detectable and, with the exception of GATA-6, their levels increased gradually thereafter (Fig. 1). It should be emphasized that even at this earliest stage, the absolute amount of all mRNAs, except that of FXRα, was ≥0.1% of GAPDH mRNA levels. Taking into account that the mRNA levels of the general transcription factor TFIIB in adult liver are ∼0.25% of that of GAPDH mRNA and that the protein products of the hepatic factors can be readily detected in early fetal liver preparations by ChIP, Western blot, and immunofluorescence analysis, we consider that the transcripts detected in fetal livers reflect substantially high expression levels (see below). COUP-TFII mRNA was the most abundantly expressed, but the least induced, transcript between E12.5 and adult (postnatal day 45 [P45]) stage. Compared with other transcripts, C/EBPα and HNF-4α mRNAs were also very highly expressed in adult liver (14% and 9% of GAPDH, respectively). The HNF-4α gene can produce two transcript variants by the use of alternative promoters, which are separated by a distance of 40 kb on chromosome 20 (Nakhei et al. 1998). In agreement with a previous report (Briancon et al. 2004), we were able to detect some HNF-4α7 mRNA in embryonic but not in adult liver. The functional significance of this variant in the liver is dubious, since its mRNA levels were >10-to 40-fold below those detected for HNF-4α1, even at the embryonic stages (Fig. 1). The closely related POU homeodomain-containing factors HNF-1α and HNF-1β were expressed at comparable levels in embryonic livers. HNF-1α mRNA further increased, while the HNF-1β transcript sharply decreased in adults (Fig. 1). Because HNF-1β is known to be expressed in biliary epithelium (Clotman et al. 2002; Coffinier et al. 2002), the observed drop in its expression in adult liver may reflect selective loss of HNF-1β from hepatocytes. Confirming this, histological analyses showed that in E18.5 livers HNF-1β is expressed in both hepatocytes and the developing biliary epithelium, while in the adult liver its expression is restricted to cholangiocytes (Supplemental Fig. 1). As expected from previous reports, HNF-6 protein was detected in both hepatocyte and cholangiocyte nuclei. A similar distribution was observed for HNF-3β, while HNF-4α and HNF-1α expression were restricted to parenchymal hepatocytes (Supplemental Fig. 1). The fetal liver contains a number of other nonhepatic cell types, as it is a main hematopoetic organ. The adult liver also contains endothelial cells and sinusoidal macrophages, besides hepatocytes and cholangiocytes. The different cellular composition of the fetal and adult liver has to be taken into account when comparing the total mRNA levels or occupancy data (see below). One way to eliminate the noise coming from nonhepatic cells would be to isolate primary hepatocytes. We noticed, however, that during a standard hepatocyte isolation procedure the expression levels of some HNFs are reduced as much as 10-fold, which makes the approach unreliable (data not shown). Instead, we estimated the percentage of cells corresponding to hepatocytes by staining with an HNF-4α antibody, which is a widely used hepatocyte marker. Microscopic examination of a number of fields revealed positive staining in 40%–60% of the nuclei in E18.5 livers and in 60%–70% of the nuclei in adult livers (Supplemental Fig. 1). No significant differences could be observed between E14.5 and E18.5 livers (data not shown).
Figure 1.
Changes in mRNA levels of hepatic transcription factors in liver tissues between E12.5 and P45. Graphs show the mean values and standard errors of absolute amounts of mRNAs of the transcription factors compared with the absolute amounts of GAPDH at the indicated stages from three different pools of livers. Numbers above double arrows depict the fold difference between E14.5 and E18.5, or the difference between E18.5 and P45 data. Numbers in parentheses show fold differences normalized to the estimated relative ratios of hepatocytes in E18.5 and P45 livers. Statistical analysis was performed by an unpaired Student's t-test. (*) P < 0.05; (**) P < 0.01. Note the different scales of the Y-axes.
The degree of increase in the levels of most mRNAs was higher between E14.5 and E18.5 d (3.4-fold to 10.5-fold), compared with the increases between E18.5 and P45 d (twofold to 3.6-fold). Taking into account an average of 1.3-fold difference in the percentage of cells corresponding to hepatocytes between E18.5 and P45 time points, the results suggest that the majority of hepatic factors reach near-maximal levels of expression at the late fetal stages of development (Fig. 1, numbers in parentheses).
Recruitment of hepatic transcription factors to the regulatory regions of their genes
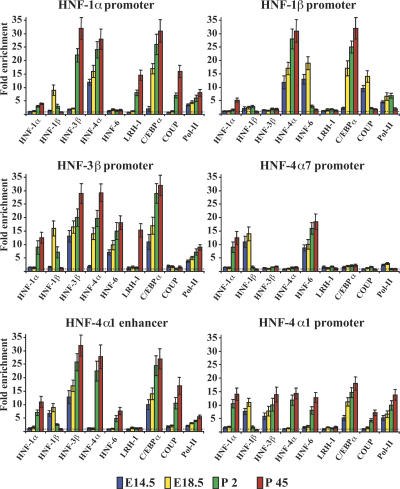
In order to draw a comprehensive cross-occupancy map of the regulatory regions of each of the main hepatic transcription factors, we performed chromatin immuno-precipitation (ChIP) assays in liver tissues derived from E14.5, E18.5, P2, and P45 mice. Soluble cross-linked chromatin was immunoprecipitated with antibodies against HNF-1α, HNF-1β, HNF-3β, HNF-4α, HNF-6, LRH-1, C/EBPα, COUP-TFII, GATA-6, and RNA pol-II, and the presence of sequences corresponding to the regulatory regions of all factors in the precipitated material was then assessed by real-time PCR. After normalization to input, ChIP signals were calculated as fold enrichments over the signals obtained with a nonspecific antibody. To obtain a stringent assessment, we considered recruitment of a factor to be significant if the ChIP signals were at least threefold above the control in all independent experiments. Our analysis did not include the so far unidentified regulatory region of COUP-TFII. Because we were studying the recruitment of factors in normal tissues, the occupancies by FXRα and PXR, which are ligand-dependent, were not tested. The C/EBPα promoter was not bound by any of the core factors (data not shown).
