Abstract
The σ28 protein is a member of the bacterial σ70-family of transcription factors that directs RNA polymerase to flagellar late (class 3) promoters. The σ28 protein is regulated in response to flagellar assembly by the anti-σ28 factor FlgM. FlgM inhibits σ28-dependent transcription of genes whose products are needed late in assembly until the flagellar basal motor structure, the hook-basal body (HBB), is constructed. A second function for the σ28 transcription factor has been discovered: σ28 facilitates the secretion of FlgM through the HBB, acting as the FlgM Type III secretion chaperone. Transcription-specific mutants in σ28 were isolated that remained competent for FlgM-facilitated secretion separating the transcription and secretion-facilitation activities of σ 28. Conversely, we also describe the isolation of mutants in σ28 that are specific for FlgM-facilitated secretion. The data demonstrate that σ28 is the Type III secretion chaperone for its own anti-sigma factor FlgM. Thus, a novel role for a σ70-family transcription factor is described.
Keywords: Flagella, gene regulation, Type III secretion
The bacterial flagellum is a self-assembling macromolecular machine anchored in the bacterial membrane that allows bacteria to move through liquid environments or crawl along surfaces (Macnab 1992). Flagellar assembly and function is a complex process, which in Salmonella typhimurium involves over 60 genes (Frye et al. 2006). The construction of individual flagella requires an ordered assembly pathway (Macnab 1992). The assembly process involves the secretion of individual subunits through the hollow core of the growing flagellum where they assemble at the tip of the structure. In addition, multi-flagellated bacteria can have individual flagella in the same cell at different stages of assembly (Bardy et al. 2003). Consequently, the assembly process is tightly regulated at the levels of substrate selection by the associated secretion apparatus and coupled gene regulation. This paper reports an unexpected discovery related to the current understanding of how a cell regulates flagellar gene expression in response to intermediate stages of flagellar assembly. A flagellar-specific transcription factor, σ28, plays a dual role as both a regulator of gene expression and a facilitator of flagellar-specific Type III secretion.
The bacterial flagellum consists of three major substructures: (1) the basal body, which acts as motor anchoring the flagellum within the cell membranes; (2) the hook, which acts as a flexible, universal joint between the basal body; and (3) the long external filament, which acts as a propeller when rotated (Berg and Anderson 1973; Macnab 1999). Self-assembly of a flagellum begins at the inner membrane and proceeds out of the cell with construction of the basal structure. A flagellar-specific Type III secretion (T3S) apparatus is assembled within the cytoplasmic membrane at the base of the basal structure (Aldridge and Hughes 2002). Individual structural subunits are then secreted from the cytoplasm into the growing flagellum by the flagellar T3S system. Efficient flagellar assembly requires that the T3S apparatus distinguish between different secretion substrates at different stages of assembly. Recent studies have shown that there is a multi-layered regulatory network in place that couples temporal expression and delivery of flagellar subunits at the right moment to the growing flagellar structures (Aldridge and Hughes 2002).
In S. typhimurium, the flagellar genes are organized into a transcriptional hierarchy composed of three promoter classes (class 1, 2, and 3) (Kutsukake et al. 1990). The class 1 promoters drive the expression of the flagellar master operon, which encodes the transcription factors FlhD and FlhC. A FlhD2C2 heterodimer directs σ70-RNA polymerase to initiate transcription from class 2 promoters (Liu and Matsumura 1994). Genes transcribed from class 2 promoters encode the structural subunits of the hook-basal body (HBB) and a number of regulatory factors that include the flagellar-specific transcription factor σ28 and its inhibitor, the anti-σ28 factor FlgM. Class 3 promoters require σ28-RNA polymerase for transcription (Ohnishi et al. 1992). Flagellar class 3 operons encode genes whose products are required late in flagellar assembly, including the flagellin filament subunit and genes of the chemosensory system (Chilcott and Hughes 2000).
A major checkpoint in flagellar assembly is the completion of the hook-basal body (HBB) structure. Associated with HBB completion are two crucial regulatory steps that couple flagellar gene expression to flagellar assembly. First, upon HBB completion, the flagellar specific T3S apparatus undergoes a substrate specificity switch from hook-rod secretion substrate specificity to late secretion substrate specificity, including flagellin filament subunits (Minamino et al. 1999; Muramoto et al. 1999). The anti-σ28 factor FlgM is also a late secretion substrate, and FlgM secretion is the signal that the HBB is complete (Gillen and Hughes 1991a). Prior to HBB completion, σ28 activity is inhibited by direct interaction with FlgM. The physical binding of σ28 to FlgM prevents σ28 from interacting with DNA or core RNA polymerase (Daughdrill et al. 1997; Chadsey et al. 1998; Sorenson et al. 2004). Secretion of FlgM from the cell by completed HBB structures (Hughes et al. 1993) releases σ28 to transcribe the late flagellar genes, which are needed only after HBB completion. Thus, FlgM secretion is coupled to HBB completion, which releases σ28 to transcribe the class 3 flagellin subunit genes, but only when completed HBB structures are present onto which flagellin must polymerize outside the cell (Karlinsey et al. 2000b).
The relative cellular levels of FlgM and σ28 are tightly controlled. Both the flgM gene and the σ28 structural gene, fliA, are transcribed from class 2 and class 3 flagellar promoters. Upon HBB completion FlgM is secreted and both flgM and fliA genes are transcribed from their class 3 promoters until a steady state is reached between FlgM and σ28 that balances expression and FlgM-inhibition of σ28-dependent class 3 transcription. Upon HBB completion, FlgM secretion will result in a reduction of the intracellular concentration of FlgM. However σ28-dependent flgM transcription has the ability to immediately compensate for this drop by increasing class 3 flgM gene expression.
The flagellar-associated T3S apparatus is not restricted to flagellar assembly. Virulence-associated T3S systems facilitate the secretion of virulence factors either into the surrounding environment or directly into host cells (Stebbins and Galan 2003). The secretion of all T3S substrates is influenced by at least one of three factors. All T3S substrates possess an N-terminal secretion signal that is strictly required for recognition and secretion by any given T3S apparatus (Namba 2001). Two other factors, Type III secretion-facilitators, referred to as T3S-chaperone proteins, and mRNA signals play a greater role in regulating the timing and location of secretion rather than secretion per se (Karlinsey et al. 2000a; Lee and Galan 2004).
T3S-chaperones are defined as a family of proteins that bind specific T3S substrates and are required for their efficient secretion (Parsot et al. 2003). Unfortunately, the nomenclature leads to confusion between the T3S-chaperone protein family and with the families of molecular chaperones such as the chaperonin GroEL, whose major role is to assist the folding of proteins (Ellis 2005; Swain and Gierasch 2005). In contrast, the T3S-chaperones facilitate the secretion of their bound substrates by guiding them to the associated T3S apparatus and even hold these substrates in an unfolded state prior to secretion. Recent evidence suggests that the T3S-chaperones play a crucial role in T3S-pathway specificity in cells possessing both flagellar and virulence-associated T3S systems. Removal of a specific T3S-chaperone from the cell by mutation allows the cognate secretion substrate to be secreted by either flagellar or virulence T3S systems (Lee and Galan 2004; Lilic et al. 2006). In S. typhimurium there are three T3S-chaperones, FlgN, FliT, and FliS, which are only required for efficient secretion of their cognate substrates (Fraser et al. 1999; Auvray et al. 2001; Aldridge et al. 2003). To date, the only flagellar late secretion substrate without a defined T3S-chaperone is the anti-σ28 factor FlgM.
In this paper we demonstrate that σ28 acts as the FlgM T3S-chaperone. Using a genetic screen based on the current knowledge of the σ28:FlgM interaction, two classes of σ28 mutant proteins were isolated. One class is defective as a transcription factor but functional as a facilitator of FlgM secretion. The second class is defective in FlgM secretion but still functional as a transcriptional factor. The major class of FlgM secretion-defective σ28 mutant proteins is altered in the ability to interact with FlgM. The implications these findings have for general substrate secretion by T3S systems are discussed.
