Abstract
Cells respond to DNA damage by activating a network of signaling pathways that control cell cycle progression and DNA repair. Genetic studies in yeast suggested that several checkpoint proteins, including the RFC-related Rad17 protein, and the PCNA-related Rad1–Rad9–Hus1 protein complex might function as sensors of DNA damage. In this study, we show that the human Rad17 protein recruits the Rad9 protein complex onto chromatin after damage. Rad17 binds to chromatin prior to damage and is phosphorylated by ATR on chromatin after damage but Rad17's phosphorylation is not required for Rad9 loading onto chromatin. The chromatin associations of Rad17 and ATR are largely independent, which suggests that they localize to DNA damage independently. Furthermore, the phosphorylation of Rad17 requires Hus1, suggesting that the Rad1–Rad9–Hus1 complex recruited by Rad17 enables ATR to recognize its substrates. Our data are consistent with a model in which multiple checkpoint protein complexes localize to sites of DNA damage independently and interact to trigger the checkpoint-signaling cascade.
Keywords: Rad17, Rad1–Rad9–Hus1 complex, ATR, Chk1, checkpoint
ATM and ATR kinases are central components of the DNA damage response (Zhou and Elledge 2000). Upon DNA damage, they activate a network of damage-response pathways by phosphorylating proteins such as p53, Brca1, Nbs1, Chk1, and Chk2. However, how ATM and ATR are activated by DNA damage and how they recognize their substrates is not understood.
In Schizosaccharomyces pombe, the phosphorylation of Chk1 and Cds1 by Rad3, a homolog of ATR and ATM, requires Rad1, Rad9, Rad17, Rad26, and Hus1 (O'Connell et al. 2000). It was therefore postulated that this group of proteins might function as sensors of DNA damage. Among these proteins, Rad17 shares homology with all five subunits of replication factor C (RFC), and it associates with the four small RFC subunits in humans and budding yeast (Griffiths et al. 1995; Green et al. 2000; Naiki et al. 2000; Lindsey-Boltz et al. 2001). In addition, Rad1, Rad9, and Hus1, which are all structurally related to PCNA (Thelen et al. 1999; Venclovas and Thelen 2000), form a complex in human cells (St Onge et al. 1999; Volkmer and Karnitz 1999). During DNA replication, RFC recognizes the primer–template junction and loads PCNA onto DNA (Tsurimoto and Stillman 1991). The analogy between Rad17 and the RFC subunits raises the possibility that a Rad17-containing complex might be involved in recognizing certain DNA structures during the damage response. Furthermore, the Rad17 complex might recruit the Rad1–Rad9–Hus1 complex to DNA lesions in a manner similar to PCNA loading by RFC. Indeed, Rad1, Rad9, and Hus1 become less extractable from the nucleus after damage, implying that the Rad1–Rad9–Hus1 complex might associate with damaged DNA (Burtelow et al. 2000). However, whether Rad17 is required for the recruitment of the Rad1–Rad9–Hus1 complex onto DNA, a key question for the model, has not been addressed.
Rad17 and the Rad1–Rad9–Hus1 complex are not the only candidates for sensors of DNA damage. Rad26, a protein tightly bound to Rad3, is phosphorylated by Rad3 after ionizing radiation (IR) even in the absence of Rad17 and the Rad1–Rad9–Hus1 complex (Edwards et al. 1999). Similarly, Ddc2/Lcd1/Pie1, the budding yeast counterpart of Rad26, can be phosphorylated independently of the RFC- and PCNA-like checkpoint proteins after damage (Paciotti et al. 2000; Rouse and Jackson 2000; Wakayama et al. 2001). These findings suggest that the Rad3–Rad26 complex might directly respond to certain types of DNA damage without involving the other putative sensors. The recent identification of ATRIP (ATR-interacting protein), a human homolog of Rad26, indicates that a similar mechanism for damage detection might also operate in human cells (Cortez et al. 2001). Given that Rad17 and the Rad1–Rad9–Hus1 complex are required for the phosphorylation of Rad3 substrates and the activation of the checkpoint, they are clearly important for the transduction and possibly amplification of the damage signal. However, whether Rad17 and the Rad1–Rad9–Hus1 complex also function in parallel with the Rad3–Rad26 complex in damage detection remains unknown.
In this study, we find that the human Rad17 protein associates with chromatin in vivo. Furthermore, Rad17 is required for the recruitment of the Rad9 complex onto chromatin and the activation of Chk1 after damage. The chromatin associations of Rad17 and ATR are largely independent, suggesting that these proteins localize to sites of DNA damage independently. Importantly, Rad17 is capable of recruiting the Rad9 complex even in the absence of ATR, indicating that Rad17 functions in parallel with ATR in damage detection. Both Rad17 and ATR bind to chromatin prior to DNA damage, and Rad17 is phosphorylated by ATR on chromatin after UV irradiation. The phosphorylation of Rad17 by ATR requires Hus1, which suggests that the Rad1–Rad9–Hus1 complex recruited by Rad17 enables ATR to phosphorylate its substrates on chromatin. Together, our data suggest that Rad17 and ATR might be involved in two separate sensory pathways and that they function in concert to fully activate the damage response in human cells.
