Abstract
Loss of Kuzbanian, a member of the ADAM family of metalloproteases, produces neurogenic phenotypes in Drosophila. It has been suggested that this results from a requirement for kuzbanian-mediated cleavage of the Notch ligand Delta. Using transgenic Drosophila expressing transmembrane Notch proteins, we show that kuzbanian, independent of any role in Delta processing, is required for the cleavage of Notch. We show that Kuzbanian can physically associate with Notch and that removal of kuzbanian activity by RNA-mediated interference in Drosophila tissue culture cells eliminates processing of ligand-independent transmembrane Notch molecules. Our data suggest that in Drosophila, kuzbanian can mediate S2 cleavage of Notch.
Keywords: Notch, Kuzbanian, processing, Drosophila
Notch (N) is an ∼3000-amino-acid transmembrane protein that is found in a wide variety of organisms ranging from Caenorhabditis elegans to humans. It is a receptor in a signal transduction pathway that mediates intercellular communication (for recent reviews, see Greenwald 1998; Artavanis-Tsakonas et al. 1999; Mumm and Kopan 2000). Upon binding its ligands, members of the DSL (Delta, Serrate, Lag2) family of transmembrane ligands, N is cleaved in its extracellular domain at a site 11 amino acids amino terminal to the transmembrane domain (Brou et al. 2000; Mumm et al. 2000). In vitro this S2 cleavage of mammalian N can be mediated by TNF-α converting enzyme (TACE; Brou et al. 2000), a member of the ADAM family of metalloproteases (for recent reviews, see Schlondorff and Blobel 1999; Primakoff and Myles 2000). Following S2 cleavage, N undergoes an intramembranous cleavage (S3) to release the soluble cytoplasmic domain, which, in conjunction with a member of the CSL (CBF1, Suppressor of Hairless, Lag1) family of DNA-binding proteins, enters the nucleus and activates transcription (Kidd et al. 1998; Lecourtois and Schweisguth 1998; Schroeter et al. 1998; Struhl and Adachi 1998). This S3 cleavage requires Presenilin (Psn) activity (De Strooper et al. 1999; Struhl and Greenwald 1999; Ye et al. 1999).
Mammalian N, but not Drosophila N, is also constitutively cleaved as part of its maturation process, in its extracellular domain at amino acid 1654, so that it is presented on the cell surface as a heterodimer (Blaumueller et al. 1997; Kidd et al., in prep.). This S1 cleavage was originally thought to be carried out by Kuzbanian (Pan and Rubin 1997), another member of the ADAM family, but has since been shown to be mediated by a furin-like enzyme (Logeat et al. 1998). More recently kuzbanian (kuz) has been shown to mediate the cleavage of Delta (Dl; Qi et al. 1999), yet in both Drosophila and C. elegans, kuz has been shown to be cell-autonomous, being required in the receiving cell (Rooke et al. 1996; Sotillos et al. 1997; Wen et al. 1997).
In this paper we show that Kuz can physically associate with N. This association led us to reexamine the role of kuz in the cleavage of N. We generated transgenic Drosophila expressing transmembrane N proteins that can act independently of Dl and assayed the function of these proteins in embryos, both phenotypically and biochemically. The function of a N protein whose activity is completely independent of Dl is almost completely dependent on kuz. Using RNA-mediated interference in Drosophila S2 cells, which do not express any known N ligands, we show that the cleavage of N proteins that can function independently of Dl requires kuz. This kuz activity acts upstream of Psn activity. Our data suggest that in Drosophila, kuz can mediate S2 cleavage of N.