Results
Known flagellar T3S-chaperones do not affect FlgM secretion
The flagellar system of Salmonella includes six late secretion substrates, which are secreted only after HBB completion. These are the hook-filament junction proteins FlgK and FlgL, the filament cap FliD, the flagellins FliC and FljB, and the anti-σ28 factor FlgM (Aldridge and Hughes 2002). Of these six proteins only two lack a defined T3S-chaperone, FljB and FlgM. In S. typhimurium only one flagellin gene is expressed at a given time due to phase variation (Bonifield and Hughes 2003). The flagellin T3S-chaperone, FliS, binds a peptide fragment containing the last 186 amino acids of FliC (Auvray et al. 2001). This fragment is very homologous to the C terminus of FljB with 82% amino acid identity (McClelland et al. 2001), suggesting that FljB can also interact with the T3S-chaperone FliS. This leaves FlgM with no obvious T3S-chaperone candidate.
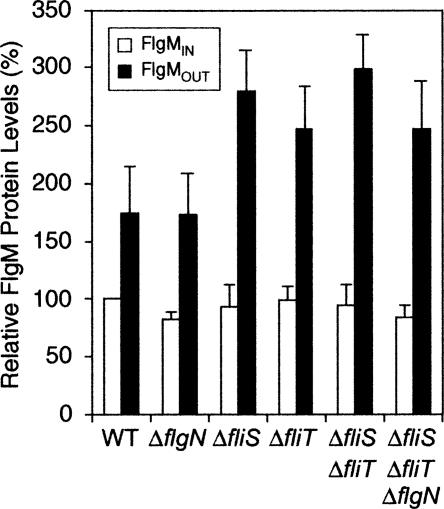
All flagellar late secretion substrates besides FlgM require a T3S-chaperone for efficient secretion. To determine if a known T3S-chaperone (FlgN, FliT, or FliS) acts on FlgM, secretion of FlgM in single, double, and triple flgN, fliT, and fliS null mutant strains was compared with secretion in a wild-type strain (Fig. 1). In the FlgM secretion assays, we compared the internal concentration of FlgM, defined as FlgMIN, with the external concentration defined as FlgMOUT. FlgMIN was the level of FlgM detected in whole cell lysates, while FlgMOUT was the level of FlgM present in supernatants of mid-log phase cultures (see Materials and Methods). No combination of flgN, fliT, and fliS null mutants resulted in a defect in FlgM secretion. In fact, FlgM secretion was enhanced in the T3S-chaperone mutant strains probably because of reduced competition for the secretion apparatus by substrates that were less abundant in the absence of their cognate T3S-chaperone. These results demonstrate that FlgM secretion is not dependent on the known flagellar T3S-chaperones.
Figure 1.
Known flagellar T3S-chaperones do not facilitate FlgM secretion. A graph showing the relative amount of intracellular FlgM (FlgMIN—unshaded bars) compared with extracellular FlgM levels (FlgMOUT—black bars). Secretion assays were performed as described in Materials and Methods. FlgMIN is defined as the level of FlgM present in whole cell lysates of mid-log phase cultures, while FlgMOUT is defined as the amount of FlgM detected in the supernatant of the same culture. The secretion assays were performed in a wild-type background (HBB+ fliC +) using null mutants of all three known T3S-chaperones compared with the wild-type strain LT2 (TH437): ΔflgN = TH5937; ΔfliS = TH5737; ΔfliT = TH5831; ΔfliS ΔfliT = TH5935; ΔfliS ΔfliT ΔflgN = TH5999. All data, including error bars, are shown relative to LT2 FlgMIN levels. The data shown are an average of three independent repeats of the secretion assays.
FlgM secretion is dependent upon σ28
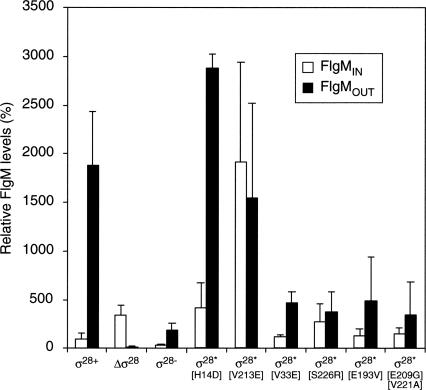
The fact that σ28 interacts with FlgM and that known T3S-chaperones do not promote FlgM secretion suggests that σ28 itself may be a candidate T3S-chaperone for FlgM. FlgM secretion was tested in the absence of σ28 using a strain deleted for the σ28 structural gene, fliA (designated as ΔΔσ28 in Fig. 2). For clarity, σ28 is referred to by its protein name (σ28) rather than by its gene name (fliA). In contrast to null mutations in flgN, fliT, and fliS, the absence of σ28 led to a significant reduction in FlgM secretion (Fig. 2). Compared with wild type, levels of FlgM outside the cell (FlgMOUT) in the absence of σ28 (Δσ28) were down 117-fold, while FlgMIN increased threefold. These data suggest that either σ28 itself or a protein dependent on σ28 for transcription facilitates FlgM secretion.
Figure 2.
FlgM secretion is dependent upon the σ28 protein. A graph showing the FlgM secretion profiles for different σ28 mutant alleles compared with σ28+. All secretion assays were performed in a strain that had a HBB+ FliC− Hin− phenotype (σ28+) where fliC expression is “ON” (Bonifield and Hughes 2003). The data shown are an average of three independent repeats and are represented as relative to FlgMIN levels for σ28+. Strains used are highlighted in Table 3 with a full description of each geno-type.
To distinguish between the two possibilities that either σ28 directly facilitates FlgM secretion or another protein dependent on σ28 for its production, σ28 derivatives that were unable to bind FlgM, but still able to bind RNA polymerase and direct flagellar class 3 promoter transcription, were tested for the ability to facilitate FlgM secretion. Previously, single amino acid substitutions in σ28 were isolated that bypass the inhibitory effect of FlgM (Chilcott and Hughes 1998; Chadsey and Hughes 2001). A bypass mutant works in the presence of an inhibitor or activator to give the same phenotype as loss of that inhibitor or activator. A mutation in σ28 that allows σ28-dependent flagellar class 3 transcription in the presence of inhibitory concentrations of FlgM (such as in HBB mutants unable to secrete FlgM) is a FlgM-bypass mutation. Such mutations in σ28 are defined as σ28* (FlgM-bypass) mutant proteins (Chadsey and Hughes 2001). Two classes of σ28* proteins were obtained. One class is defective in FlgM binding, while the second class of σ28* mutant proteins was more stable to proteolysis. Both σ28* mutant types result in an excess of free σ28 in the cell and accompanying σ28-dependent transcription under FlgM inhibitory conditions. If σ28 directly facilitates FlgM secretion, then a σ28* protein defective in FlgM binding (such as the σ28*[V213E] mutant protein) would be defective in FlgM secretion, while the more stable σ28* protein that binds FlgM similar to wild-type σ28* (such as σ28*[H14D]) would facilitate FlgM secretion. If another protein dependent on σ28 for its production facilitates FlgM secretion, then FlgM secretion should occur at high levels in strains with either σ28* mutant type (FlgM-binding defective σ28 or more stable σ28 protein) expressed. The σ28*[V213E] mutant protein is 70-fold reduced in its ability to bind FlgM, while the σ28*[H14D] mutant binds FlgM with wild-type efficiency but is a more stable σ28 protein (Chadsey and Hughes 2001). The secretion of FlgM in the presence of σ28*[V213E] and σ28*[H14D] was compared with secretion in the presence of wild-type σ28 (Fig. 2). For σ28*[H14D], no significant change in the secretion profile of FlgM was detected apart from a fourfold increase in FlgMIN presumably due to increased class 3 flgM gene transcription by σ28*[H14D]-RNA polymerase (Chadsey and Hughes 2001).