On the HNF-1α promoter, we observed a progressive build-up of factors at the different stages. After the initial recruitment of HNF-4α in E14.5 livers, which was sufficient to recruit RNA pol-II and activate transcription, HNF-1β and C/EBPα were added at E18.5, followed by the postnatal association of HNF-1α, HNF-3β, LRH-1, and COUP-TFII, with concomitant dissociation of HNF-1β (Fig. 2; Supplemental Table 1A). The HNF-1β promoter was initially bound by HNF-4α, HNF-6, and COUP-TFII, followed by the recruitment of C/EBPα at E18.5. In adult livers, where the gene is active only in biliary epithelial cells, HNF-6 and COUP-TFII were dissociated from the promoter, while HNF-1α associated with it. At this stage the signal obtained with RNA pol-II antibody was at background levels, suggesting that the contribution of potential recruitments in biliary epithelial cells, where the gene is active, to the ChIP signals detected is negligible (Supplemental Fig. 1).
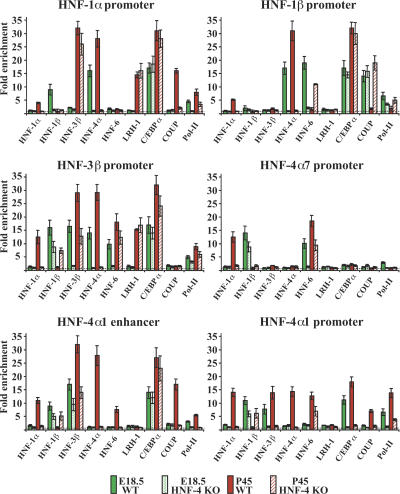
Figure 2.
Analysis of transcription factor recruitment to the HNF-1α, HNF-1β, HNF-3β, and HNF-4α regulatory regions. Chromatin immunoprecipitations with antibodies against the different transcription factors, shown below the X-axis, were performed in cross-linked chromatin prepared from E14.5, E18.5, P2, and P45 livers. The data from qPCR reactions (with primer sets shown in Supplemental Table 3) were first normalized to the input and expressed as fold enrichment over those obtained with nonimmune serum, which were set at value 1 (dashed horizontal line). Bars show mean values and standard errors from experiments performed with three different pools of livers. Qualitative assessment of the occupancy data is also presented in Supplemental Table 1.
An expanding occupancy pattern was observed at the HNF-3β regulatory region, which was bound by HNF-3β, HNF-6, and C/EBPα in E14.5 livers and recruited HNF-1β and HNF-4α at E18.5 d. In adult hepatocytes, LRH-1 was also associated with it, while HNF-1β was replaced by HNF-1α.
On the HNF-4α7 promoter, some RNA pol-II could be detected in fetal livers along with HNF-1β and HNF-6. In postnatal samples, where the gene is inactive, HNF-6 remained bound to the promoter, and HNF-1β was replaced by HNF-1α.
In fetal liver samples, the DNA immunoprecipitated by antibodies against factors that specifically bind to the HNF-4α1 enhancer (C/EBPα and HNF-3β) or the HNF-4α1 proximal promoter (RNA pol-II) contained sequences encompassing both the distant enhancer and proximal promoter region. The simultaneous presence of the two DNA fragments in the immunoprecipitates of these factors demonstrates that the enhancer and the promoter regions are in close proximity and form a higher order complex by looping out the intervening DNA (Hatzis and Talianidis 2002). Recruitment of HNF-1β, but not HNF-1α, which has binding sites at both regions, was also evident at these stages. In postnatal livers, additional factors, including HNF-6, HNF-4α, and COUP-TFII, were recruited and an exchange of HNF-1α for HNF-1β occurred (Fig. 2; Supplemental Table 1).
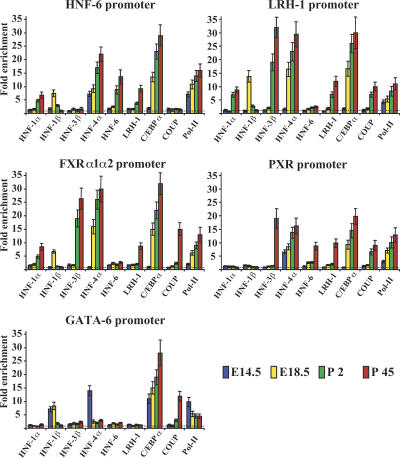
HNF-4α was the only factor occupying the HNF-6 promoter in E14.5 livers, joined by HNF-1β and C/EBPα at E18.5 and postnatally by HNF-6 and LRH-1. In this case, too, we observed an exchange of HNF-1α for HNF-1β (Fig. 3; Supplemental Table 1).
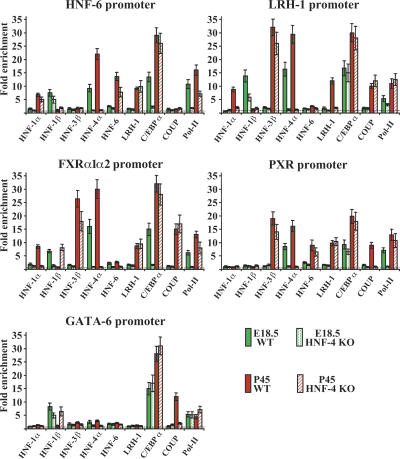
Figure 3.
Analysis of transcription factor recruitment to the HNF-6, LRH-1, FXRα1α2, PXR, and GATA-6 promoter regions. The graphs show the data of chromatin immunoprecipitation assays with the indicated antibodies and are presented as in Figure 2.
At E14.5, the LRH-1 promoter was not occupied by any of the studied factors. RNA pol-II recruitment was already detectable at this stage, suggesting that other regulators govern the initial activation of this gene. Recruitment of HNF-1β, HNF-4α, and C/EBPα occurred at E18.5, followed by the replacement of HNF-1β with HNF-1α and the recruitment of HNF-3β, LRH-1, and COUP-TFII in the postnatal periods.
RNA pol-II recruitment along with HNF-1β, HNF-4α, and C/EBPα was first detected at the FXRα1α2 promoter at E18.5. After birth, HNF-1α replaced HNF-1β, and HNF-3β was added, while LRH-1 and COUP-TFII association could also be detected in adults.
The PXR promoter was initially occupied by HNF-4α. Recruitment of C/EBPα and COUP-TFII occurred at E18.5 and P2, respectively. Association of additional factors, including HNF-3β, HNF-6, and LRH-1, was observed only in adult livers. HNF-1β, HNF-4, and C/EBPα occupied the GATA-6 promoter at E14.5, when the gene was most active. HNF-4α and HNF-1β dissociated from the promoter at E18.5 and P2, respectively. In adult liver we could also detect association by COUP-TFII (Fig. 3; Supplemental Table 1).