Results
Human Rad17 associates with chromatin in vivo
To investigate whether Rad17 is part of the protein complex that associates with DNA and has the potential to detect DNA damage, we tested if Rad17 could associate with chromatin in vivo. By fractionating extracts of asynchronously growing 293T cells (Mendez and Stillman 2000), we obtained fractions of cytoplasmic proteins (S1), nuclei (P1), soluble nuclear proteins (S2), and a fraction enriched for chromatin-bound proteins (P2). A cytoplasmic protein (Grb2) and a nuclear protein (Orc2) were well separated by the fractionation (Fig. 1A). Like Orc2, a subunit of the origin recognition complex, a portion of Rad17 was detected in the chromatin fraction (Fig. 1A). After chromatin was digested with micrococcal nuclease, Orc2 and Rad17 could no longer be sedimented, showing that these proteins were, indeed, associated with chromatin.
Figure 1.
Association of human Rad17 with chromatin. (A) Extracts of 293T cells were fractionated as described in Materials and Methods. The resultant fractions were resolved on SDS-PAGE and immunoblotted with the indicated antibodies. (WCE) Whole-cell extracts; (S1) cytoplasmic proteins; (P1) intact nuclei; (S2) soluble nuclear proteins; (P2) chromatin-enriched sediment; (S2‘, P2‘) soluble fraction and sediment from micrococcal nuclease-treated nuclei, respectively; (MNase) micrococcal nuclease. (B) 293T cells were either untreated or treated with 20 Gy of γ-IR or 80 J/m2 of UV and harvested after 2 h. Fractions derived from these cells were analyzed by immunoblotting with the indicated antibodies.
To test whether various forms of DNA damage could stimulate the chromatin association of Rad17, we isolated chromatin from undamaged cells and from cells treated with ionizing radiation (IR) or UV. The amounts of Rad17 on chromatin were not significantly changed after damage (Fig. 1B). In contrast, increasing amounts of Rad9 were detected on chromatin in the damaged cells, indicating that the Rad1–Rad9–Hus1 complex was recruited onto chromatin after damage.
Rad17 is required for Rad9 binding to chromatin and Chk1 phosphorylation after damage
Because Rad17 is structurally related to RFC and was already bound to chromatin when Rad9 was recruited, we asked whether Rad17 played a role in loading Rad9 onto chromatin. We first tested if overexpression of Rad17 could enhance the association of Rad9 with chromatin. To evaluate the effects of Rad17 overexpression, we monitored the chromatin association of endogenous Rad9 and transiently expressed GFP–Rad9. Both forms of Rad9 can be recruited onto chromatin after cells are irradiated with UV (Fig. 2A). Orc2, whose chromatin binding was not affected by damage, served as the loading control for the chromatin fractions. When Flag–Rad17 was transiently expressed, a fraction of it bound to chromatin like endogenous Rad17, leading to an increase in Rad17 on chromatin. Consequently, the UV-induced chromatin association of both endogenous Rad9 and GFP–Rad9 was stimulated. Although the chromatin association of GFP–Rad9 was slightly elevated by Flag–Rad17 prior to damage, the induction by UV was still pronounced. Thus, Rad17 can promote Rad9 binding to chromatin in a damage-dependent manner.
Figure 2.
Requirement of Rad17 for loading Rad9 onto chromatin and phosphorylation of Chk1. (A) 293T cells were cotransfected with plasmids expressing GFP–Rad9 and either vectors alone or plasmids expressing Flag–Rad17. After 48 h, cells were untreated or treated with 50 J/m2 of UV and harvested after 2 h. Fractions from these cells were analyzed by immunoblotting with the indicated antibodies. (S1+S2) A combined fraction of S1 and S2. (B) HeLa cells were either transfected with siRNA targeting Rad17 or mock-transfected, and untreated or treated with UV as in A. Fractions from these cells were analyzed by immunoblotting. (C) HeLa cells transfected with siRNA targeting Rad17 or mock-transfected HeLa cells were untreated or treated with 50 J/m2 of UV and harvested after 2 h. Extracts of the cells were immnoprecipitated with an anti-p-S345 Chk1 antibody (Liu et al. 2000). Both extracts (WCE) and immunoprecipitates (lower panel) were analyzed by immunoblotting.
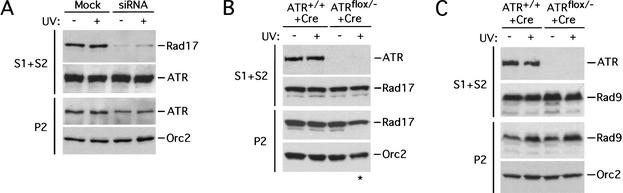
To address whether Rad17 is required for the recruitment of Rad9 onto chromatin, we used small interfering RNA (siRNA) to specifically inhibit the expression of Rad17 (Elbashir et al. 2001). Transfection of the siRNA duplexes targeting Rad17 reduced its protein level by ∼80%. Importantly, the amounts of Rad17 on chromatin were also reduced by the siRNA (Fig. 2B). The overall levels of several other checkpoint proteins, including ATR and Rad9, were not affected by the siRNA (Fig. 2B; see Fig. 5A below), showing that the inhibition was specific to Rad17. The basal level of chromatin-bound Rad9 was reduced in the siRNA-transfected cells even prior to damage. After UV irradiation, the damage-induced chromatin association of Rad9 was clearly attenuated in the siRNA-transfected cells. Furthermore, the overall levels of hyperphosphorylated Rad9 were also reduced in the siRNA-transfected cells after damage. Therefore, Rad17 is required not only for loading Rad9 onto chromatin after damage but also for its hyperphosphorylation. Because essentially all the Rad9 in human cells is present in the Rad1–Rad9–Hus1 complex (Burtelow et al. 2001), this result strongly suggests that Rad17 is required for recruiting the Rad1–Rad9–Hus1 complex onto chromatin.
Figure 5.