In contrast to FlgM secretion in the presence of σ28*[H14D], a change in FlgM secretion was observed for σ28*[V213E], which is defective in binding FlgM. No significant change to FlgMOUT was observed for σ28*[V213E], whereas FlgMIN increased 19-fold presumably resulting from increased class 3 flgM gene transcription by σ28*[V213E]-RNA polymerase (Chadsey and Hughes 2001). These data suggest that, even though FlgM is secreted in the presence of σ28*[V213E], secretion is significantly reduced and some of the secretion defect is compensated by a large increase of FlgMIN. This is consistent with previous findings that the lack of the FlgN T3S-chaperone can be overcome by the overexpression of its cognate secretion substrates (FlgK and FlgL) (Aldridge et al. 2003). Thus FlgM secretion is dependent on its ability to bind to σ28 and not on σ28-dependent transcription of an unknown FlgM T3S-chaperone gene.
FlgM and σ28 stability are interdependent
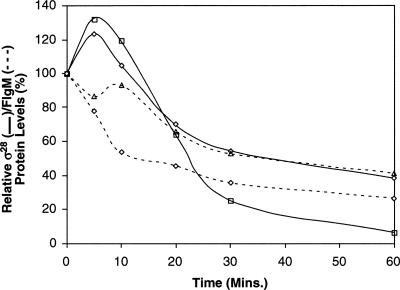
Many T3S substrates are relatively unstable prior to secretion in the absence of their cognate T3S-chaperone (Bennett and Hughes 2000; Aldridge et al. 2003). To investigate whether protein stability of FlgM and σ28 has a role in FlgM secretion, the stabilities of both proteins were tested in wild-type, ΔflgM, and Δσ28 strains (Fig. 3; see Supplemental Material for protein half-life calculations). σ28 was slightly more stable in the presence of FlgM; its half-life increased from 24 min when FlgM was absent to 30 min. Significantly, after 60 min ∼5% of σ28 is detected in the ΔflgM strain whereas ∼40% of the detectable σ28 protein is present in the flgM + strain. In contrast, FlgM was more stable in the absence of σ28 increasing from 18 min in the wild-type strain to 32 min in the Δσ28 strain. However, these strains are competent for FlgM secretion (HBB+ backgrounds), and therefore FlgM turnover also includes FlgM secretion for σ28+ strains as only whole cell lysates were assayed. Recently FlgM was demonstrated to be a stable protein (Aldridge et al. 2006). This suggests that the interaction of FlgM with σ28 facilitates its secretion and not its degradation. Thus, for FlgM, the changes observed are a reflection of FlgM accumulation in cells when σ28 is absent, consistent with our hypothesis that σ28 facilitates its secretion.
Figure 3.
The stabilities of FlgM and σ28 are interdependent. The stabilities of σ28 (solid lines) and FlgM (dashed lines) were followed after growth was stopped at mid-log (OD600 = 0.6–0.8) by the addition of spectinomycin to inhibit protein synthesis (Aldridge et al. 2003). Stability assays were performed for three independent repeats. Average protein levels were calculated as relative values of the T0 time point for each sample. σ28 was more stable over time in the flgM+ strain (solid-line diamonds [strain TPA368]) when compared with the ΔflgM strain (solid line-squares [strain TPA378]). In contrast, FlgM was much more stable in the absence of σ28 (cf. σ28+ background, dashed-line diamonds [strain TPA368], with the Δσ28 mutant, dashed-line triangles [TPA376]). A degree of the change in FlgM stability in all these backgrounds is due to secretion rather than stability. Average OD600 values for all strains used over the 60 min plus the calculations used for the protein half-lives can be found in the Supplemental Material.
Separation of σ28-dependent FlgM secretion from σ28 transcription
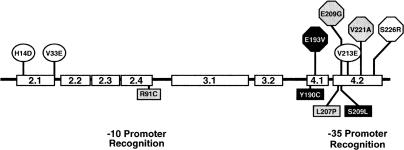
We wanted to determine if the transcription activity of σ28 could be genetically separated from its FlgM secretion-facilitator activity. The first step was the isolation of mutants in σ28 specific to its flagellar class 3 transcription activity, defined as σ28 −. To obtain σ28 − mutants, a deletion of the σ28 structural gene, fliA (ΔfliA5805::tetRA), was replaced with PCR mutagenized fliA-containing DNA fragments using the λ-RED recombination system (see Materials and Methods; Karlinsey and Hughes 2006). The ΔfliA5805::tetRA allele is a deletion of the entire fliA gene replaced with a tetracycline-resistance element, tetRA, of transposon Tn10. A positive selection for loss of tetracycline resistance allows for the direct selection of recombinants in which the ΔfliA5805::tetRA allele was replaced with the mutagenized DNA that includes fliA (Maloy and Nunn 1981). Tetracycline-sensitive (TcS) recombinants were screened, in the absence of flgM, for those defective in transcription of the σ28-dependent (flagellar class 3) motA gene using a lac operon transcriptional fusion vector in motA (motA5461::MudJ). From 697 TcS transformants, two Lac− isolates that had wild-type levels of σ28 protein were isolated. Subsequent DNA sequence analysis revealed that both were double mutants: σ28 −[R91C L207P] and σ28 −[Y190C S209L]. The σ28 −[R91C L207P] mutant had an amino acid substitution located in each of the –10 and –35 promoter recognition regions of σ28 while σ28 −[Y190C S209L] had two amino acid substitutions located in the –35 promoter recognition region (Fig. 4). Interestingly no single amino acid changes were isolated, suggesting that complete loss of transcriptional activity required more than one change in σ28. Furthermore, because the screen demanded that the transcription-defective σ28 mutants had a wild-type protein stability, it is likely that all transcription-defective mutants resulting from defects in interactions with RNA polymerase were unstable since none was isolated.
Figure 4.
σ28 mutants isolated or used during this study. Circled residues: σ28 mutants isolated previously shown biochemically to have wild-type H14D (σ28*[H14D]) or altered FlgM binding properties V33E and V213E (σ28*[V213E]) (Chadsey and Hughes 2001). Squares: σ28− alleles used to show that the two activities of σ28 are independent—σ28−[R91C L207P] shaded in gray; σ28−[Y190C S209L] shaded in black. Hexagons: characterized σ28* mutants isolated during the FlgM-secretion mutant screen—σ28*[E193V], black hexagon; σ28*[S226R], white hexagon; σ28*[E209G, V221A], gray hexagons.
The transcription-defective σ28 proteins were found to be competent at facilitating FlgM secretion. FlgM secretion assays using σ28−[R91C L207P] when compared with wild type and Δσ28 showed that, even though FlgMIN was significantly reduced (presumably due to loss of flgM gene transcription from its σ28-dependent class 3 promoter), σ28−[R91C L207P] did facilitate FlgM secretion (Fig. 2). Thus, we were able to separate the transcription and FlgM T3S-chaperone activities of σ28 by mutation. Therefore, the transcriptional function of σ28 is not essential for σ28 to facilitate FlgM secretion.
Genetic screen rationale for the isolation of FlgM secretion-defective mutants in σ28
The ability to isolate transcription-specific mutants in σ28 demonstrated that its two functions could be separated. This suggested that a region of σ28 might be specific to the facilitation of FlgM secretion (T3S-chaperone activity). It is known that a twofold increase in the intracellular FlgM concentration results in a 100-fold reduction of σ28-dependent class 3 transcription due to the stoichiometric nature of the interaction between σ28 and FlgM (Karlinsey et al. 2000a). The above data predict that when σ28 is unable to facilitate FlgM secretion, then FlgM would accumulate in the cell (Figs. 2,3) and result in reduced σ28-dependent class 3 promoter activity. Unfortunately, this phenotype (decreased class 3 transcription) is the same as σ28 mutants that are defective in transcription. Therefore, we needed to develop a genetic screen that we could use to separate transcription mutants from FlgM-secretion mutants.