Results
Kuzbanian can physically associate with Notch
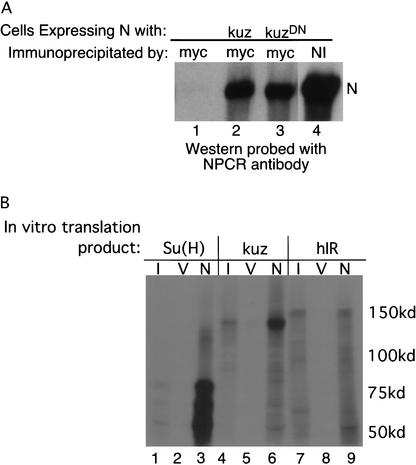
We added 6 myc epitope tags to the carboxyl termini of both Kuz and a dominant-negative form of Kuz, lacking the protease domain KuzDN (Pan and Rubin 1997). Myc-tagged Kuz or KuzDN was coexpressed with N in S2 cells, and the extracts were immunoprecipitated with anti-myc antibodies. As can be seen in Figure 2A, lanes 2 and 3 (below), N is coimmunoprecipitated by anti-myc antibodies only when it is coexpressed along with Kuz or KuzDN (Fig. 2, cf. lanes 2 and 3 with lane 1; these cells were transfected with N alone). To address whether this interaction is direct, we generated in bacteria a GST–N fusion protein encoding amino acids 1623–1893 of N (BD in Fig. 1A). In vitro translated Suppressor of Hairless [Su(H)] and Kuz can be pulled down by this GST–N fusion (Fig. 2B, lanes 3,6), but in vitro translated human insulin receptor cannot (Fig. 2B, lane 9). None of the in vitro translation products associates with GST alone (Fig. 2B, lanes 2,5,8). Thus there is a direct interaction between Kuz and N.
Figure 2.
Kuz can associate with N. (A) S2 cells transfected with actin-driven N alone (lanes 1,4) or with actin N plus actin-driven myc-tagged kuz (lane 2) or kuzDN (lane 3) were immunoprecipitated with anti-myc (lanes 1–3) or anti-NI (lane 4) antibodies, and the Western probed with anti-NPCR antisera. See Figure 1A for location of the epitopes recognized by the antibodies. (B) Bacterially produced GST–N fusion protein encoding amino acids 1623–1893 of N (from the end of the LNG repeats to the start of the ankyrin repeats, BD in Fig. 1A) was used in pull-down assays of in vitro translated Su(H) (lane 3), Kuz (lane 6) and human insulin receptor (hIR, lane 9) labeled with 35S methionine. (I) 1% of the input in vitro translation product; (V) GST alone; (N) GST–N fusion.
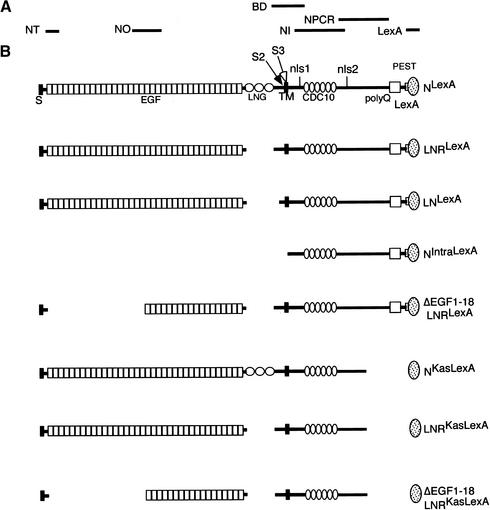
Figure 1.
Diagram of N constructs and localization of epitopes recognized by antibodies. (A) The antibodies used in this work, above the regions of N used to generate them, and the region of N fused to GST (BD) used in the pull-down assays in Figure 2B. The top molecule in B is wild-type N tagged with the DNA-binding domain of LexA, NLexA. (S) Signal sequence; (EGF) epidermal growth factor-like repeats; (LNG) Lin-12, N, Glp-1 repeats; (S2) location of TACE cleavage site in mammals; (S3) location of Psn-dependent cleavage site; (TM) transmembrane domain; (nls1, nls2) nuclear localization signals; (CDC10) cdc10 or ankyrin repeats; (polyQ) polymeric glutamines; (PEST) PEST sequence thought to be involved in protein stability. Shown beneath NLexA are the various deletions used in this work.
Whereas Pan and Rubin originally proposed that kuz is responsible for cleavage of N (Pan and Rubin 1997), more recently it has been suggested that the phenotypes resulting from loss of kuz are attributable to its role in the processing of Dl, and no effect of the loss of kuz on N proteins was seen in flies (Qi et al. 1999) or in mammalian cells (Brou et al. 2000; Mumm et al. 2000). Because of the association we observed between N and Kuz, we reinvestigated the role of kuz in the cleavage of N. Because it has been proposed that Kuz cleaves Dl, we worked with N molecules that can function independently of Dl.