Rad17 associates with chromatin and recruits Rad9 independently of ATR. (A) HeLa cells transfected with siRNA targeting Rad17 or mock-transfected HeLa cells were untreated or treated with 80 J/m2 of UV, harvested after 2 h. The cells were then fractionated as in Figure 1A, and the resultant fractions were analyzed by immunoblotting with the indicated antibodies. (B,C) ATR+/+ and ATRflox/− cells infected with Ad-Cre were untreated or treated with UV as in A. Fractions from these cells were analyzed by immunoblotting with the indicated antibodies. (*) This lane was underloaded as shown by Orc2 signal.
To determine where Rad17 functions in the checkpoint pathway, we tested if Rad17 was required for the activation of Chk1. In response to UV and hydroxyurea (HU), Chk1 is phosphorylated at serines 317 and 345 in an ATR-dependent manner (Liu et al. 2000; Zhao and Piwnica-Worms 2001). The phosphorylation of these serines is required for the HU-induced activation of Chk1 (Zhao and Piwnica-Worms 2001). As shown in Figure 2C, the UV-induced phosphorylation of Chk1 on Ser 345 was attenuated when the expression of Rad17 was inhibited, showing that the ATR-dependent phosphorylation of Chk1 is mediated by Rad17. This is consistent with previous studies on the homologous genes in Schizosaccharomyces pombe (O'Connell et al. 2000).
Damage-induced phosphorylation of Rad17 in ATR−/− and A-T cells
The role of Rad17 in mediating the phosphorylation of Chk1 by ATR indicates that Rad17 itself might be present in the same complex as ATR and Chk1 after damage. It was recently reported that Rad17 was phosphorylated on serines 635 and 645 after damage and that the phosphorylation could be inhibited by overexpression of kinase-inactive ATR (Bao et al. 2001). To address more directly whether ATR is responsible for the damage-induced phosphorylation of Rad17, we examined the phosphorylation of Rad17 in ATR−/− cells (Cortez et al. 2001). Given that ATR is essential for cell viability (Cortez et al. 2001), ATR−/− cells could only be obtained by excising the conditional ATR allele from ATRflox/− cells. In the ATRflox/− cells, exon 2 of one ATR allele was disrupted, and exon 2 of the second ATR allele was flanked by two loxP sites (Cortez et al. 2001). The removal of exon 2 leads to a frame shift at amino acid 20 followed by an in-frame Stop codon in exon 3. To produce ATR−/− cells, ATRflox/− cells were infected with Cre-expressing adenovirus (Ad-Cre). Because ATR−/− cells underwent apoptosis after 5 d, UV, HU, and IR treatments were conducted 3–4 d after infection, when the level of ATR protein was reduced by >90% (Fig. 3A,B). As controls, ATRflox/− cells infected with adenovirus expressing GFP (Ad-GFP) and parental cells (HCT116) infected with Ad-Cre were also treated. To monitor the phosphorylation of Rad17 on Ser 635, we generated an antibody that specifically recognized phosphorylated Ser 635 (p-Ser 635). The UV-induced phosphorylation of Rad17 was readily detected in control cells but not in ATR−/− cells (Fig. 3B). The residual signal of phosphorylated Rad17 is likely caused by the cells that did not delete the conditional allele of ATR. In contrast, the HU-induced phosphorylation of Rad17 was reduced but not abolished in ATR−/− cells (Fig. 3B). Interestingly, the IR-induced phosphorylation of Rad17 in ATR−/− cells was only slightly reduced during the first 2 h but quickly diminished thereafter, indicating a biphasic response, the latter of which is ATR-dependent (Fig. 3B).
Figure 3.
Phosphorylation of Rad17 by ATR and ATM. (A) HCT116 (ATR+/+) or its ATRflox/− derivative cells infected with adenovirus expressing Cre (Ad-Cre) or adenovirus expressing GFP (Ad-GFP) were collected at the indicated times after infection. Extracts were prepared from these cells and analyzed by immunoblotting with antibodies to ATR and ATM. (B) ATR+/+ and ATRflox/− cells were infected with Ad-Cre or Ad-GFP as indicated. Four days after infection, the cells were treated with 50 J/m2 of UV, 20 Gy of γ-IR, or 1 mM of HU and collected at the indicated times after the treatments. Extracts of the cells were analyzed by immunoblotting with the indicated antibodies. (C) A-T cells (AT22IJE-T) or A-T cells complemented with ATM (AT22IJE-T/YZ5) were treated with 20 Gy of γ-IR and collected at the indicated times after irradiation. Extracts of the cells were analyzed by immunoblotting with the indicated antibodies. (*) A protein nonspecifically recognized by the anti-ATM antibody.
HU and IR could induce phosphorylation of Rad17 even in the absence of ATR, suggesting that ATM might also phosphorylate Rad17 in vivo. To test whether ATM plays a major role in Rad17 phosphorylation when ATR is present, we compared the phosphorylation of Rad17 in A-T cells and in A-T cells complemented with wild-type ATM. In response to IR, UV, and HU, Rad17 was efficiently phosphorylated in A-T cells (Fig. 3C; data not shown). The expression of ATM in A-T cells had no apparent effects on the UV- and HU-induced phosphorylation of Rad17 (data not shown). However, ATM slightly enhanced the phosphorylation of Rad17 during the first hour after IR irradiation (Fig. 3C), implicating ATM in the initial phosphorylation of Rad17 in response to double-strand breaks.