The above patterns of associations and dissociations of factors with the regulatory regions demonstrate the dynamic nature of the processes that generate a highly complex interconnected network in developing hepatocytes. Careful examination of the experimental data reveals two important general features. First, we note that RNA pol-II occupancy of promoters was always positively correlated with the transcript levels of the genes. Second, the absolute values of the ChIP signals exhibited promoter-specific and factor expression-specific variations. Variations in the absolute signals between different antibodies and different promoter regions were expected, given the varying IP efficiencies of the different antibodies, the differential accessibility of the epitopes, or the differences in the cross-linking efficiency of the proteins to the individual promoter regions and also the small differences (average 1.3-fold) in the proportion of hepatocytes in the total cell populations, as explained in the previous section. We also noted a qualitative, but not quantitative, correlation between the expression of the transcription factors and the occupancy data. For example, CEBPα expression increased 12.6-fold between E14.5 and P45, but the difference in the absolute occupancy values on the HNF-3β promoter was only about threefold. This suggests that the levels of the factors in fetal livers are sufficient to occupy one or both alleles of the regulators in the majority of the cells. Although, as described above, variations can be explained by the differential accessibility or cross-linking of the protein in a more complex PIC, we are tempted to speculate that the large increases in expression of the individual factors may also play a role in providing a factor supply for the increasing number of downstream targets activated during liver development.
The role of HNF-4 α in the stability of the hepatic cross-regulatory network
Because of the well documented central role of HNF-4α in hepatocyte differentiation after the specification stage (Parviz et al. 2003), we examined how its loss would affect the hepatic regulatory network in embryonic and adult liver. We crossed HNF-4loxP mice (Hayhurst et al. 2001), in which exons 4 and 5 are flanked by loxP sites, with Alfp-Cre (Coffinier et al. 2002) or Alb-Cre (Hayhurst et al. 2001) transgenic mice to obtain liver-specific HNF-4-deficient mice in embryonic or adult stages, respectively. RT-PCR, Western blot, and histological analysis revealed loss of HNF-4α at E15.5 in the livers of HNF-4lox/lox/Alfp-Cre mice and between P35 and P40 in the livers of HNF-4lox/lox/Alb-Cre mice (see also Supplemental Fig. 1D).
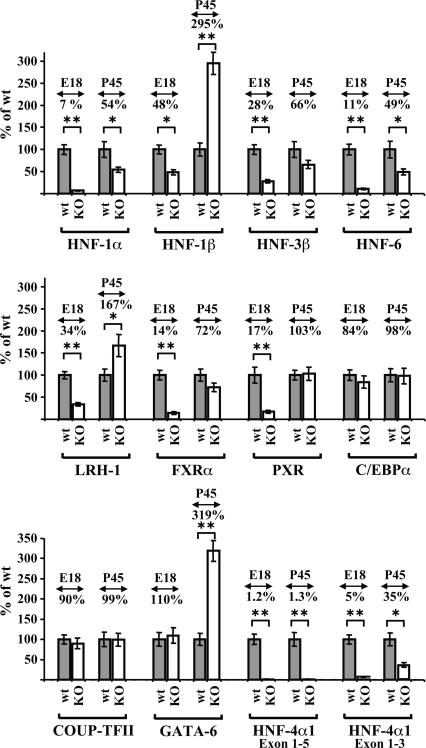
Loss of HNF-4α in adult livers either did not influence at all, or only marginally affected (at most twofold) the expression of the other hepatic factors (Fig. 4). Interestingly, HNF-1β and GATA-6 expression was significantly induced in the adult HNF-4α-deficient livers. In the case of HNF-1β, the above activation reflects re-expression of the gene in hepatocytes, rather than an increase of its expression in biliary epithelium (Supplemental Fig. 1A). In contrast with adults, inactivation of HNF-4α in embryonic livers resulted in a dramatic decrease of the mRNA levels of most hepatic factors (Fig. 4). With the exception of C/EBPα, COUP-TFII, and GATA-6, whose mRNA levels were not significantly changed, the expression of HNF-1α, HNF-1β, HNF-3β, HNF-6, LRH-1, FXRα, and PXR was reduced to 7%, 48%, 28%, 11%, 34%, 14%, and 17% of the control, respectively.
Figure 4.
Effects of HNF-4α inactivation on the expression of hepatic regulators in fetal and adult livers. Quantitative RTPCR reactions using pooled total RNAs from embryonic (E18.5) and adult (P45) livers as in Figure 1. The liver tissues were from wild-type (wt) animals (gray bars) or HNF-4α-deficient (KO) animals (white bars). HNF-4α KO livers were from HNF-4lox/lox/ Alfp-Cre mice (E18.5) and HNF-4lox/lox/Alb-Cre (P45) mice, as indicated. The bars represent mean values, expressed as a percentage of those obtained with wild-type samples, and standard errors from three different pools. Numbers above the bars depict the average percentages of mRNA levels in HNF-4α KO livers compared with wild-type livers. Statistical analysis was performed by an unpaired Student's t-test. (*) P < 0.05; (**) P < 0.01.
In both animal models, deletion of exons 4 and 5 results in the splicing of exon 3 to exon 6, which may encode a truncated mRNA with a premature stop codon at amino acid position 127. This enabled us to evaluate the effect of HNF-4α inactivation on its own transcription by using appropriate primer sets. Figure 4 shows that while the full-length HNF-4α1 mRNA dropped to basal levels in both animal models, some truncated transcript, corresponding to 35% of the control, could be detected in adult HNF-4α KO livers. It should be noted that reduced stability of the truncated transcript due to nonsense-mediated decay might also contribute partly to the above decrease.
The above differential effects prompted us to study how the promoter occupancy patterns of the hepatic regulators are affected by the depletion of HNF-4α. In adult livers, loss of HNF-4α led to partial changes in the array of factors occupying the different promoters. Several factors, despite their relatively lower levels of expression, could still be identified on promoters by a chromatin immunoprecipitation assay.