The N molecules we have used to analyze the role of kuz are depicted in Figure 1. All the N proteins are tagged at their carboxyl termini with the DNA-binding domain of LexA so that their cleavage can be monitored in vivo, and all the Western blots of embryonic extracts described below were probed with anti-LexA antibodies. NLexA, NIntraLexA, which encodes the soluble cytoplasmic domain of N, and NΔLNrptsLexA (LNLexA; deleted for amino acids 1469–1625) have been described previously (Lieber et al. 1993; Kidd et al. 1998). The deletion in NΔLNRLexA (LNRLexA; deleted for amino acids 1482–1593) is a subset of NΔLNrptsLexA and encompasses just the LNG (Lin12/N/Glp1) repeats. NΔEGF1–18 and LNRLexA is missing EGF repeats 1–18, as well as amino acids 1482–1593.
When expressed in S2 cells along with myc-tagged Kuz or KuzDN, both LN and LNR can be coimmunoprecipitated by anti-myc antibodies. We did not observe a difference in the degree of association of N, LN, or LNR with Kuz or with KuzDN (data not shown).
The strength of the antineurogenic phenotype correlates with both the levels of S3 cleavage and nuclear localization
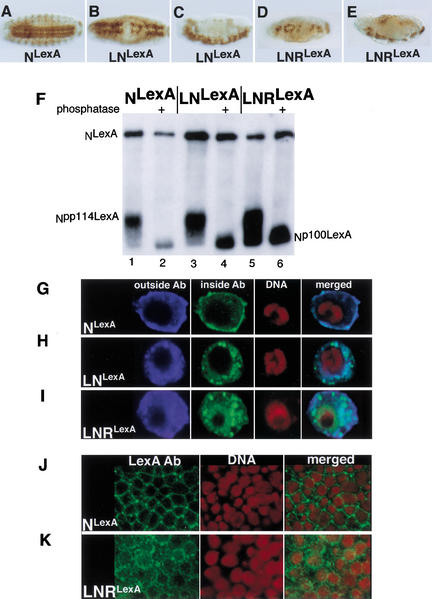
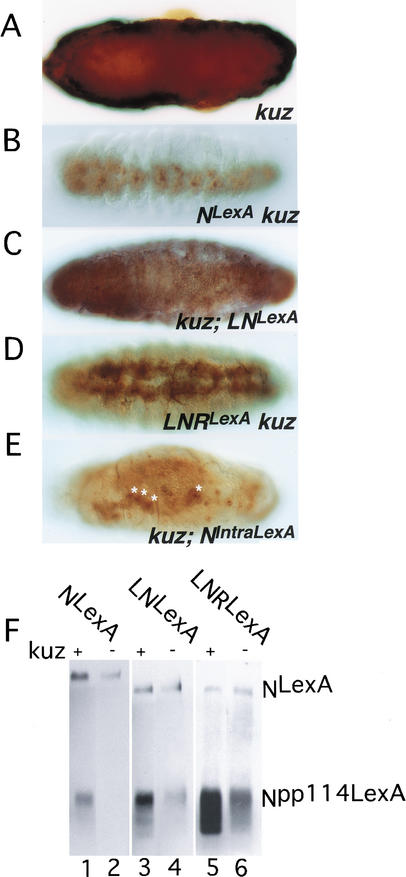
In Drosophila embryos loss of members of the N signal transduction pathway causes overproduction of neuroblasts at the expense of epidermoblasts, resulting in hypertrophy of the embryonic central and peripheral nervous systems. Conversely, expression of gain-of-function N molecules generates antineurogenic phenotypes in which neuroblasts are underproduced, resulting in the loss of central and peripheral nervous system elements. Figure 3, A–E, shows the effect on the morphology of the embryonic central nervous system (CNS) of ectopically expressing NLexA, LNLexA, or LNRLexA using daughterless (da) GAL4 as a driver. Although expression of NLexA does not grossly affect the structure of the CNS (Fig. 3A), the expression of LNLexA (Fig. 3B,C) or LNRLexA (Fig. 3D,E) produces gaps in the architecture of the CNS characteristic of an antineurogenic phenotype (Lieber et al. 1993). However, the antineurogenic phenotype produced by LNRLexA (Fig. 3D,E) is stronger than that produced by LNLexA (Fig. 3B,C). It is possible that this difference is due to the insertion sites of the transgenes, but we think this is unlikely for two reasons. First, we immunoprecipitated extracts of embryos expressing NLexA, LNLexA, and LNRLexA with anti-Su(H) antibodies and probed the Western with anti-LexA antibodies. We could then control for the expression level of the transgenes by determining the ratio of S3-cleaved N bound to Su(H) to uncleaved N associated with Su(H). An example is shown in Figure 3F. Although the ratio of S3-cleaved N to N (Npp114LexA or Np100LexA:NLexA) is approximately the same in extracts of embryos expressing NLexA and LNLexA, the ratio is at least 7-fold greater in embryos expressing LNRLexA. (Six separate experiments were quantitated.) Second, as can be seen in Figure 3, G–I, when transiently expressed in Drosophila S2 cells, the cytoplasmic domain of LNR (Fig. 3I) unlike that of N (Fig. 3G) and LN (Fig. 3H) can readily be detected in nuclei. Using anti-LexA antibodies, we were able to detect the cytoplasmic domain of LexA-tagged LNR in the nuclei of embryos as well (Fig. 3K). Thus, the strength of the antineurogenic phenotype correlates with both the levels of S3 cleavage and nuclear localization. This suggests that the function of a N molecule as assayed phenotypically in different genetic backgrounds can be correlated with the activity of this N molecule as assayed biochemically and provides a basis for us to interpret the effects of loss of elements of the N pathway on N function.