Damage-induced phosphorylation of Rad17 on chromatin
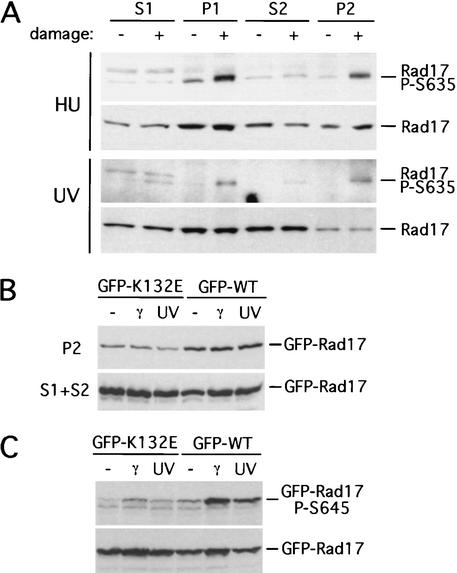
Although the association of Rad17 with chromatin is not regulated by damage, its ability to recruit Rad9 and to mediate Chk1 activation is damage-dependent. To test whether the chromatin-bound Rad17 is regulated by phosphorylation, we fractionated the extracts of HU- or UV-treated mouse embryonic fibroblasts (MEFs) and examined the distribution of phosphorylated Rad17 among the fractions (Fig. 4A). Most of the phosphorylated Rad17 was detected on chromatin. Because the amounts of Rad17 on chromatin did not change significantly after damage, the pool of Rad17 already on chromatin was most likely phosphorylated after damage. Unlike the phosphorylated Rad17, hyperphosphorylated Rad9 was detected both on and off chromatin (Fig. 1B). The release of hyperphosphorylated Rad9 from chromatin suggests that the association of the Rad1–Rad9–Hus1 complex with chromatin is dynamic.
Figure 4.
Phosphorylation of Rad17 by ATR on chromatin. (A) MEF cells (BS-1/pCaggs-Hus1) were untreated or treated with HU (1 mM for 24 h) or UV (80 J/m2) and harvested after 2 h and fractionated as in Figure 1A. Fractions from these cells were analyzed by immunoblotting with the indicated antibodies. (B,C) 293T cells transfected with plasmids expressing either GFP–rad17K132E or GFP–Rad17 were untreated or treated with 20 Gy of γ-IR or 80 J/m2 of UV and harvested after 2 h. Fractions (B) and extracts (C) were prepared from these cells and analyzed by immunoblotting with antibodies to Rad17 and p-Ser645 of Rad17.
The association of RFC with DNA requires ATP (Tsurimoto and Stillman 1991). Like all five RFC subunits, Rad17 possesses an ATP-binding domain (Parker et al. 1998b). GFP–rad17K132E, a mutant that lacks the conserved lysine in the Walker A motif, exhibited poor association with chromatin compared with GFP–Rad17 (Fig. 4B). Consistent with the idea that the efficient chromatin association of Rad17 might be a prerequisite for its phosphorylation, GFP–rad17K132E was not efficiently phosphorylated in the cells treated with IR or UV (Fig. 4C).
Rad17 associates with chromatin and recruits Rad9 independently of ATR
If Rad17 functions as a sensor of DNA damage, one role it might play is to recruit ATR to DNA lesions. To assess this possibility, we first tested if ATR bound to chromatin in a damage-dependent manner. Surprisingly, as in the case of Rad17, ATR associated with chromatin both before and after UV treatment (Fig. 5A). Furthermore, the amounts of ATR on chromatin did not increase in damaged cells. The chromatin association of ATR was verified by micrococcal-nuclease digestion and solubilization (data not shown). To address whether ATR required Rad17 to bind to chromatin, we analyzed the chromatin association of ATR in the cells transfected with the siRNA targeting Rad17. ATR still associated with chromatin when the level of Rad17 was significantly reduced (Fig. 5A), suggesting that ATR could bind to chromatin independently of Rad17. However, the amounts of ATR on chromatin were slightly reduced when the abundance of Rad17 was low, which indicates that Rad17 might help to stabilize the association of ATR with chromatin. We also tested if Rad17 needed ATR to associate with chromatin. In ATR−/− cells, Rad17 was still bound to chromatin before and after UV irradiation (Fig. 5B). Moreover, the amounts of Rad9 on chromatin increased similarly in ATR+/+ and ATR−/− cells after UV treatment (Fig. 5C). We therefore conclude that Rad17 can bind to chromatin and recruit Rad9 in the absence of ATR.
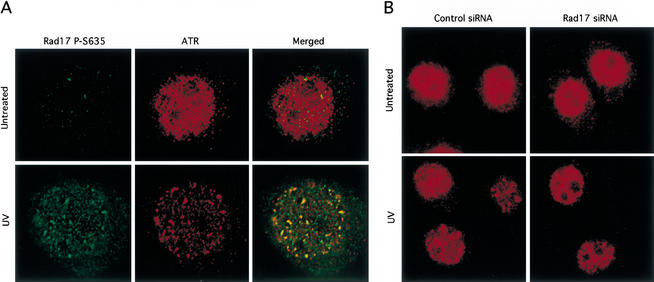
ATR is localized to nuclear foci in response to replication blocks or DNA damage (Tibbetts et al. 2000), indicating a damage-induced redistribution of ATR on chromatin. By using the antibody to p-Ser 635, we detected nuclear foci of phosphorylated Rad17 in UV-treated cells (Fig. 6A). Importantly, the foci of phosphorylated Rad17 colocalized with ATR foci, indicating the presence of both proteins at sites of DNA damage. To determine whether Rad17 is required for the formation of ATR foci, we examined the localization of ATR in the cells in which Rad17 expression was inhibited (Fig. 6B). Upon UV irradiation, ATR foci were detected in both the cells transfected with the siRNA targeting Rad17 and the cells transfected with control siRNA. Given that the residual Rad17 in the cells transfected with the siRNA targeting Rad17 could not efficiently support Rad9 loading and Chk1 activation, this observation suggests that the formation of ATR foci is largely independent of Rad17. However, we cannot exclude the possibility that the low residual amounts of Rad17 play a role in the redistribution of ATR.