In the absence of HNF-4α, the HNF-1α promoter was still bound by HNF-3β, LRH-1, and C/EBPα, but HNF-1α and COUP-TFII were dissociated from it (Fig. 5; Supplemental Table 1). These factors are perhaps necessary for optimal RNA pol-II recruitment, but not sufficient for it. In contrast, C/EBPα alone, the only factor remaining associated with the HNF-1α promoter in embryonic HNF-4α KO hepatocytes, was unable to recruit RNA pol-II in the absence of the other regulators. We detected rerecruitment of HNF-6 and COUP-TFII to the reactivated HNF-1β promoter in adult HNF-4α KO mice, with concomitant release of HNF-1α, which together with HNF-4α acts as a repressor on this gene (Supplemental Fig. 2). This could constitute the molecular basis of re-expression of HNF-1β in adult HNF-4-deficient hepatocytes.
Figure 5.
Transcription factor recruitment to the HNF-1α, HNF-1β, HNF-3β, and HNF-4α regulatory regions in wild-type and HNF-4-deficient mouse livers. Chromatin immunoprecipitations with the indicated antibodies were performed in pooled liver extracts prepared from wild-type E18.5 embryos (E18.5 WT), HNF-4lox/lox/Alfp-Cre E18.5 embryos (E18.5 HNF-4 KO), wild-type 45-d-old mice (P45 WT) and HNF-4lox/lox/Alb-Cre 45-d-old mice (P45 HNF-4 KO), as indicated. Bars show mean values and standard errors from experiments performed with three different pools of livers. Qualitative assessment of the occupancy data is also presented in Supplemental Table 1
In adult livers, HNF-1β replaced HNF-1α on the HNF-3β promoter, while all other factors remained associated with it. In contrast, only HNF-1β and C/EBPα bound this regulatory region in embryonic HNF-4α KO livers. The HNF-4α7 promoter retained either HNF-6 or HNF-1β, but neither of them was able to support RNA pol-II recruitment.
The selective recruitment of HNF-1β, HNF-3β, and C/EBPα to the enhancer region and the selective recruitment of HNF-1β and HNF-6 to the promoter region of the HNF-4α gene in adult HNF-4α KO mice indicate that these regulatory regions adopted a linear conformation. Although in this case we could observe some RNA pol-II association and transcription, a similar configuration adopted in embryonic HNF-4α KO livers that lacked HNF-6 binding did not recruit RNA pol-II.
In adult HNF-4α KO livers, only HNF-4α was absent from the HNF-6 promoter. In embryonic liver, however, both HNF-4α and C/EBPα were dissociated from it (Fig. 6; Supplemental Table 1).
Figure 6.
Transcription factor recruitment to the HNF-6, LRH-1, FXRα1α2, PXR, and GATA-6 promoters in wild-type and HNF-4-deficient mouse livers. The graphs show the data of chromatin immunoprecipitation assays with the indicated antibodies and are presented as in Figure 5.
The LRH-1 promoter retained HNF-3β, C/EBPα, and COUP-TFII and became somewhat more active in adult liver. On the other hand, loss of HNF-4α in embryonic liver did not affect the binding of the other two factors (HNF-1β and C/EBPα). In this case the gene was less active, pointing to a dual function of HNF-4α on this gene.
The FXRα promoter was cleared of all factors in embryonic liver, but retained HNF-3β, LRH-1, C/EBPα, and COUP-TFII along with the reassociation of HNF-1β in adult HNF-4 KO livers.
Inactivation of HNF-4α did not affect HNF-3β, HNF-6, LRH-1, and C/EBPα recruitment to the PXR promoter in adult livers. In fetal livers, however, binding of C/EBPα was not sufficient to recruit RNA pol-II in the absence of HNF-4α.
On the GATA-6 promoter, the only difference observed was the dissociation of COUP-TFII in adult HNF-4α KO liver, suggesting that this factor has a negative modulatory effect, since GATA-6 expression was induced in this animal model.
Taken together, comparison of the data obtained with wild-type and the two HNF-4α KO mice shows a general correlation between the extent of the multiplicity of factors binding to the different regulatory regions and the extent of RNA pol-II recruitment, or mRNA levels. Furthermore, the actual combination of the specific HNFs associated with the promoters can influence the transcriptional status of the genes in both positive and negative directions.
Discussion
The overall gene expression profile of hepatic cells is subject to continuous changes during liver development and in response to environmental stimuli. During development, hepatocyte differentiation is controlled by the same small set of transcription factors, which are involved in the regulation of genes in the fully differentiated adult liver (Duncan 2000; Zaret 2002). The timing of the initial expression and the functional properties of the main hepatic regulators, however, indicate that hierarchical regulatory events should be involved during the early phases of liver development. For example, HNF-3 (FoxA) proteins are already expressed in endodermal progenitor cells and play a role in the specification of the hepatic lineage (Bossard and Zaret 2000; Lee et al. 2005). A characteristic feature of the HNF-3 proteins is their intrinsic ability to open highly condensed nucleosomal arrays, which is consistent with their “pioneer” activity in establishing a competent state on the regulatory regions of their targets at the specification phase of liver development (Cirillo et al. 2002). HNF-4α is activated later and required for hepatoblast differentiation, while HNF-1α is induced in a subsequent step of hepatocyte differentiation (Cereghini et al. 1992; Duncan 2000; Zaret 2002). Such hierarchical cascades, in which one hepatic factor induces the expression of the other, may eventually culminate into a cross-regulatory network. Analysis of the promoter sequences of the main hepatic regulators with the Genomatix MatInspector library identifies binding sites for many of the other regulators (Supplemental Table 2). The functional importance of many of these sites has been previously demonstrated by reporter assays in cultured cells. Furthermore, some of the potential interactions between the hepatic regulators and their promoter regions have been confirmed in vivo by chromatin immunoprecipitation approaches (Hatzis and Talianidis 2002; Odom et al. 2004). The results of this study provide a complete map of cross-regulatory interactions between the 12 main hepatic transcription factors. Combined with the analysis of the temporal and spatial expression patterns of the regulators during liver development and the studies in mice lacking one of the key hepatic regulators (HNF-4α), the maps provide novel insights into the biological role of the hepatic transcription factor network.