Figure 3.
A comparison of the activities of LNLexA and LNRLexA. (A–E) The effect that expressing NLexA (A), LNLexA (B,C), or LNRLexA (D,E) under control of daughterless (da) GAL4 (a ubiquitous driver) has on the nervous system of wild-type embryos. (A,B,D) Ventral views; (C,E) lateral views. The embryos were stained with an anti-horseradish peroxidase (HRP) antibody that reacts with the nervous system. (F) Western blot showing increased production of Npp114LexA in embryos expressing LNRLexA. Extracts of embryos expressing NLexA (lanes 1,2), LNLexA (lanes 3,4), or LNRLexA (lanes 5,6) under the control of daGAL4 were immunoprecipitated with anti-Su(H) antisera, and the Western was reacted with anti-LexA antisera. The immunoprecipitates in lanes 2, 4, and 6 were treated with phosphatase prior to electrophoresis. NLexA is the full-length N protein that coimmunoprecipitates with Su(H); Npp114LexA is the phosphorylated cleaved cytoplasmic domain that associates with Su(H); and Np100LexA is the phosphatased cytoplasmic domain that associates with Su(H) (Kidd et al. 1998). (G–I) The increase in nuclear entry of the cytoplasmic domain derived from LNRLexA. S2 cells were transfected with UAS constructs encoding NLexA, LNLexA, or LNRLexA, and a plasmid encoding heat shock (hs) GAL4 at a ratio of 1:10. Cells were heat-shocked for 30 min and allowed to recover for 2 h prior to fixation. They were then reacted with the NT antibody, which reacts with the extracellular domain; the NPCR antibody, which reacts with the intracellular domain; and sytox green to label the DNA. See Figure 1A for location of the epitopes recognized by the antibodies. (J,K) embryos expressing NLexA (J) or LNRLexA (K) under control of daGal4 were reacted with anti-LexA antibody to detect the N protein derived from the transgene and propidium iodide, which reacts with the DNA.