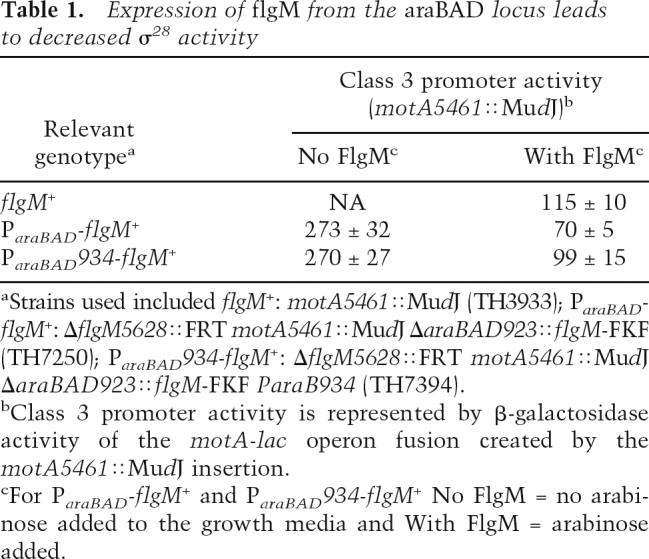
Mutants in σ28 defective in FlgM secretion (defined here as σ28†) are expected to show reduced class 3 transcription only in the presence of FlgM while transcription-specific mutants should exhibit reduced class 3 transcription in the presence and absence of FlgM. This required a method that allowed for the isolation of σ28 mutants exhibiting lower class 3 promoter activity in the presence of wild-type FlgMIN levels followed by a screen for the restoration of class 3 transcription in the absence of FlgM. For this screen, a strain background was constructed where flgM gene expression was independent of σ28 transcription. The flgM+ coding region was placed under the control of the arabinose promoter ParaBAD at the araCBAD locus (see Materials and Methods; Supplemental Material). This allowed for the induction of flgM + expression independent of flagellar assembly by the addition of arabinose to the growth medium. In this construct, expression of flgM+ from the arabinose promoter resulted in too much intracellular FlgM. Compared with wild-type expression of flgM+ from its normal chromosomal location, expression of ParaBAD-flgM+ resulted in reduced class 3 promoter expression by 40% (Table 1). Unfortunately, this was already the phenotype expected for FlgM-secretion σ28 mutants.
Table 1.
Expression of flgM from the araBAD locus leads to decreased σ28 activity
In order to express the flgM+ gene from the ParaBAD promoter to levels comparable with what is expressed from its native promoters, the ParaBAD promoter was mutagenized and screened for FlgM anti-σ28 activity using the motA5461::MudJ as a reporter of class 3 promoter activity (see Materials and Methods). Four ParaBAD promoter mutants were obtained that exhibited a Lac-phenotype from the motA5461::MudJ reporter similar to what we observed in a wild-type flgM + strain. These were further characterized by DNA sequence analysis (see Supplemental Material). One mutant, ParaBAD934-flgM+, resulted from a TA to CT double change between the −35 and −10 boxes and exhibited class 3 transcription activity that was comparable with wild type and was subsequently used in all further experiments.
Isolating σ28 mutants defective in FlgM-secretion
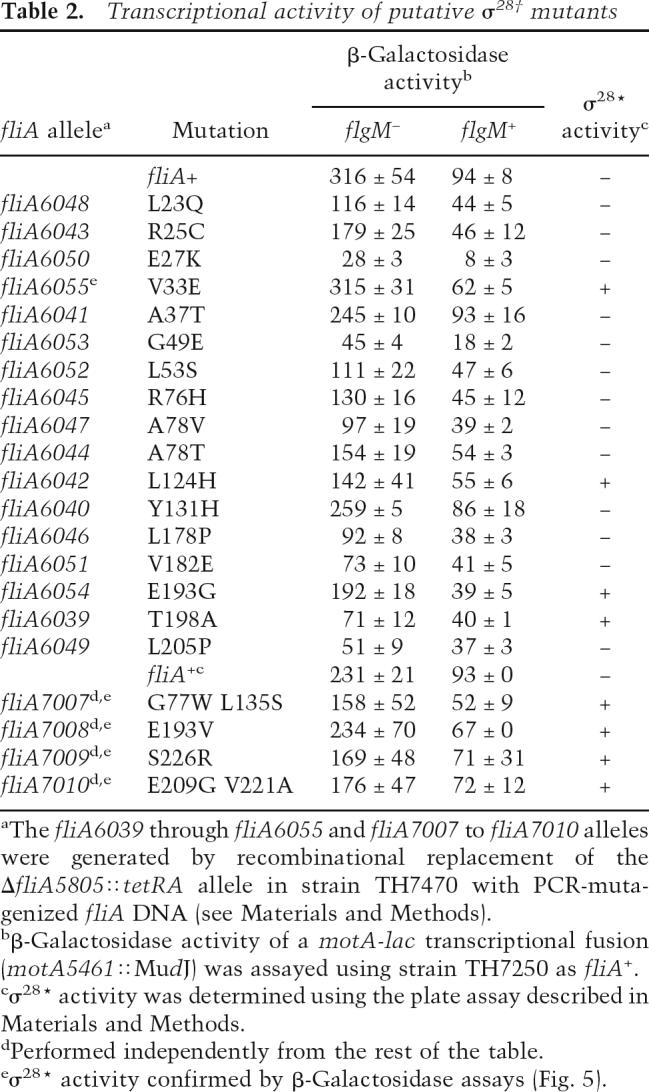
Using the ParaBAD934-flgM+ expression construct, σ28† mutants were isolated that had reduced σ28-dependent class 3 transcription in the presence of FlgM (with arabinose inducer present) again by recombinational replacement of the ΔfliA5805::tetRA allele with PCR-mutagenized fliA-containing DNA (Karlinsey and Hughes 2006). These mutants were then screened for normal σ28-dependent class 3 transcription in the absence of FlgM (no arabinose inducer present) (see Materials and Methods). From a total of 3050 TcS recombinants, 96 (3.1%) putative σ28 FlgM secretion-defective (σ28†) mutants were isolated. DNA sequence analysis identified 24 mutants that resulted from single amino acid substitutions, eight had two amino acid changes, and seven resulted from greater than two amino acid substitutions. The rest possessed either truncation mutations or mutations outside the coding sequence including promoter region mutations. The amino acid changes did not cluster to a single region of σ28 but were located throughout the protein (Table 2). Twenty-one mutants were assayed for β-galactosidase activity (Table 2). The majority of the putative σ28† mutants had reduced class 3 promoter activity in the presence of FlgM. However, many exhibited a reciprocal reduction in class 3 promoter activity in the absence of FlgM (false-positive mutants) and were discarded. Five mutants were found to have the predicted phenotype, i.e., wild-type activity in the absence of FlgM and reduced activity in the presence of FlgM. One of these mutations, V33E, had been previously identified as a σ28* mutant that was defective in binding to FlgM (Chadsey and Hughes 2001). The σ28* proteins allow class 3 transcription in a HBB− strain that does not secrete FlgM.
Table 2.