Figure 6.
Colocalization of ATR and phosphorylated Rad17 in UV-induced nuclear foci. (A) HeLa cells were untreated or treated with 50 J/m2 of UV. After 4 h, the cells were immunostained with antibodies to ATR and p-Ser635 of Rad17. (B) HeLa cells transfected with siRNA targeting Rad17 or control siRNA were untreated or treated with UV as in A. After 4 h, the cells were immunostained with antibodies to ATR.
Hus1 mediates the phosphorylation of Rad17 by ATR
Although both Rad17 and ATR associate with chromatin prior to damage, the phosphorylation of Rad17 is clearly stimulated by damage. These findings suggest that the phosphorylation of Rad17 by ATR might be mediated by other proteins that bind to chromatin in a damage-dependent manner. Because the Rad1–Rad9–Hus1 complex is recruited onto chromatin by Rad17 after damage, it emerges as a likely candidate for such a mediator.
The existence of Hus1-null MEF cells made it possible to analyze the phosphorylation of Rad17 in the absence of the Rad1–Rad9–Hus1 complex (Weiss et al. 2000). Compared with Hus1−/− p21−/− cells complemented with wild-type Hus1, Hus1−/− p21−/− cells showed much reduced levels of Rad9 (Fig. 7A). Furthermore, the UV- or HU-induced hyperphosphorylation of Rad9 was not seen in Hus1-null cells (Fig. 7A,B). Finally, Rad9 was only detected in the cytoplasmic fraction of Hus1−/− p21−/− cells before and after HU treatment, indicating that the nuclear import and chromatin binding of Rad9 relied on Hus1 (Fig. 7B). Consistently, the nuclear localization of the fission yeast Hus1 requires Rad1 and Rad9 (Caspari et al. 2000). Both the chromatin association and the damage-induced hyperphosphorylation of Rad9 were rescued by the expression of Hus1 in Hus1−/− p21−/− cells (Fig. 7A,B). Taken together, these results suggest that the deletion of Hus1 abolished the integrity and function of the Rad1–Rad9–Hus1 complex.
Figure 7.
Requirement of Hus1 for Rad17 phosphorylation. (A) Mouse Hus1−/− p21−/− cells (BS-1/pCaggs) or Hus1−/− p21−/− cells complemented with Hus1 (BS-1/pCaggs-Hus1) were untreated or treated with HU (1 mM for 24 h) or UV (80 J/m2, harvested after 2 h). Extracts of these cells were analyzed by immunoblotting with the antibodies to Rad9 and actin. (B) Hus1−/− p21−/− cells or Hus1−/− p21−/− cells complemented with Hus1 were untreated or treated with HU as in A. Fractions from these cells were analyzed by immunoblotting. (C) The extracts described in A were analyzed by immunoblotting with antibodies to Rad17, p-S635 of Rad17, and actin. (D) Hus1−/− p21−/− cells or Hus1−/− p21−/− cells complemented with Hus1 were untreated or treated with 20 Gy of γ-IR and collected at the indicated times after irradiation. Extracts of these cells were analyzed by immunoblotting as in C.
Although the level of Rad17 was slightly reduced in Hus1-null cells (Fig. 7C,D), its association with chromatin was not affected (data not shown). The UV-induced phosphorylation of Rad17 was abolished in Hus1−/− p21−/− cells just as in ATR−/− cells (Fig. 7C). The HU-induced phosphorylation of Rad17 was also considerably reduced in Hus1-null cells (Fig. 7C). Like ATR, Hus1 is not essential for the IR-induced phosphorylation of Rad17. After Hus1−/− p21−/− cells were treated with IR, Rad17 was initially phosphorylated but quickly lost this modification, indicating that ATM is rapidly and transiently activated in the absence of Hus1 (Fig. 7D). Thus, the damage-induced phosphorylation of Rad17 exhibits remarkably similar dependencies on Hus1 and ATR. This indicates strongly that the phosphorylation of Rad17 by ATR is mediated by the Rad1–Rad9–Hus1 complex.
We noticed that the HU-induced phosphorylation of Rad17 was more severely reduced in Hus1−/− cells than in ATR−/− cells. It is possible that HU causes the accumulation of DNA damage in the absence of ATR that results in ATM activation. The amount or kind of DNA damage caused by HU might be less severe in Hus1−/− cells than in ATR mutants, resulting in less activation of ATM. Alternatively, it is possible that the activation of ATM by HU is partially compromised in Hus1−/− cells. The Rad1–Rad9–Hus1 complex might be involved in the processing of HU-induced damage that facilitates the activation of ATM. It is also possible that the presence of ATR in Hus1−/− cells might prevent the efficient activation of ATM by HU by carrying out a limited response to replication blocks.