LN and LNR are Delta- and kuzbanian-independent but Delta- and kuzbanian- responsive
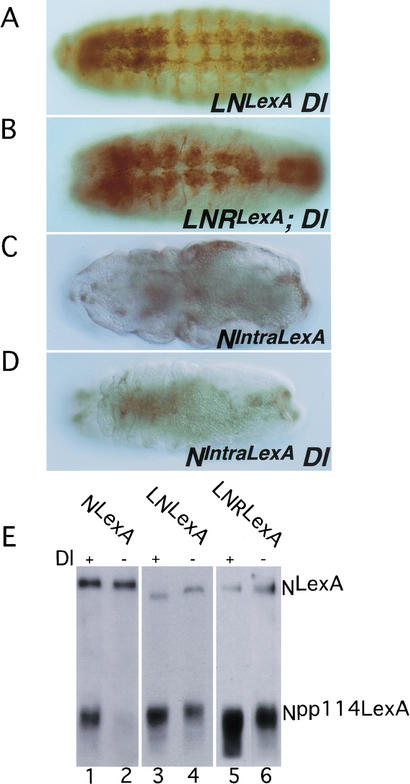
We then assayed the function of the mutated N proteins in Dl and kuz backgrounds. Because Dl is required in the germ line for proper development of follicle cells (López-Schier and St. Johnston 2001), we were unable to generate embryos that were maternally Dl null, and our experiments were therefore carried out in embryos that were zygotic Dl nulls. As can be seen in Figure 4, A and B, both LN and LNR can function independently of Dl, although in neither case was the antineurogenic phenotype produced in a Dl background as strong as that produced in a wild-type background (Fig. 4, cf. 4A with 3B and 4B with 3D). This contrasts with what we observe with NIntraLexA, where the antineurogenic phenotypes produced in Dl+ (Fig. 4C) and Dl− (Fig. 4D) backgrounds are comparable. This indicates that although LNLexA and LNRLexA do not require Delta for function, they do respond to Delta and have greater activity when Delta is present. The Dl independence but Dl responsiveness of LNLexA and LNRLexA is also apparent when we compare the ratios of S3-cleaved N bound to Su(H) to uncleaved N associated with Su(H) (Npp114LexA:NLexA) in extracts of Dl+ and Dl− embryos (Fig. 4E). Almost no Npp114LexA is detectable in the Dl NLexA extracts (Fig. 4E, lane 2) in accord with the neurogenic phenotype of Dl NLexA embryos (data not shown). Given that the phenotypes of LNLexA and LNRLexA are Dl-independent, it is not unexpected that Npp114 associated with Su(H) can still be detected in Dl LNLexA (Fig. 4E, lane 4) and Dl LNRLexA (Fig. 4E, lane 6) extracts, and this is further confirmation that the gain-of-function phenotypes observed are due to the unregulated production of the soluble cytoplasmic domain. However, although LNLexA and LNRLexA are ligand-independent, they are ligand-responsive. In a Dl background the ratio of Npp114:N in LNLexA and LNRLexA extracts is reduced by 40%–70% compared with that found in a wild-type background (three separate experiments were quantitated). This would account for the weaker antineurogenic phenotype produced by these proteins in the absence of Dl. We show below that these deleted N proteins do, indeed, have ligand-independent activity and that the residual activity we see in Dl zygotic nulls is not solely due to maternally contributed Dl.
Figure 4.
Signaling from LNLexA and LNRLexA is decreased in the absence of Dl. (A–D) Embryos expressing LNLexA (A), LNRLexA (B), and NIntraLexA (C,D) under control of daGAL4, reacted with anti-HRP antibody. The embryo in C is Dl+, the other three are zygotic Dl nulls. The same transgenic lines were used in wild-type (Fig. 3B–E; Fig. 4C) and Dl backgrounds. Of the three antineurogenic N proteins, NIntraLexA produces the strongest antineurogenic phenotype in a wild-type background. (E) Extracts of embryos expressing NLexA (lanes 1,2), LNLexA (lanes 3,4), and LNRLexA (lanes 5,6) under control of daGAL4, in either wild-type (lanes 1,3,5) or zygotic Dl backgrounds (lanes 2,4,6), were immunoprecipitated with anti-Su(H) antibody and the Western reacted with anti-LexA antibody. To ensure that the protein being characterized is derived from Dl embryos, both the N transgenes and daGAL4 were recombined onto Dl chromosomes.
We then assayed the phenotypes produced by LNLexA and LNRLexA in embryos that were maternally and zygotically kuz null. As can be seen in Figure 5A, using an antibody that reacts with nervous tissue, embryos lacking kuz show hypertrophy of nervous system elements resulting in the lack of an organized CNS (cf. Figs. 5A and 3A). Both LNLexA (Fig. 5C) and LNRLexA (Fig. 5D) can function independently of Kuz, as can be seen by the suppression of the neural hypertrophy and the presence of an identifiable CNS. But as we observe in Dl embryos, the phenotypes produced in kuz− embryos are weaker than those produced in kuz+ embryos (cf. Fig. 5C with 3B and 5D with 3D). Although there is suppression of the neurogenic phenotype in the case of LNLexA, no antineurogenic phenotype is manifest, and the antineurogenic phenotype of LNRLexA is weaker in a kuz background. Again, this contrasts with what we observe with NIntraLexA, which produces comparable phenotypes in kuz+ and kuz− backgrounds (cf. Fig. 5E with 4C). Therefore, both LNLexA and LNRLexA are kuz-responsive. Surprisingly, although expression of NLexA cannot suppress the Dl neurogenic phenotype (data not shown), it is able to suppress the kuz neurogenic phenotype (Fig. 5B). In fact, the size of the nervous system is smaller than in kuz LNRLexA embryos (Fig. 5, cf. B with D).