Increasing complexity of the hepatic regulatory network during liver development
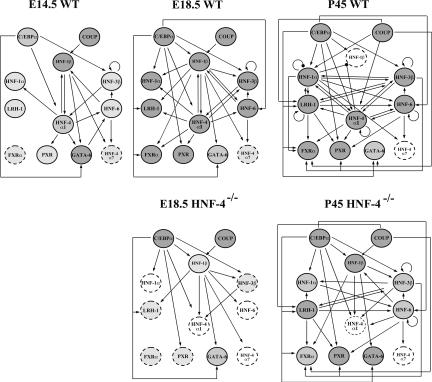
The results concerning the promoter occupancy and expression patterns of the main hepatic regulators during embryonic and postnatal liver development are summarized schematically in Figure 7. Some of the recruitment events have also been observed in human and mouse liver (Odom et al. 2004; Tan et al. 2006), while some others have been predicted by transfection assays in cultured cells, or the phenotypic analysis of mouse knockout models. At first glance, it is apparent that the number of cross-regulatory interactions between the studied factors increases progressively after E14.5, a stage at which the cells are already committed to differentiation toward the hepatocyte fate. Already at the E14.5 stage, several regulatory motifs, including single-input (e.g., HNF-4α1-HNF-1α or HNF-4α1-PXR), double-input (e.g., HNF1β/HNF-3β-HNF-4α1 or HNF-4α1/GATA-6-HNF-6), multi-input (e.g., HNF-4α1/HNF-6/COUP-TFII-HNF-1β), and autoregulatory (HNF-3β) motifs can be drawn. At the E18.5 stage, single-input motifs that may be integrated into regulatory chains evolve to multi-input motifs and reach a much higher complexity in adult liver, dominated by multicomponent circuits and auto-regulatory loops. This suggests that during liver development hierarchical interactions are progressively replaced by combinatorial regulatory circuits, which build up a highly complex cross-regulatory network. Within this network we identify a core group of six transcription factors (HNF-1α, HNF-1β, HNF-3β, HNF-4α1, HNF-6, and LRH-1), which occupy the regulatory regions of each other and peripheral members (FXRα, PXR, GATA-6, HNF-4α7), which are occupied by the core group, or factors that associate with the promoters of the core group, but are controlled independently of them (C/EBPα and COUP-TFII).
Figure 7.
Schematic presentations of cross-regulatory interactions between the hepatic regulators. Maps of promoter occupancies in livers of the indicated animals were drawn based on the chromatin immunoprecipitation data. Relative expression levels of the individual regulators are depicted as follows: (white dashed circles) no expression; (light-gray dashed circles) low expression; (gray full circles) increased expression; (dark-gray full circles) high expression.
The hepatic regulators can influence each other's expression in both positive and negative manners
The increased complexity in promoter occupancies was in most cases in good correlation with the increased expression of the individual transcription factors. However, exceptions to this rule could be identified, which sheds light into important biological phenomena. For example, in the adult liver the HNF-4α7 promoter was occupied by HNF-1α and HNF-6, but this promoter was inactive as evidenced by the lack of RNA pol-II recruitment and the absence of the mRNA coding for this transcript variant. On the other hand, some, albeit very low, HNF-4α7 mRNA could be observed in embryonic livers, where the promoter was occupied by HNF-1β and HNF-6, suggesting that minor changes in the composition of factors can alter the transcription status of the gene. Although we do not know why the recruitment of HNF-1α and HNF-6 is not sufficient to support an active preinitiation complex on this gene, the above finding demonstrates that relying solely on promoter occupancy data could lead to erroneous interpretations concerning regulatory actions. Factor occupancy in this and other cases may have functions other than transcriptional activation, such as keeping the locus competent for recruiting other activators under certain physiological conditions.
The above scenario also applies to the inactivation of the HNF-1β gene in adult hepatocytes, in which case our results provide a better mechanistic explanation. HNF-1α and HNF-1β share extensive homology in their DNA-binding and dimerization domains and can bind to the same DNA sequence as homo-or heterodimers (Cereghini 1996). The sequential activation of their genes and the more widespread expression of HNF-1β suggested that the two proteins may have different developmental roles (Cereghini et al. 1992). In agreement with this we found that the two factors are coexpressed in fetal but not in adult hepatocytes. Interestingly, however, HNF-1α was not recruited to the promoters of any of the other regulators in fetal liver, although it was associated with several of its downstream targets (data not shown). Instead, HNF-1β occupied most of the regulatory regions in this study. In adult hepatocytes, where HNF-1β is not expressed, HNF-1α had replaced it on all promoters except that of GATA-6. These switches in occupancy patterns suggest that the two factors can complement the function of each other. Corroborating this scenario are the results obtained in adult HNF-4 KO livers, where HNF-1β is re-expressed in hepatocytes and replaced HNF-1α on the regulatory regions of HNF-3β and HNF-4α1. In addition, ectopic expression of HNF-1α in HNF-1β-deficient embryoid bodies could restore visceral endoderm formation, suggesting that such complementation can also occur on downstream targets (Haumaitre et al. 2003).
Our results also provide insights into the intriguing mechanism involved in HNF-1β repression in adult hepatocytes. We have previously shown that HNF-1α can negatively modulate HNF-4α-regulated genes, including its own gene, via physical interaction with HNF-4α (Ktistaki and Talianidis 1997a). We have demonstrated that such a mechanism can operate only on promoters that lack a functional HNF-1-binding site. Our ChIP analysis in adult liver shows that HNF-1α may indeed modulate its own expression, as it was recruited to its own promoter. HNF-1α, however, is expressed in adult hepatocytes, suggesting that it fine tunes, rather than completely represses, its own expression. Lack of such indirect negative autoregulatory feedback in HNF-4α-deficient hepatocytes may in part be responsible for the small changes in its expression. The progressive recruitment of factors on the HNF-1α promoter, starting with HNF-4α at or before E14.5, and the differential increase of HNF-1α mRNA levels confirm that HNF-4α plays a dual role on its control. During the initial developmental period HNF-4α is an essential activator of the HNF-1α gene, while in the adult liver it may also provide a platform of recruitment of HNF-1α, which has a negative modulatory effect.