Discussion
Rad17 controls loading of the Rad1–Rad9–Hus1 complex onto chromatin
In this study we provide evidence that the human Rad17 protein acts to promote the loading of the PCNA-related Rad1–Rad9–Hus1 complex onto chromatin in response to DNA damage. This is supported by two observations. The first is that increasing Rad17 levels stimulates both basal and UV-inducible loading of the Rad1–Rad9–Hus1 complex onto chromatin. Second, reducing Rad17 levels in vivo reduces the amount of Rad1–Rad9–Hus1 complex loaded onto DNA in response to damage. Rad17 is structurally related to RFC1, a DNA-binding subunit of the RFC complex that acts to load PCNA onto primed templates during DNA replication (Tsurimoto and Stillman 1991). RFC1 binds to the smaller RFC2-5 complex of proteins to form the RFC complex. Human Rad17 associates with all four small RFC subunits in vivo and comigrates with at least one of them on glycerol gradients (Lindsey-Boltz et al. 2001), suggesting that it might form an RFC-like complex with RFC2-5 as shown in budding yeast. Our results are consistent with a model in which Rad17 acts to replace RFC1 in the RFC complex to confer different recognition properties on the complex. The Rad17–RFC complex might now recognize damaged DNA and act as a DNA-damage-regulated machine that loads the Rad1–Rad9–Hus1 complex onto sites of damage.
Chromatin association of Rad17 correlates with function
The ATP-binding motif in RFC1 is required for the replication and ATPase activities of RFC (Cai et al. 1998). We find that a Rad17 mutant lacking the ATP-binding motif associates poorly with chromatin and is unable to efficiently promote Rad9 binding to chromatin when overexpressed (data not shown). These data further support the model in which the Rad17–RFC complex associates with damaged chromatin and recruits the Rad1–Rad9–Hus1 complex in a manner similar to PCNA loading by RFC.
Rad17 is phosphorylated in response to DNA damage, and overexpression of the nonphosphorylatable mutant of Rad17 has been shown to interfere with DNA-damage-induced cell-cycle arrest (Bao et al. 2001). We found that the amounts of Rad17 on chromatin do not change in response to DNA damage but that the pool of phosphorylated Rad17 is almost exclusively chromatin-bound, suggesting that the phosphorylation occurs on chromatin. Consistent with this observation, we found that the Rad17 mutant that binds poorly to chromatin shows significantly less DNA-damage-induced phosphorylation.
Regulation of Rad17 by ATR
Rad17 is phosphorylated on serines 635 and 645 after damage (Bao et al. 2001). We have provided conclusive evidence that Rad17 phosphorylation is regulated by ATR, using a conditional ATR mutant cell line. Our findings are consistent with a recent report that arrived at a similar conclusion using a less definitive technique, the overproduction of a kinase-defective ATR mutant that could potentially interfere with multiple pathways (Bao et al. 2001). Our findings indicate that ATR is primarily, if not entirely, responsible for the UV-induced phosphorylation of Rad17. In addition, we observed that Rad17 was still phosphorylated in HU-treated ATR−/− cells, indicating that another kinase, most likely ATM, responds to HU-induced damage in the absence of ATR. In the absence of DNA replication checkpoint function in yeast, HU treatment has been shown to activate the DNA-damage checkpoint pathway (Alcasabas et al. 2001). Finally, in response to double-strand breaks, we found that ATR is not essential for the initial phosphorylation of Rad17, but it is important for the maintenance of this phosphorylation. The initial phosphorylation is likely to be due to ATM. The function of the Rad17 phosphorylation at the molecular level remains to be determined.
ATR and Rad17, two complexes independently recruited to sites of DNA damage
The properties one would associate with a sensor of DNA damage would be the ability to associate with damaged DNA and a genetic requirement for signaling the presence of damaged DNA (Zhou and Elledge 2000). The ATR–ATRIP, Rad17–RFC, and Rad1–Rad9–Hus1 complexes all have such properties and were candidate sensors. ATR–ATRIP complexes localize to foci in response to DNA damage that are also sites of Nbs1 and Brca1 localization (Tibbetts et al. 2000). Human Mre11 was previously shown to only form foci in portions of the nucleus that had incurred DNA damage (Nelms et al. 1998), providing an indirect but compelling argument that the chromatin-localized foci containing these proteins corresponded to DNA damage. The organization of this pathway was unclear. The structural resemblance of Rad17 to RFC made it a strong candidate for a sensor that would work upstream of ATR–ATRIP to control its function. On the other hand, there was evidence that the ATRIP homolog Rad26 in S. pombe was phosphorylated by the ATR homolog Rad3 independently of Rad17 (Edwards et al. 1999), suggesting that ATR–ATRIP proteins might be upstream.
Our data indicate that the chromatin associations of Rad17 and ATR are largely independent, suggesting that both of these proteins have the potential to associate with DNA independently. Furthermore, phosphorylated Rad17 colocalizes with ATR in nuclear foci after UV irradiation, indicating that both proteins are present at sites of DNA damage. Consistent with a direct role of the ATR–ATRIP complex in damage detection, the formation of the UV-induced ATR foci is independent of Rad17. In the absence of ATR, Rad17 can not only associate with chromatin but also recruit Rad9 onto chromatin after UV irradiation. These data unambiguously implicate the Rad17 and Rad1–Rad9–Hus1 complexes in an ATR-independent sensory pathway in human cells. The two groups of sensors might have different structural specificities. It is possible that they work in concert to reinforce the specificity for damage detection and prevent inappropriate activation of the checkpoint. Alternatively, these sensors might function jointly to sensitize the detection of certain types of DNA damage.