Transcriptional activity of putative σ28† mutants
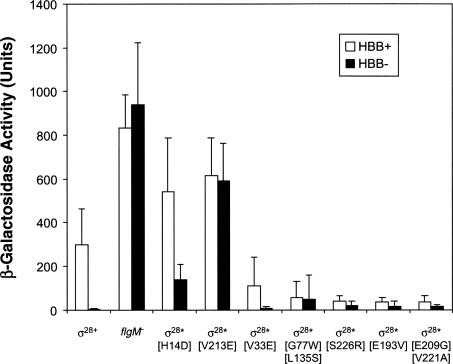
The four other mutants were assayed for σ28* activity by measuring class 3 promoter activity using a different class 3 reporter gene construct (fliC5050::MudJ) in a HBB− flgM + strain. β-Galactosidase assays showed that three of the four mutants resulted in a three-to fourfold increase in class 3 transcription in the HBB− background, which was comparable to that seen for σ28*[V33E] (Fig. 5). In comparison, the σ28*[V213E] mutant, which is 70-fold reduced in its ability to bind FlgM, and σ28*[H14D] mutant, which exhibited increased stability, possessed a 148-fold or 36-fold increase in class 3 promoter activity, respectively. The fourth mutant [G77W L135S] showed a much stronger σ28* phenotype (12.5-fold increase in class 3 transcription in the HBB− background) and was not characterized further. This suggests that the three new mutants could be classed as σ28* mutants that had defects in their ability to bind FlgM. This assumption is based on the fact that all four mutants, V33E, S226R, E193V, and [E209G, V221A], are still responsive to FlgM inhibition (cf. HBB+ with HBB− in Fig. 5) but still exhibit some σ28* activity, whereas σ28*[V213E] was completely insensitive to FlgM inhibition.
Figure 5.
Putative σ28† mutants exhibit weak σ28* activity. β-Galactosidase activity of a fliC-lacZ transcriptional fusion (fliC5050::MudJ) in various flagellar mutant backgrounds. The activities of σ28 mutants were compared with σ28+, flgM −, σ28*[H14D] (increased-stability σ28 with wild-type FlgM-binding), and σ28*[V213E] (severe FlgM binding defective σ28) in strains with either a HBB+ (white bars) or HBB− (black bars) phenotype. Surprisingly, even though σ28*[V33E], σ28*[G77W L135S], σ28*[E193V], σ28*[S226R], and σ28*[E209G V221A] all exhibited lower σ28 transcriptional activity for HBB+ strains compared with HBB+ σ28+, they all showed a weak σ28* phenotype in the HBB− strains. In contrast, significant increases in fliC-lacZ transcription for both HBB+ and HBB− strains were observed for the σ28*[H14D] and σ28*[V213E] mutants. A complete description of the genotypes of all HBB+ strains used in this analysis is to be found in Table 3. HBB− strains were constructed by transduction of the flgG574::Tn10 allele.
The σ28* mutants isolated in a σ28† screen are defective for FlgM secretion
In the above screen for FlgM secretion-defective mutants in σ28 (σ28†), the mutants isolated appeared to have a slight defect in binding to FlgM (weak σ28*). However, all exhibited a significant reduction in class 3 transcription in a strain competent for FlgM secretion (HBB+), consistent with a σ28† phenotype (Table 2; Fig. 5). In contrast, σ28*[V213E] and σ28*[H14D] mutants exhibited a significant increase in class 3 promoter activity in the HBB+ strain. Even though all mutants isolated possessed weak σ28* activity, the strong inhibition of class 3 promoter activity for the HBB+ strain suggested a defect in FlgM secretion. Thus, there appears to be an overlap between the σ28* (FlgM-binding) and σ28† (FlgM-secretion) activities. Furthermore, the class 3 promoter activity in the absence of FlgM for the four mutants was comparable with wild type (Table 2), indicating that the lower class 3 transcription in the presence of FlgM did not result from reduced σ28 transcriptional activity. FlgM secretion assays were therefore performed for the four mutants possessing overlapping σ28* and σ28† phenotypes (V33E, S226R, E193V, and [E209G, V221A]) (Fig. 2). Compared with σ28+, all four mutants exhibited a strong reduction in FlgM secretion (FlgMOUT). Significantly, intracellular FlgM levels (FlgMIN) were increased compared with wild type (V33E: 1.2-fold; S226R: 2.7-fold; E193V: 1.3-fold; and [E209G, V221A]: 1.4-fold), indicating that the reduced FlgMOUT levels did not result from reduced flgM gene expression. As all four σ28 mutants isolated during this study possessed a minor σ28* phenotype, the data are consistent with an overlap in the FlgM binding and FlgM secretion-facilitator activities of σ28.
Discussion
Efficient flagellar assembly requires tight control of protein subunit production, secretion, and self-assembly. Bacteria that possess multiple flagella per cell might also require mechanisms to allow the different flagella within a single cell to develop independently. A major checkpoint for flagellar assembly in all bacteria is the completion of the intermediate structure known as the hook-basal body (HBB). In S. typhimurium, HBB completion coincides with FlgM secretion and the initiation of σ28-dependent flagellar class 3 transcription of genes that encode late flagellar subunits and the chemosensory machinery. Prior to secretion, FlgM binds and inhibits the activity of σ28. The interaction between FlgM and σ28 allows the cell to couple flagellar gene expression to the assembly process.
Prior to HBB completion, the flagellar T3S system is specific for hook and rod-type secretion substrates. Upon HBB completion, the specificity of the T3S apparatus is switched to late secretion substrate recognition. Five out of the six late secretion substrates have been shown to require a T3S-chaperone for efficient secretion coupled to assembly. The only flagellar late secretion substrate without a defined chaperone was FlgM. Evidence from the T3S field suggested that all substrates secreted after the substrate-specificity switch required a T3S-chaperone for efficient secretion. Based on this evidence, the identity of the T3S-chaperone for FlgM secretion was investigated. Surprisingly, σ28 itself facilitates FlgM secretion defining a novel role for a σ70-family transcription factor as a T3S-chaperone. The following results demonstrate that FlgM secretion is facilitated by σ28, but is independent of σ28 transcriptional activity: (1) A strain deleted for the σ28 gene (complete null mutant) is defective for FlgM secretion; (2) a σ28* mutant that is defective in binding FlgM (σ28*[V213E]) is defective in FlgM secretion; (3) a σ28* mutant with increased stability that shows normal binding to FlgM (σ28*[H14D]) is proficient in FlgM secretion; and (4) σ28 mutants inactive for transcription activity, but still produce the σ28 protein, are proficient for FlgM secretion. Thus σ28 is defined as the FlgM T3S-chaperone based on the strong evidence obtained from the analysis presented here.
The conclusions drawn from these data have come from experiments performed using a system in which extra care has been taken not to deviate from a physiologically relevant condition. Only the genetic screen employed σ28-independent flgM expression. However, all subsequent analysis of FlgM protein levels, σ28 activity, and FlgM-secretion for σ28 mutants with the required phenotype were performed using a wild-type background with σ28-dependent flgM expression.
The conclusions drawn here are also in agreement with the physical characteristics that define a T3S-chaperone. The majority of T3S-chaperones identified to date have a low molecular size and a low pI, usually in the range of pI = 3.9–5.3 (Parsot et al. 2003); σ28 has a theoretical pI of 5.12 and a molecular weight of 28 kDa. The accepted flagellar T3S-chaperones FlgN, FliT, and FliS all have a molecular weight of 14 kDa and pIs that also fall in the range defined by Parsot et al. (2003). One difference is that the known flagellar T3S-chaperones act as dimers whereas σ28 acts as a monomer.
Recent studies of a growing number of T3S-chaperones associated with both flagellar and virulence T3S systems have identified a subset of T3S-chaperones to possess multiple functions: facilitation of T3S substrate recognition, secretion, and regulation of gene expression. Some examples include SicA (SPI-1) (Darwin and Miller 2001), FlgN (Flagella) (Karlinsey et al. 2000a), and FliT (Flagella) (Kutsukake et al. 1999) from S. typhimurium, Spa15 and IpgC from Shigella flexneri (Parsot et al. 2005), and SycD and SycH from Yersinia spp (Feldman and Cornelis 2003). Apart from FlgN and FliT whose regulatory mechanisms are still poorly understood the majority of T3S-chaperones to be studied regulate gene expression via secondary interactions with transcriptional activators. By comparison, σ28 being itself a transcriptional activator, secretion of its partner FlgM has the immediate effect of freeing σ28 to interact with core RNA polymerase and activate σ28-dependent flagellar gene transcription. With a growing number of T3S-chaperones possessing a gene regulatory function, the question arises to whether all or the majority of the protein family are bi-functional.