Figure 5.
Signaling from LNLexA and LNRLexA is decreased in the absence of Kuz. All the embryos (A–E) were reacted with anti-HRP antibody. (A) Ventral view of an embryo that is maternally and zygotically kuz null. (B–E) The effect of expressing in a maternal and zygotic kuz background NLexA (B), LNLexA (C), LNRLexA (D), and NIntraLexA (E). All the N proteins were expressed under control of daGAL4. The white stars in E point out the remnants of the nervous system in kuz; NIntraLexA embryos. The same transgenic lines were used in wild-type (Fig. 3A–E; Fig. 4C) and kuz backgrounds. (F) Extracts of embryos expressing NLexA (lanes 1,2), LNLexA (lanes 3,4), and LNRLexA (lanes 5,6) under control of daGAL4, in either wild-type (lanes 1,3,5) or maternal and zygotic kuz backgrounds (lanes 2,4,6) were immunoprecipitated with anti-Su(H) antibody and the Western reacted with anti-LexA antibody. To ensure that the protein being characterized is derived from kuz embryos, the N transgenes were recombined onto kuz chromosomes.
We next asked what effect the loss of Kuz has on S3 cleavage as measured by coimmunoprecipitation of Npp114LexA with Su(H). As was the case in Dl embryos, the ratio of Npp114:N in LNLexA and LNRLexA extracts is reduced by 50%–70% in kuz− embryos compared with in kuz+ embryos (Fig. 5F, lanes 3–6). The loss of Kuz is therefore affecting the phenotypes produced by LNLexA and LNRLexA by reducing the amount of S3-cleaved N. Although expression of NLexA is able to suppress the kuz neurogenic phenotype, there is a drastic reduction in the amount of Npp114 associated with Su(H) (Fig. 5F, lanes 1,2). As we have shown above that the strength of the antineurogenic phenotype correlates with the level of S3 cleavage, this suggests the phenotypic suppression is not caused by the canonical Dl/Su(H) pathway. We discuss this curious result below.
Delta and kuzbanian embryos differ in the cleavage products immunoprecipitated by anti-N antibodies
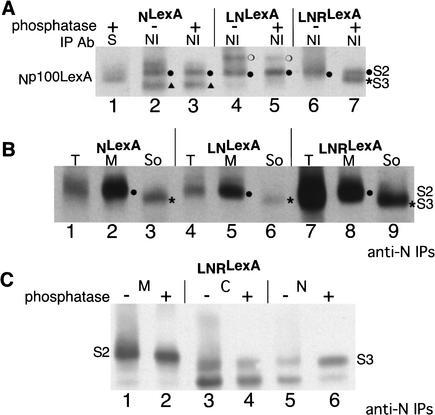
We are able to identify the band that corresponds to S3-cleaved N, the cytoplasmic form of N that associates with Su(H), in phosphatased anti-N immunoprecipitations of embryonic extracts based on its comigration with Np100LexA, the phosphatased soluble form of N that coimmunoprecipitates with Su(H) (Fig. 6A, lanes 1,7), and based on its localization in the soluble fraction when embryonic extracts are fractionated (Fig. 6B, cf. lanes 2 and 3, 5 and 6, 8 and 9). In LNRLexA extracts, S3-cleaved N is also found in nuclei (Fig. 6C, lanes 5,6). We are able to identify the band that corresponds to S2-cleaved N in phosphatased anti-N immunoprecipitations based on its accumulation in embryos that lack Psn (Fig. 6D, lane 4) and based on its localization in membranes (Fig. 6B, cf. lanes 2 and 3, 5 and 6, 8 and 9). Both S3 and S2 products are phosphorylated, although the level of phosphorylation of S2 is much lower than that of S3. (Compare the large shift in mobility upon phosphatase treatment of S3-cleaved N evident in anti-Su(H) immunoprecipitations of Figure 3F with the slight shift in mobility upon phosphastase treatment of S2-cleaved N in anti-N immunoprecipitates of the membrane fraction in Figure 6C, lanes 1 and 2. This shift is more easily visible on shorter exposures.)