Recently, two studies in yeast have shown that the counterparts of the Rad1–Rad9–Hus1 and ATR–ATRIP complexes can be recruited to an HO-induced double-strand break independently (Kondo et al. 2001; Melo et al. 2001). Our data concerning IR are completely consistent with their observations, and our studies on UV and HU indicate that this is a general response to all types of damage. Our conclusion that the localization of ATR is not Rad17-dependent is based on the reduction in Rad17 levels by siRNA but not the complete elimination of Rad17 by mutation. However, both yeast studies showed no difference in the loading of Mec1–Ddc2 complexes in strains deleted for RAD24, the gene encoding the Rad17 homolog.
A two-step model for checkpoint activation
As noted above, Rad17 phosphorylation in response to DNA damage is ATR-dependent. In vitro, ATR has been shown to phosphorylate the relevant Rad17 sites, suggesting that Rad17 is directly phosphorylated by ATR (Kim et al. 1999). Furthermore, phosphorylated Rad17 colocalizes with ATR in discrete nuclear foci after UV irradiation. These findings suggest that Rad17 is phosphorylated by ATR at sites of DNA damage. Although Rad17 and ATR might localize to DNA lesions independently, Rad17 cannot be phosphorylated by ATR after UV irradiation in the absence of Hus1. This supports the argument that the Rad1–Rad9–Hus1 complex is important for mediating the phosphorylation of Rad17 by ATR, suggesting a two-step model for checkpoint activation. One step is the loading of the ATR–ATRIP complex onto sites of damaged chromatin, and the other step is loading of the Rad1–Rad9–Hus1 complex.
The Rad1–Rad9–Hus1 complex could serve to organize the higher-order structure of the entire checkpoint protein complex to facilitate signaling. Once loaded, the Rad1–Rad9–Hus1 complex could act to bring ATR–ATRIP to Rad17 for phosphorylation, a substrate-recruitment role. A damage-induced association between Rad17 and ATR was recently reported (Bao et al. 2001). The Rad1–Rad9–Hus1 complex might be involved in bridging this interaction. Alternatively, the Rad1–Rad9–Hus1 complex could activate the ATR–ATRIP complex when it encounters Rad17 on chromatin. For example, the Rad1–Rad9–Hus1 complex might promote the phosphorylation of Rad17 by bringing in additional factors and/or processing the DNA lesions. Rad9 and Hus1 were shown to interact with the histone deacetylase HDAC1 (Cai et al. 2000), and Rad9 was shown to interact with TopBP1, a protein containing multiple BRCT domains (Makiniemi et al. 2001). In addition, both Rad1 and Rad9 were reported to have 3′ to 5′ exonuclease activity (Parker et al. 1998a; Bessho and Sancar 2000), which might influence ATR activity if it responds to DNA structure.
We have previously shown that the damage-induced phosphorylation of Chk1 is dependent on ATR (Liu et al. 2000). We show here that the Chk1 phosphorylation induced by UV also requires Rad17. These findings suggest that the two parallel sensory pathways converge before the phosphorylation of Chk1. It is possible that Chk1 is recruited to chromatin by the interaction of these two complexes to become a substrate of ATR.
At present, it is not clear if the ATR–ATRIP and Rad17–RFC complexes are the primary sensors of DNA damage, or if they are merely independently recruited to sites of damage. Resolution of this issue will require biochemical reconstitution of the activities of these complexes and direct testing of the recruitment model for substrate recognition. Another important issue is the relative roles of these complexes in response to DNA damage versus roles during DNA replication. Are they activated during normal replication and superactivated in response to DNA damage? As ATR is essential for cellular viability, it must have a role in every cell cycle. These issues will require future scrutiny to unravel the function of these important complexes.
Materials and methods
Chromatin fractionation
Chromatin fractionations were performed essentially as described by Mendez and Stillman (2000). A total of ∼3 × 106 cells were washed with PBS and resuspended in 200 μL of solution A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 10 mM NaF, 1 mM Na2VO3, protease inhibitors). Triton X-100 was added to a final concentration of 0.1%, and the cells were left on ice for 5 min. Cytoplasmic proteins (S1) were separated from nuclei (P2) by low-speed centrifugation (1300g for 4 min). Isolated nuclei were washed once with solution A and lysed in 200 μL of solution B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT). After a 10-min incubation on ice, soluble nuclear proteins (S2) were separated from chromatin (P2) by centrifugation (1700g for 4 min). Isolated chromatin was washed once with solution B and spun down at high speed (10,000g for 1 min). Finally, chromatin was resuspended in 200 μL of SDS sample buffer and sheared by sonication. To digest chromatin with micrococcal nuclease, nuclei (P1) were resuspended in solution A containing 1 mM CaCl2 and 50 units of micrococcal nuclease (Sigma). After 2 min of incubation at 37°C, nuclei were lysed and fractionated as above.
Plasmids, oligonucleotides, and transfections
The Univector Plasmid-fusion System was used to generate the expression plasmids for Rad17 and Rad9 (Liu et al. 1998). Briefly, the Rad17 and Rad9 genes were first cloned into the univector pUNI50 and then fused to expression vectors pHM202–Flag and pHM202–GFP, respectively. The transfections were carried out with Lipofectamine Plus reagent (Life Technologies).
The siRNA duplex targeting Rad17 was prepared by annealing two 21-ribonucleotide oligonucleotides. The sequences of the oligonucleotides were: CAGACUGGUUGACCCAUCTT and GAUGGGUCAACCCAGUCUGTT. HeLa cells were transfected twice 24 h apart with the siRNA duplex and Oligofectamine (Life Technologies), and were analyzed 72 h after the first transfection.