Several lines of evidence suggest that a similar mechanism maybe responsible for the hepatocyte-specific inactivation of the HNF-1β gene during postnatal liver development. First, the HNF-1β promoter does not contain a functional HNF-1-binding site, and, unlike in embryos, it was co-occupied by HNF-1α and HNF-4α1 in adult livers. Second, in transient transfection assays performed in Hepa 1–6 cells, which endogenously express HNF-4α1, HNF-1α, and HNF-1β, overexpression of either HNF-4α1 or HNF-1α reduced the activity of the HNF-1β promoter. The repression was more pronounced when both HNF-4α1 and HNF-1α were overexpressed (Supplemental Fig. 2). Third, HNF-1α was not recruited to the HNF-1β promoter in HNF-4α1-deficient adult hepatocytes, where HNF-1β expression persists. Fourth, in the livers of HNF-1α KO mice, increased levels of HNF-1β mRNA have been observed (Pontoglio et al. 1996).
COUP-TFII has also been known to exert positive and negative effects on transcription depending on promoter context (Ktistaki and Talianidis 1997b). It can coactivate target genes, such as HNF-1α, via interaction with HNF-4 (Ktistaki and Talianidis 1997b), or HNF-1β, via interactions with Oct-1 (Power and Cereghini 1996), but when bound to DNA (e.g., in the case of the GATA-6 promoter) it inhibits transcription. Taking these findings into account, the correlation between COUP-TFII recruitment onto the HNF-1β, HNF-1α, and GATA-6 genes and their altered transcription status provides an in vivo confirmation of the dual roles of this factor on its targets.
Based on the above, we propose that the promoter-specific dual function of certain factors plays an important role in controlling the overall expression pattern of the hepatic regulators during liver development.
The complexity of the hepatic cross-regulatory network correlates with the stability of expression of the individual factors
One possible outcome of an increasing number of transcription factors occupying a given gene is the generation of a preinitiation complex that can better support multiple rounds of RNA pol-II recycling, which is manifested by an increased rate of transcription. High-level expression of the individual regulators, in turn, is a prerequisite for an increased number of recruitments to the promoters. Although, as discussed above, we found in most cases a good correlation between the number of factors occupying the studied genes and their steady-state mRNA levels during hepatic development, such a correlation was more evident between E14.5 and E18.5 d embryos. The less than twofold increases observed in the expression of several regulators between the E18.5 and P45 stages points to additional biological significance of the enhanced complexity of the network.
Our results suggest that the multiple regulatory interactions within the hepatic network also contribute to the stability of expression of the individual regulators. A complex network may provide selective advantages to ensure reduced responses to different environmental stimuli, which may transiently affect the expression levels, or the activity of a single transcription factor, thus contributing to the maintenance of the overall phenotype. This scenario is clearly supported by our results from liver-specific HNF-4 KO mice. Inactivation of HNF-α in E18.5 livers resulted in a much more dramatic decrease in the expression of the other hepatic factors compared with that observed upon HNF-4α inactivation at the adult stage. Differential sensitivity in expression was correlated with the complexity of the promoter occupancy maps at the two stages.
Our studies in HNF-4α KO mice models also provide a number of interesting mechanistic insights into the regulation of the individual factors. One regulatory event that caught our attention concerned the mechanism involved in the autoregulation of the HNF-4α gene. Both embryonic and adult stage-specific inactivation of HNF-4α led to the disruption of enhancer–promoter communication, although the recruitment of most of the other factors was not affected. The linear configuration was able to support some transcription in adult hepatocytes, where HNF-6 was still recruited into the proximal promoter region, but not in fetal hepatocytes that lacked HNF-6. This points to a novel autoregulatory mechanism in which the product of a gene influences the higher order structure of its distant regulatory regions required for maximal transcription.
The relationship between the complexity of the cross-regulatory network and the stability of the hepatic gene expression program also provides explanations for previous observations concerning the functional redundancy of hepatic regulators in several animal models. Inactivation of most of the hepatic factors at stages after hepatic lineage specification did not affect the overall liver phenotype. For example, inactivation of HNF-3β in hepatocytes affects the expression of only very few genes (Sund et al. 2000; Rubins et al. 2005). In HNF-1α KO mice the liver develops normally and, although the expression of several downstream metabolic genes is affected, the overall liver phenotype is preserved (Pontoglio et al. 1996; Lee et al. 1998; Shih et al. 2001a). Importantly, with the exception of HNF-6 and FXRα, the expression of other hepatic regulators is not reduced by the absence of HNF-1α (Shih et al. 2001a, b). Similarly, hepatocyte-specific inactivation of HNF-4α in the liver causes altered expression of several downstream genes, but the overall liver phenotype is less affected (Hayhurst et al. 2001), compared with the much more complex phenotype, including defective liver architecture caused by embryonic inactivation of HNF-4α, where the hepatic cross-regulatory network is less complex and the genes of several hepatic regulators are repressed (Parviz et al. 2003). HNF-6 and HNF-1β inactivation has been shown to affect the development of the biliary epithelium, but not the expression of the other hepatic regulators and the majority of hepatic genes (Clotman et al. 2002; Coffinier et al. 2002). Last but not least, haploinsufficiency caused by heterozygous mutations of HNF-1α and HNF-4α in MODY patients affects mainly pancreatic b-cell function and less liver function (Servitja and Ferrer 2004).
In summary, the results of this study point to the progressive build-up of a highly interconnected transcription factor network during hepatocyte development. The network exhibits an unusually high level of plasticity in terms of regulatory effects, which provides the molecular basis for observations pertaining to dominant and redundant functions of its individual members.
Materials and methods
Animals and histological analysis
HNF-4loxP, Alb-Cre (Hayhurst et al. 2001), and Alfp-Cre (Kellendonk et al. 2000) mice were backcrossed to CBA-CAxC57Bl/ 10 background, maintained in grouped cages in a temperature-controlled virus-free facility on a 12-h light/dark cycle, and fed by standard chow diet and water ad libitum. Further breedings were performed in order to obtain HNF-4lox/lox/Alfp-Cre and HNF-4lox/lox/Alb-Cre mice. The excision of exons 4 and 5 of the HNF-4α gene was verified by PCR analysis of DNA prepared from livers. Time course analysis by RT-PCR revealed loss of HNF-4α mRNA from the E15.5 d fetal liver in HNF-4lox/lox/ Alfp-Cre mice and between P35 and P40 in the livers of HNF-4lox/lox/Alb-Cre mice. Unaltered expression of HNF-4α in other organs, including kidney, intestine, and pancreas in both animal models was verified by RT-PCR and immunohistochemistry. All the data presented for wild-type animals were obtained with the CBA-CAxC57Bl/10 strain. In most cases our analysis included HNF-4lox/lox, HNF-4lox/wt, Alfp-Cre, or Alb-Cre mice and obtained the same results as those of wild-type animals.