The bi-functionality of T3S-chaperones allows for gene regulation to be coupled to secretion of the bound substrate. Thus, the dual nature of T3S-chaperones provides a molecular device that coordinates gene regulation in a checkpoint manner, whether the checkpoint is in structural assembly or in pathogenesis. The main objective of the interaction between σ28 and FlgM is to couple flagellar gene expression to flagellar assembly. Therefore, it is not surprising that the σ28:FlgM complex has evolved into a T3S-chaperone:substrate partnership in order to “sense” HBB completion.
How does the flagellar secretion apparatus dissociate FlgM from σ28?
The fact that the σ28† mutants isolated in this study as defective in FlgM secretion possessed at least some σ28* activity (FlgM-binding defective) indicates that, even though σ28− mutants still secreted FlgM in a σ28-dependent manner, the two functions of σ28 have overlap within the σ28 protein. This is not surprising since σ28 must bind FlgM in order to facilitate its secretion, and conversely FlgM secretion must be concomitant with its release from σ28. Given the remarkable strength of the σ28:FlgM interaction (Kd = 0.2 nM) (Chadsey and Hughes 2001), the dissociation of the σ28:FlgM complex by the flagellar T3S system must occur by a mechanism that is able to unravel this tight complex. FlgM is a very small protein of 97 amino acids. A cocrystal of the Aquifex aeolicus σ28:FlgM complex demonstrates that FlgM essentially wraps around σ28 (Sorenson et al. 2004).
While FlgM binds σ28 at multiple sites throughout its three structural regions (conserved regions 2, 3, and 4) mutants of σ28 most defective in binding FlgM are all in region 4 (Fig. 4; Chadsey and Hughes 2001). Recently, it was shown that the T3S-chaperone, FlgN, associates with FliI in the presence of its substrate, FlgK (Thomas et al. 2004). The exact nature of the interaction with FliI has not yet been determined. In the case of the σ28:FlgM complex, we propose that the binding of σ28 to FlgM exposes the N-terminal secretion domain of FlgM and facilitates interaction of the σ28:FlgM complex to the flagellar T3S apparatus. If the flagellar T3S apparatus is competent for FlgM secretion (HBB+), the action of FlgM secretion would effectively peel FlgM from σ28, resulting in secreted FlgM and σ28 that is free to interact with RNA polymerase and transcribe flagellar class 3 promoters. Our model is consistent with the disassociation of SptP from the SicP:SptP complex by the ATPase InvC of the SPI-1 virulence-associated T3S in Salmonella (Akeda and Galan 2005). From our understanding of the σ28/ FlgM interaction and our genetic data we conclude that the structural conformation of the T3S-chaperone:substrate complex is crucial for efficient substrate recognition by the ATPase. With the σ28 mutants used during this study we are now in a position to determine if and how the σ28:FlgM complex interacts with the T3S apparatus.
The evolution of the multifunctional T3S-chaperone and would T3S-facilitator be a better name?
It appears that T3S-chaperones can play multiple roles as general facilitators of secretion either through protein stabilization or targeted delivery (or both) and as regulators of gene expression, but in a manner such that the regulatory function coincides with the timing of secretion for a given substrate. Such a mechanism would determine which T3S-chaperones evolve to bind a given substrate and for T3S-chaperones with regulatory functions. The cognate secretion substrate would be selected according to when it is secreted in the assembly process or in the virulence pathway so that the regulatory function can be appropriately timed with the secretion and assembly of a specific protein.
Finally we would like to highlight once more the confusion that can occur with the current nomenclature of proteins deemed to have “chaperone” functions. T3S-chaperones can be confused with other molecular chaperones most of which are involved in folding or degrading proteins that never leave the cell. Even though proteins that guide protein folding, like GroEL, are occasionally now referred to as chaperonins (Ellis 2005; Swain and Gierasch 2005), the term “chaperone” is being used, even if correctly, too often. One solution would be to rename the T3S-chaperones as T3S-facilitators. Data is available suggesting that this family of accessory proteins is in fact facilitating the secretion of cognate substrates. Furthermore, many, if not all, T3S-facilitator substrates can be secreted via a T3S secretion system in the absence of their T3S-facilitator (Aldridge et al. 2003; Lee and Galan 2004; Lilic et al. 2006). However, if current terminology is to be kept, then we stress that whenever mentioning chaperones associated with T3S systems they should be referred to as “T3S-chaperones” always prefixed with T3S.
Materials and methods
Bacterial strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are shown in Table 3. Cultivation of bacteria and the concentration of antibiotics used throughout this study have been previously described (Bonifield and Hughes 2003). The generalized transducing phage of S. typhimurium P22 HT105/1 int-201 was used in all transductional crosses (Davis et al. 1980). Media used during this study included Luria Bertin (LB) broth, MacConkey-lactose agar (Difco), and Triphenyltetrazolium chloride (TTC) lactose agar (Gillen and Hughes 1991a,b). Tetracycline sensitivity (TcS) selection was performed as described (Maloy 1990). All experiments involving the growth of strains containing the plasmid pKD46 were performed at 30°C. Loss of pKD46 from given strains was achieved simply by growth at 37°C after electroporation of PCR products. All FlgM secretion assays and β-galactosidase assays were performed on cultures grown in LB at 37°C with shaking to an OD600 = 0.6–0.8.
Table 3.
Strains and plasmids used
Recombinant DNA techniques
Standard molecular DNA manipulation techniques have been previously described (Aldridge and Jenal 1999). The plasmid pJOY was constructed as follows. The FRT-kan-FRT (FKF) cassette from pKD4 was amplified using primers BamHIP1 (5′-GCGGGATCCGTGTAGGCTGGAGCTGCTTC-3′) and HindIIIP2 (5′-GGGAAGCTTCATATGAATATCCTCC TTAG-3′) digested with BamHI and HindIII and cloned into pUC19 creating plasmid pPA122. The flgM coding sequence was amplified with primers FlgAkpn1F (5′-CGCGGTACCC AGCCTTCAGCATGACGG-3′) and FlgMstpkpn (5′-CCGG TACCTTATTTACTCTGTAAGTAGC-3′) and subcloned into pPA122 upstream of the FKF cassette by restriction digestion with KpnI, resulting in pJOY.
Obtaining arabinose-controlled flgM expression at the araBAD locus
To achieve arabinose-inducible expression of flgM, the primer ARAFLGM (5′-ACTGTTTCTCCATACCTGTTTTTCTGGAT GGAGTAAGACGATGAGCATTGACCGTACCTC-3′) was used with AraGTR (5′-GTTATGCACTGCATCCTCGGCATTTT TACCCCAGGCAAACTGACCATGATTACGCCAAGC-3′) to amplify the flgM-FKF insert of pJOY. These primers possessed 5′ 40-nucleotide overhangs complementary to the start codon of araB (ARAFLGM) and codons 174–162 of araD for AraGTR.
The design of the primers meant that ARAFLGM produced a transcriptional fusion of flgM, resulting in a transcript with the araBAD 5′ untranslated region. The ARAFLGM–AraGTR PCR product was electroporated into strain TH437 (pKD46), grown at 30°C in Luria Broth with 0.2% arabinose as previously described (Aldridge et al. 2003). ParaBAD-flgM + (strain TH6599 (ΔaraBAD923::flgM +-FKF)) was isolated by screening kanamycin-resistant transformants for a Ara− phenotype and confirmed by PCR. The ParaBAD-flgM + construct was moved by cotransduction using leu-1151::Tn10 selecting for Leu− Ara− TcR transductants into a ΔflgM motA5461::MudJ (TH6662) background. The leu-1151::Tn10 allele was replaced with a leu+ allele from wild-type strain LT2 by P22-mediated transduction. Confirmation that ParaBAD-flgM + had been retained was possible by plating transductants on MacConkey Lactose plates ±0.2% arabi-nose and screening for arabinose-dependent inhibition of class 3 promoter activity using the motA5461::MudJ reporter construct. The ParaBAD-flgM + construct showed reduced class 3 promoter activity compared with TH3933 in the presence of arabi-nose (Table 1).