Antibodies
The phospho-specific antibody to pS635 of Rad17 was raised against the peptide CLPL(pS)QNSGS. Generation of the phospho-specific antibody to pS345 of Chk1 was previously described (Liu et al. 2000). The rabbit polyclonal antibodies to Rad17 and Rad9, the goat polyclonal antibody to ATR/FRP1, and the mouse monoclonal antibody to Chk1 were purchased from Santa Cruz Biotechnology. The mouse antibody to Orc2 was obtained from Pharmingen.
Adenovirus infection
Adenoviruses expressing Cre (Ad-Cre) or GFP (Ad-GFP) were obtained from the Tissue Culture Facility at Baylor College of Medicine. A total of ∼1 × 106 ATR+/+ (HCT116) or ATRflox/− cells were seeded on 10-cm plates 24 h before infection. The amounts of adenovirus used were empirically determined for maximum deletion of the conditional allele of ATR and low toxicity (Cortez et al. 2001). The viruses were removed after 24 h, and the cells were allowed to grow in virus-free medium for 3 days.
Immunofluorescence
HeLa cells were grown on glass coverslips in 35-mm dishes. Fixation and permeablization were performed with 3% paraformaldehyde for 10 min followed by 0.5% triton for 10 min. Samples were blocked in 1% BSA at room temperature for 10 min and then incubated with anti-Rad17 S635P (5 μg/mL) or anti-ATR (2 μg/mL) antibodies at 37°C for 1 h. After extensive washing, samples were incubated in FITC- or Cy3-conjugated secondary antibodies (Jackson Immnuoresearch Laboratories) at 37°C for 45 min. Images were obtained on either a Nikon microscope with Delta Vision deconverlution software or a Zeiss LSM510 confocal microscope.
Acknowledgments
We thank R. Weiss and P. Leder for the Hus1-null cells and L. Karnitz for plasmids and antibodies. This work was supported by a grant from NIH to S.E. (GM44664). L.Z. is a Fellow of the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation. S.E. is an Investigator with the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL selledge@bcm.tmc.edu; FAX (713) 798-8717.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.950302.
References
- Alcasabas A, Osborn A, Bachant J, Hu F, Werler P, Bousset K, Furuya K, Diffley J, Carr AM, Elledge SJ. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A, Chen SM, Abraham RT, Wang XF. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature. 2001;411:969–974. doi: 10.1038/35082110. [DOI] [PubMed] [Google Scholar]
- Bessho T, Sancar A. Human DNA damage checkpoint protein hRAD9 is a 3′ to 5′ exonuclease. J Biol Chem. 2000;275:7451–7454. doi: 10.1074/jbc.275.11.7451. [DOI] [PubMed] [Google Scholar]
- Burtelow MA, Kaufmann SH, Karnitz LM. Retention of the human Rad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J Biol Chem. 2000;275:26343–26348. doi: 10.1074/jbc.M001244200. [DOI] [PubMed] [Google Scholar]
- Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9–hHus1–hRad1 (9-1-1) DNA damage responsive checkpoint complex. J Biol Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- Cai J, Yao N, Gibbs E, Finkelstein J, Phillips B, O'Donnell M, Hurwitz J. ATP hydrolysis catalyzed by human replication factor C requires participation of multiple subunits. Proc Natl Acad Sci. 1998;95:11607–11612. doi: 10.1073/pnas.95.20.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai RL, Yan-Neale Y, Cueto MA, Xu H, Cohen D. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J Biol Chem. 2000;275:27909–27916. doi: 10.1074/jbc.M000168200. [DOI] [PubMed] [Google Scholar]
- Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: Partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Edwards RJ, Bentley NJ, Carr AM. A Rad3–Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- Griffiths DJ, Barbet NC, McCready S, Lehmann AR, Carr AM. Fission yeast rad17: A homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Recruitment of mec1 and ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- Lindsey-Boltz LA, Bermudez VP, Hurwitz J, Sancar A. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc Natl Acad Sci. 2001;98:11236–11241. doi: 10.1073/pnas.201373498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Li MZ, Leibham D, Cortez D, Elledge SJ. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Chk1 is an essential kinase that is regulated by ATR and required for the G(2)/M DNA damage checkpoint. Genes & Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Makiniemi M, Hillukkala T, Tuusa J, Reini K, Vaara M, Huang D, Pospiech H, Majuri I, Westerling T, Makela TP, et al. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J Biol Chem. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes & Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5888–5896. doi: 10.1128/mcb.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- O'Connell MJ, Walworth NC, Carr AM. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 2000;10:296–303. doi: 10.1016/s0962-8924(00)01773-6. [DOI] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes & Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Parker AE, Van de Weyer I, Laus MC, Oostveen I, Yon J, Verhasselt P, Luyten WH. A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J Biol Chem. 1998a;273:18332–18339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- Parker AE, Van de Weyer I, Laus MC, Verhasselt P, Luyten WH. Identification of a human homologue of the Schizosaccharomyces pombe rad17+ checkpoint gene. J Biol Chem. 1998b;273:18340–18346. doi: 10.1074/jbc.273.29.18340. [DOI] [PubMed] [Google Scholar]
- Rouse J, Jackson SP. LCD1: An essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 2000;19:5801–5812. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge RP, Udell CM, Casselman R, Davey S. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol Biol Cell. 1999;10:1985–1995. doi: 10.1091/mbc.10.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen MP, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes & Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer–template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer E, Karnitz LM. Human homologs of Schizosaccharomyces pombe rad1, hus1, and rad9 form a DNA damage-responsive protein complex. J Biol Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Kondo T, Ando S, Matsumoto K, Sugimoto K. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:755–764. doi: 10.1128/MCB.21.3.755-764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes & Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]