For histological analysis, livers were fixed in either 4% paraformaldehyde or 100% methanol and embedded in paraffin. Liver sections (5-to 6-μm thick) were boiled in 10 mM Nacitrate for 20 min and after washings with PBS were blocked with either normal goat serum or 1% BSA in PBS. Incubations with primary antibodies and AlexaFluor 568 or AlexaFluor 555 secondary antibodies and counterstaining with DAPI were performed as described (Boulias et al. 2005). The primary antibodies used for staining were anti-HNF-1β polyclonal mouse serum raised against a bacterially expressed His-tagged C-terminal (amino acids 374–557) fragment of the HNF-1β protein, rabbit HNF-4α polyclonal antibody described in Hatzis and Talianidis (2001), and antibodies from Santa Cruz Biotechnology (HNF-1α, H-140; HNF-3β, M-20; HNF-6, H-100).
mRNA analysis and chromatin immunoprecipitation assays
Total RNAs were prepared from livers by the guanidinium isothiocyanate method and, after digestion with DNAse I, were further purified using the RNeasy kit (Qiagen). Reverse transcription and quantitative RT-PCR analysis was performed as described (Hatzis and Talianidis 2001). Each experiment was performed with three different RNA preparations from different pools of livers. The number of livers in each pool was as follows: 20 for E12.5, E13.5, and E14.5; 15 for E15.5 and E16.5; 10 for E17.5 and E18.5; five for P2 and P45. The nucleotide sequences of primer sets used for PCR are shown in Supplemental Table 3. In order to obtain an estimate of the absolute amounts of mRNAs and eliminate variations in amplification efficiencies of the different primer sets, we first purified the amplified products after separation by agarose gels, and after quantitation and serial dilutions, real-time PCR reactions were performed to obtain a standard curve. The amounts of each mRNA from the experimental data were then calculated using a linear regression curve equation and expressed as a percentage of total amounts of GAPDH mRNA. The average GAPDH mRNA levels did not show variations during liver development, which was further confirmed by comparisons with two additional controls (β-actin and acidic ribosomal phosphoprotein 36B4).
Chromatin immunoprecipitation assays were performed as described (Soutoglou and Talianidis 2002; Boulias et al. 2005) with the following modifications. Liver tissue was minced to small pieces in PBS and after addition of formaldehyde to a 1% final concentration immediately was subjected to 10 strokes of dounce homogenization. Cross-linking was continued for 10 min and stopped by the addition of glycine at 0.125 M final concentration. Cross-linked nuclei were purified by centrifugation through a sucrose gradient (Kouskouti and Talianidis 2005) and after extraction were sonicated to obtain medium-length DNA fragments ranging from 500 bp to 2000 bp. The antibodies used for immunoprecipitations were the rabbit polyclonal antibodies against HNF-4α, COUP-TFII, and LRH-1 described in Hatzis and Talianidis (2001), Boulias et al. (2005), and Chalkiadaki and Talianidis (2005) and antibodies from Santa Cruz Bio-technology against HNF-1α (H-140), HNF-1β (H-85), HNF-3β (M-20), HNF-6 (H-100), C/EBPα (14AA), and RNA pol-II (H-224). The specificity of each antibody was verified by IP-Western blot assays using cross-linked extracts. Each assay was performed with three different pools of liver tissue. The number of livers in each pool was the same as described for RNA preparations. Quantitative PCR reactions were performed with at least two primer sets for each promoter amplifying adjacent regions. Because of the intentionally chosen low resolution that comes from the large fragment sizes of the sonicated DNA, we ob-tained similar results with each consecutive primer set; for simplicity only the results obtained with one set of primers are shown. All of the amplification data were first normalized to input (nonimmunoprecipitated chromatin) and expressed as fold enrichment over those obtained with immunoprecipitations using a nonimmune serum.
Acknowledgments
We thank S. Cereghini for providing the HNF-1β promoter-containing reporter plasmid. This work was supported by a fellowship from the Onassis Foundation to I.K. and grants from GSRT (PENED-03ED542) and EU (LSHG-CT-2004-502950 and MTKD-CT2005 029610).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.390906.
Supplemental material is available at http://www.genesdev.org.
References
- Bailly A., Spath G., Bender V., Weiss M.C. Phenotypic effects of the forced expression of HNF4 and HNF1α are conditioned by properties of the recipient cell. J. Cell Sci. 1998;111:2411–2421. doi: 10.1242/jcs.111.16.2411. [DOI] [PubMed] [Google Scholar]
- Bossard P., Zaret K.S. Repressive and restrictive mesodermal interactions with gut endoderm: Possible relation to Meckel's Diverticulum. Development. 2000;127:4915–4923. doi: 10.1242/dev.127.22.4915. [DOI] [PubMed] [Google Scholar]
- Boulias K., Katrakili N., Bamberg K., Underhill P., Greenfield A., Talianidis I. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J. 2005;24:2624–2633. doi: 10.1038/sj.emboj.7600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briancon N., Bailly A., Clotman F., Jacquemin P., Lemaigre F.P., Weiss M.C. Expression of the α7 isoform of hepatocyte nuclear factor (HNF) 4 is activated by HNF6/ OC-2 and HNF1 and repressed by HNF4α1 in the liver. J. Biol. Chem. 2004;279:33398–33408. doi: 10.1074/jbc.M405312200. [DOI] [PubMed] [Google Scholar]
- Bulla G.A. Hepatocyte nuclear factor-4 prevents silencing of hepatocyte nuclear factor-1 expression in hepatoma x fibroblast cell hybrids. Nucleic Acids Res. 1997;25:2501–2508. doi: 10.1093/nar/25.12.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;10:267–282. [PubMed] [Google Scholar]
- Cereghini S., Ott M.O., Power S., Maury M. Expression patterns of vHNF1 and HNF1 homeoproteins in early postimplantation embryos suggest distinct and sequential developmental roles. Development. 1992;116:783–797. doi: 10.1242/dev.116.3.783. [DOI] [PubMed] [Google Scholar]
- Chalkiadaki A., Talianidis I. SUMO-dependent compartmentalization in promyelocytic leukemia protein nuclear bodies prevents the access of LRH-1 to chromatin. Mol. Cell. Biol. 2005;25:5095–5105. doi: 10.1128/MCB.25.12.5095-5105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., Zaret K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Clotman F., Lannoy V.J., Reber M., Cereghini S., Cassiman D., Jacquemin P., Roskams T., Rousseau G.G., Lemaigre F.P. The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development. 2002;129:1819–1828. doi: 10.1242/dev.129.8.1819. [DOI] [PubMed] [Google Scholar]