To obtain ParaBAD-flgM + expression levels comparable with PflgM-flgM + expression levels, recombinational replacement PCR mutagenesis was used (Karlinsey and Hughes 2006). First, the araC–araBAD intergenic region in TH7250 was replaced with a PCR product amplified using ParaBtetR (5′-GTCT TACTCCATCCAGAAAAACAGGTATGGAGAAACAGTAT TAAGACCCACTTTCACA-3′) and ParaBtetA (5′-GTCCATAT CGACCAGGACGACAGAGCTTCCGTCTCCGCAACTAAG CACTTGTCTCCTG-3′) and a chromosomal Tn10dTc insert as a template. Second, ΔParaBAD-927::tetRA in TH7253 was then replaced with PCR products amplified using 1, 2.5, or 5 Units Taq DNA polymerase (Promega) with primers FlgM+34R (5′-CGGGTTTCAAAGGTGAGG-3′) and AraC+5R (5′-CCATGA TTTCTCTACCCC-3′) using TH7250 chromosomal DNA as a template. PCR products were transformed by electroporation into TH7253 (pKD46) and plated on Tc-sensitive plates. To screen for an arabinose inducible Lac-phenotype comparable with TH3933, a total of 300 TcS transformants were picked onto Tc-sensitive plates and replica-plated onto LB + tetracycline, LB-agar, MacConkey-lactose (Difco) ±0.2% arabinose, and TTC-lactose ±0.2% arabinose. Transformants exhibiting the correct Lac-phenotype were purified on LB-agar plates before sequence analysis of the araC–araBAD intergenic region and confirmation of the Lac phenotype by β-galactosidase assay of liquid LB cultures. This analysis resulted in the isolation of TH7393, TH7394, TH7395, and TH7396 with TH7394 being chosen for subsequent experiments.
Isolation of fliA mutants by PCR mutagenesis
To isolate mutations in fliA the natural error rate of Taq DNA polymerase was exploited in conjunction with recombinational replacement mutagenesis using the mutant ΔfliA5805::tetRA (Karlinsey and Hughes 2006). All mutagenic PCR reactions were performed in triplicate where the individual reactions had either 1, 2.5, or 5 Units of Taq DNA polymerase per reaction. Primers FliA−118F (5′-GGCGCTACAGGTTACATAAG-3′) and FliA+765R (5′-TAGTCTATACGTTGTGCGGC-3′) were used to amplify the coding sequence of fliA from a chromosomal DNA preparation of LT2. PCR products were purified using the PCR clean up kits from either Qiagen or Sigma before being concentrated down to 10 μL. Three aliquots of freshly prepared electroporation-competent cells of TH7470 (pKD46) were electroporated with 3 μL of each PCR reaction. Cells were allowed to recover in 1 mL LB broth for 1 h before plating out 5 × 200 μL of both a 10−1 and 10−2 dilution for each electroporation on Tc-sensitive plates. To screen for class 3 promoter activity using motA5461::MudJ, TcS transformants were replicapicked onto MacConkey-lactose ±0.2% arabinose and TTC-lactose ±0.2% arabinose plates.
Analysis of σ28* activity of σ28 mutant alleles
σ28* alleles are able to drive expression of class 3 promoters in a HBB− background where FlgM secretion is inhibited. To determine whether putative σ28† mutants exhibited σ28* activity, two HBB− mutations, flgG574::Tn10 and ΔflgG-L2157, were used. First, P22 phage stocks of σ28† mutants were prepared and used as donors in transduction of TPA598. This allowed the σ28† mutants to be tested for class 3 promoter activity in HBB+/− strains with σ28-dependent flgM expression rather than flgM + expression from ParaBAD. TcS transductants were replica-printed onto Tc-sensitive and Tc-sensitive + kanamycin plates to iso late TcS transductants that retained the fliA-linked fliC5050::MudJ reporter insert. The HBB− mutation flgG574::Tn10 was then introduced by transduction by selecting for Tn10-encoded TcR. Transductants were purified and the activity of fliC5050::MudJ was determined by β-galactosidase assays. Alternatively, the deletion ΔflgG-L2157 was introduced by cotransduction of a linked pyrC691::Tn10 marker. Whether the alleles assayed possessed σ28* activity was determined by replica-picking 100 TcR transductants onto MacConkey-lactose + Tetracycline and TTC-lactose + Tetracycline plates. σ28 mutants were defined as σ28* mutants in this assay if on Lac indicator plates no Lac− transductants were observed, σ28* activity was confirmed for positive isolates by β-galactosidase assays.
Analysis of FlgM and σ28 by immunodetection
All samples used for secretion assays in this study were taken at mid-log phase corresponding to an OD600 = 0.6–0.8. For immunodetection of FlgM and σ28, tricine SDS polyacrylamide gels were used instead of standard glycine SDS gels. All immunodetections were performed as described by Aldridge et al. (2003). Protein stability assays and quantification of immunoblots were performed essentially as described by Aldridge et al. (2003). Secretion assays for FlgM have been previously described (Karlinsey et al. 2000b). Briefly, strains were inoculated into 3 mL of LB broth and incubated with continuous shaking until the OD600 was ∼0.6–0.8. Two-thirds of the culture was centrifuged and the cell pellet was resuspended in SDS-sample buffer (Karlinsey et al. 2000b); this whole-cell lysate sample was defined as FlgMIN. A 1.8 mL aliquot of the supernatant was centrifuged a second time, filtered through a 0.2-μm PES filter, and the proteins were extracted by filtration over a nitrocellulose filter (BA85). The filter was soaked in SDS sample buffer for 15 min at 65°C to recover the proteins present, and the sample was defined as FlgMOUT. Prior to loading onto SDS-tricine gels, all samples were normalized to the OD of the culture and volume used in preparation (Karlinsey et al. 2000b). Detection of chemiluminescence produced by the ECL plus western detection kit (GE Healthcare) was performed using either a Storm 840 phosphoimager (GE Healthcare) or a Fuji LAS3000 imageanalyzer. Statistical analysis and calculation of the half-lives of FlgM and σ28 were performed using Microsoft Excel (see Supplemental Material).
β-Galactosidase reporter gene assays
All β-galactosidase assays were performed on mid-log phase cultures. Strains were inoculated into 3 mL of LB broth and incubated with continuous shaking until the OD600 was ∼0.6–0.8. Cells were recovered by centrifugation for 10 min at 4000 rpm and resuspended in 3 mL saline buffer. β-Galactosidase assays have been described previously by Aldridge et al. (2003).
Acknowledgments
We thank the participants of the Cold Spring Harbor Laboratories “Advanced Bacterial Genetics” Course (2002) for their help in developing the σ28† screen and members of the Hughes lab for critically reading this manuscript prior to submission. Use of the FUJI LAS3000 was by kind permission of Dr. C. Jones and Dr. N. Curtin of the Northern Institute for Cancer Research, Newcastle, UK. Public Health Service grant GM56141 awarded to K.T.H. from NIH (USA) has funded this work in addition to a Nuffield Foundation (UK) “Awards to newly appointed lecturers in Science, Mathematics and Engineering” grant, no. NAL/ 00745/G, awarded to P.D.A.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.380406.