- Coffinier C., Gresh L., Fiette L., Tronche F., Schutz G., Babinet C., Pontoglio M., Yaniv M., Barra J. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1β. Development. 2002;129:1829–1838. doi: 10.1242/dev.129.8.1829. [DOI] [PubMed] [Google Scholar]
- Costa R.H., Kalinichenko V.V., Holterman A.X., Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- Duncan S.A. Transcriptional regulation of liver development. Dev. Dyn. 2000;219:131–142. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1051>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- Duncan S.A., Navas M.A., Dufort D., Rossant J., Stoffel M. Regulation of a transcription factor network required for differentiation and metabolism. Science. 1998;281:692–695. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]
- Hatzis P., Talianidis I. Regulatory mechanisms controlling human hepatocyte nuclear factor 4alpha gene expression. Mol. Cell. Biol. 2001;21:7320–7330. doi: 10.1128/MCB.21.21.7320-7330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— 2002Dynamics of enhancer-promoter communication during differentiation-induced gene activation Mol. Cell 101467–1477. [DOI] [PubMed] [Google Scholar]
- Haumaitre C., Reber M., Cereghini S. Functions of HNF1 family members in differentiation of the visceral endoderm cell lineage. J. Biol. Chem. 2003;278:40933–40942. doi: 10.1074/jbc.M304372200. [DOI] [PubMed] [Google Scholar]
- Hayhurst G.P., Lee Y.H., Lambert G., Ward J.M., Gonzalez F.J. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellendonk C., Opherk C., Anlag K., Schutz G., Tronche F. Hepatocyte-specific expression of Cre recombinase. Genesis. 2000;26:151–153. doi: 10.1002/(sici)1526-968x(200002)26:2<151::aid-gene17>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Kouskouti A., Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ktistaki E., Talianidis I. Modulation of hepatic gene expression by hepatocyte nuclear factor 1. Science. 1997a;277:109–112. doi: 10.1126/science.277.5322.109. [DOI] [PubMed] [Google Scholar]
- ———1997bChicken ovalbumin upstream promoter transcription factors act as auxiliary cofactors for hepatocyte nuclear factor 4 and enhance hepatic gene expression Mol. Cell. Biol. 172790–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.J., Conley P.B., Chen L., Sladek F.M., Darnell J.E., Jr., Crabtree G.R. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- Lee Y.H., Sauer B., Gonzalez F.J. Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1α knockout mouse. Mol. Cell. Biol. 1998;18:3059–3068. doi: 10.1128/mcb.18.5.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.S., Friedman J.R., Fulmer J.T., Kaestner K.H. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Lemaigre F., Zaret K.S. Liver development update: New embryo models, cell lineage control, and morphogenesis. Curr. Opin. Genet. Dev. 2004;14:582–590. doi: 10.1016/j.gde.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Li J., Ning G., Duncan S.A. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes & Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- Nakhei H., Lingott A., Lemm I., Ryffel G.U. An alternative splice variant of the tissue specific transcription factor HNF4α predominates in undifferentiated murine cell types. Nucleic Acids Res. 1998;26:497–504. doi: 10.1093/nar/26.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom D.T., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F., Matullo C., Garrison W.D., Savatski L., Adamson J.W., Ning G., Kaestner K.H., Rossi J.M., Zaret K.S., Duncan S.A. Hepatocyte nuclear factor 4α controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- Pontoglio M., Barra J., Hadchouel M., Doyen A., Kress C., Bach J.P., Babinet C., Yaniv M. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- Power S.C., Cereghini S. Positive regulation of the vHNF1 promoter by the orphan receptors COUP-TF1/Ear3 and COUP-TFII/Arp1. Mol. Cell. Biol. 1996;16:778–791. doi: 10.1128/mcb.16.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubins N.E., Friedman J.R., Le P.P., Zhang L., Brestelli J., Kaestner K.H. Transcriptional networks in the liver: Hepatocyte nuclear factor 6 function is largely independent of Foxa2. Mol. Cell. Biol. 2005;25:7069–7077. doi: 10.1128/MCB.25.16.7069-7077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servitja J.M., Ferrer J. Transcriptional networks controlling pancreatic development and β cell function. Diabetologia. 2004;47:597–613. doi: 10.1007/s00125-004-1368-9. [DOI] [PubMed] [Google Scholar]
- Shih D.Q., Bussen M., Sehayek E., Ananthanarayanan M., Shneider B.L., Suchy F.J., Shefer S., Bollileni J.S., Gonzalez F.J., Breslow J.L., et al. Hepatocyte nuclear factor-1α is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 2001a;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- Shih D.Q., Screenan S., Munoz K.N., Philipson L., Pontoglio M., Yaniv M., Polonsky K.S., Stoffel M. Loss of HNF-1α function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes. 2001b;50:2472–2480. doi: 10.2337/diabetes.50.11.2472. [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- Sund N.J., Ang S.L., Sackett S.D., Shen W., Daigle N., Magnuson M.A., Kaestner K.H. Hepatocyte nuclear factor 3β (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Mol. Cell. Biol. 2000;20:5175–5183. doi: 10.1128/mcb.20.14.5175-5183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Yoshida Y., Hughes D.E., Costa R.H. Increased expression of hepatocyte nuclear factor 6 stimulates hepatocyte proliferation during mouse liver regeneration. Gastroenterology. 2006;130:1283–1300. doi: 10.1053/j.gastro.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Hughes D.E., Rausa F.M., 3rd, Kim I.M., Tan Y., Darlington G.J., Costa R.H. C/EBPα and HNF6 protein complex formation stimulates HNF6-dependent transcription by CBP coactivator recruitment in HepG2 cells. Hepatology. 2006;43:276–286. doi: 10.1002/hep.21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K.S. Regulatory phases of early liver development:Paradigms of organogenesis. Nat. Rev. Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- Zhao R., Watt A.J., Li J., Luebke-Wheeler J., Morrisey E.E., Duncan S.A. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol. Cell. Biol. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]