References
- Akeda Y., Galan J.E. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–915. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- Aldridge P., Hughes K.T. Regulation of flagellar assembly. Curr. Opin. Microbiol. 2002;5:160–165. doi: 10.1016/s1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- Aldridge P., Jenal U. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 1999;32:379–391. doi: 10.1046/j.1365-2958.1999.01358.x. [DOI] [PubMed] [Google Scholar]
- Aldridge P., Karlinsey J., Hughes K.T. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol. Microbiol. 2003;49:1333–1345. doi: 10.1046/j.1365-2958.2003.03637.x. [DOI] [PubMed] [Google Scholar]
- Aldridge P., Karlinsey J.E., Becker E., Chevance F.F., Hughes K.T. Flk prevents premature secretion of the anti-sigma factor FlgM into the periplasm. Mol. Microbiol. 2006;60:630–643. doi: 10.1111/j.1365-2958.2006.05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvray F., Thomas J., Fraser G.M., Hughes C. Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 2001;308:221–229. doi: 10.1006/jmbi.2001.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy S.L., Ng S.Y., Jarrell K.F. Prokaryotic motility structures. Microbiol. 2003;149:295–304. doi: 10.1099/mic.0.25948-0. [DOI] [PubMed] [Google Scholar]
- Bennett J.C., Hughes C. From flagellum assembly to virulence: The extended family of type III export chaperones. Trends Microbiol. 2000;8:202–204. doi: 10.1016/s0966-842x(00)01751-0. [DOI] [PubMed] [Google Scholar]
- Berg H.C., Anderson R.A. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245:380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- Bonifield H.R., Hughes K.T. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J. Bacteriol. 2003;185:3567–3574. doi: 10.1128/JB.185.12.3567-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadsey M.S., Hughes K.T. A multipartite interaction between Salmonella transcription factor σ28and its anti-sigma factor FlgM: Implications for σ28holoenzyme destabilization through stepwise binding. J. Mol. Biol. 2001;306:915–929. doi: 10.1006/jmbi.2001.4438. [DOI] [PubMed] [Google Scholar]
- Chadsey M.S., Karlinsey J.E., Hughes K.T. The flagellar anti-sigma factor FlgM actively dissociates Salmonella typhimurium σ28RNA polymerase holoenzyme. Genes & Dev. 1998;12:3123–3136. doi: 10.1101/gad.12.19.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott G.S., Hughes K.T. The type III secretion determinants of the flagellar anti-transcription factor, FlgM, extend from the amino-terminus into the anti-σ28domain. Mol. Microbiol. 1998;30:1029–1040. doi: 10.1046/j.1365-2958.1998.01131.x. [DOI] [PubMed] [Google Scholar]
- ——— 2000Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli Microbiol. Mol. Biol. Rev. 64694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin K.H., Miller V.L. Type III secretion chaperone-dependent regulation: Activation of virulence genes by SicA and InvF in Salmonella typhimurium . EMBO J. 2001;20:1850–1862. doi: 10.1093/emboj/20.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughdrill G.W., Chadsey M.S., Karlinsey J.E., Hughes K.T., Dahlquist F.W. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, σ28 . Nat. Struct. Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- Davis R.W., Botstein D., Roth J.R. Advanced bacterial genetics: A manual for genetic engineering. Cold Spring Harbor Laboratory, Cold Spring Harbor; NY: 1980. [Google Scholar]
- Ellis R.J. Chaperomics: In vivo GroEL function defined. Curr. Biol. 2005;15:R661–R663. doi: 10.1016/j.cub.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Feldman M.F., Cornelis G.R. The multitalented type III chaperones: All you can do with 15 kDa. FEMS Microbiol. Lett. 2003;219:151–158. doi: 10.1016/S0378-1097(03)00042-9. [DOI] [PubMed] [Google Scholar]
- Fraser G.M., Bennett J.C., Hughes C. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol. Microbiol. 1999;32:569–580. doi: 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- Frye J., Karlinsey J.E., Felise H.R., Marzolf B., Dowidar N., McClelland M., Hughes K.T. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2006;188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillen K.L., Hughes K.T. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium . J. Bacteriol. 1991a;173:6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———1991bNegative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium J. Bacteriol. 1732301–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K.T., Gillen K.L., Semon M.J., Karlinsey J.E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- Karlinsey J.E., Hughes K.T. Genetic transplantation: Salmonella enterica serovar Typhimurium as a host to study sigma factor and anti-sigma factor interactions in genetically intractable systems. J. Bacteriol. 2006;188:103–114. doi: 10.1128/JB.188.1.103-114.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsey J.E., Lonner J., Brown K.L., Hughes K.T. Translation/secretion coupling by type III secretion systems. Cell. 2000a;102:487–497. doi: 10.1016/s0092-8674(00)00053-2. [DOI] [PubMed] [Google Scholar]
- Karlinsey J.E., Tanaka S., Bettenworth V., Yamaguchi S., Boos W., Aizawa S.I., Hughes K.T. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 2000b;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Ohya Y., Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium . J. Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K., Ikebe T., Yamamoto S. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of. Salmonella. Genes Genet. Syst. 1999;74:287–292. doi: 10.1266/ggs.74.287. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Galan J.E. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 2004;51:483–495. doi: 10.1046/j.1365-2958.2003.03840.x. [DOI] [PubMed] [Google Scholar]
- Lilic M., Vujanac M., Stebbins C.E. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol. Cell. 2006;21:653–664. doi: 10.1016/j.molcel.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Liu X., Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R.M. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- ——— 1999The bacterial flagellum: Reversible rotary propellor and type III export apparatus J. Bacteriol. 181:7149–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S.R. Experimental techniques in bacterial genetics. Jones and Bartlett; Boston, MA: 1990. [Google Scholar]
- Maloy S.R., Nunn W.D. Selection for loss of tetracycline resistance by Escherichia coli . J. Bacteriol. 1981;145::1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Sanderson K.E., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413::852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- Minamino T., Gonzalez-Pedrajo B., Yamaguchi K., Aizawa S.I., Macnab R.M. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 1999;34:295–304. doi: 10.1046/j.1365-2958.1999.01597.x. [DOI] [PubMed] [Google Scholar]
- Muramoto K., Makishima S., Aizawa S., Macnab R.M. Effect of hook subunit concentration on assembly and control of length of the flagellar hook of. Salmonella. J. Bacteriol. 1999;181:5808–5813. doi: 10.1128/jb.181.18.5808-5813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba K. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells. 2001;6:1–12. doi: 10.1046/j.1365-2443.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Kutsukake K., Suzuki H., Lino T. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: An antisigma factor inhibits the activity of the flagellum-specific sigma factor, σF . Mol. Microbiol. 1992;6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- Parsot C., Hamiaux C., Page A.L. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 2003;6:7–14. doi: 10.1016/s1369-5274(02)00002-4. [DOI] [PubMed] [Google Scholar]
- Parsot C., Ageron E., Penno C., Mavris M., Jamoussi K., d'Hauteville H., Sansonetti P., Demers B. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri . Mol. Microbiol. 2005;56:1627–1635. doi: 10.1111/j.1365-2958.2005.04645.x. [DOI] [PubMed] [Google Scholar]
- Sorenson M.K., Ray S.S., Darst S.A. Crystal structure of the flagellar sigma/anti-sigma complex σ28/FlgM reveals an intact sigma factor in an inactive conformation. Mol. Cell. 2004;14:127–138. doi: 10.1016/s1097-2765(04)00150-9. [DOI] [PubMed] [Google Scholar]
- Stebbins C.E., Galan J.E. Priming virulence factors for delivery into the host. Nat. Rev. Mol. Cell Biol. 2003;4::738–743. doi: 10.1038/nrm1201. [DOI] [PubMed] [Google Scholar]
- Swain J.F., Gierasch L.M. First glimpses of a chaperonin-bound folding intermediate. Proc. Natl. Acad. Sci. 2005;102:13715–13716. doi: 10.1073/pnas.0506510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J., Stafford G.P., Hughes C. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. 2004;101:3945–3950. doi: 10.1073/pnas.0307223101. [DOI] [PMC free article] [PubMed] [Google Scholar]