Figure 6.
Anti-N immunoprecipitates of extracts from Dl and kuz embryos differ. (A) Extracts of embryos expressing NLexA (lanes 2,3), LNLexA (lanes 4,5), and LNRLexA (lanes 6,7), under the control of daGAL4 were immunoprecipitated with anti-NI antibody (see Fig. 1A) and the Western reacted with anti-LexA antibody. Extracts of embryos expressing NLexA were also immunoprecipitated with anti-Su(H) antibody (lane 1). The immunoprecipitates in lanes 1, 3, 5, and 7 were treated with phosphatase prior to electrophoresis. Np100LexA is the phosphatased cytoplasmic domain that interacts with Su(H) (Kidd et al. 1998). (S2 ●) Migration of S2-cleaved N; (S3 *) migration of S3-cleaved N; (S) Su(H); (▴ in lanes 2,3) an ∼97-kD cleavage product present in NI immunoprecipitates of extracts expressing NLexA that does not associate with Su(H), but is dependent on Psn (see text); (○ in lanes 5,6) an ∼108-kD cleavage product found in NI immunoprecipitates of LNLexA. (B) Extracts of embryos expressing NLexA (lanes 1–3), LNLexA (lanes 4–6), and LNRLexA (lanes 7–9), under the control of daGAL4, which had been fractionated into membrane and soluble fractions, were immunoprecipitated with anti-NI antibody and the Western reacted with anti-LexA antibody. (T) Unfractionated extract; (M) membrane fraction; (So) soluble fraction. All immunoprecipitates were phosphatased prior to electrophoresis. (C) Extracts of embryos expressing LNRLexA under the control of daGAL4 were fractionated into membrane, cytoplasmic, and nuclear fractions prior to immunoprecipitation with anti-NI antibody. The immunoprecipitates in lanes 2, 4, and 6 were phosphatased. The Western was reacted with anti-LexA antibody. (M) Membrane; (C) cytoplasm; (N) nuclear. The smear representing phosphorylated S3 in the nuclear fraction (lane 5) is more easily visible on longer exposures. (D,E) Extracts of embryos expressing NLexA were immunoprecipitated with anti-NI antibody (D, lanes 1–5; E, lanes 2–7) and the Western reacted with anti-LexA antibody. The extracts in lanes 1 and 5 of D and lanes 2 and 5 of E were from wild-type embryos (WT). The extracts in lane 2 of D and lanes 3 and 6 of E were from zygotic Dl embryos. The extracts in lane 3 of D and lanes 4 and 7 of E were from maternal and zygotic kuz embryos. The extract in lane 4 of D was from maternal and zygotic Psn embryos. (D, lane 6; E, lane 1) Extracts of NLexA embryos were also immunoprecipitated with anti-Su(H) antibody (S). The immunoprecipitates in lanes 4 and 5 of D were derived from embryos expressing NLexA under the control of armadillo (arm) GAL4. All the other immunoprecipitates were derived from embryos expressing NLexA under the control of daGAL4. (▴ in D, lanes 1,2,5; E, lanes 2,3,5,6) The NLexA derived protein that is dependent on Psn but does not associate with Su(H). (E, lanes 5–7) A longer exposure of lanes 2–4. (S2* in E) The N cleavage product the size of S2-cleaved N that is found in kuz extracts but is not processed further (see text). (F,G) Comparisons of the anti-NI immunoprecipitates of WT (lane 3), Dl (lane 4), kuz (lane 5), and Psn (lane 6) embryos expressing LNLexA (F) and LNRLexA (G). In lanes 1 and 2 extracts of embryos expressing NLexA were immunoprecipitated with anti-Su(H) antibody (S) and anti-NI antibody respectively. (G, lane 7) extracts of Psn embryos expressing NLexA were immunoprecipitated with anti-NI antibody. (G*) S3-cleaved N. All the immunoprecipitates in D–G were phosphatased prior to electrophoresis. To ensure that the protein being characterized is derived from Dl embryos, both the N transgenes and daGAL4 were recombined onto Dl chromosomes and to ensure that the protein being characterized is derived from kuz and Psn embryos, the N transgenes were recombined onto kuz and Psn chromosomes, respectively. All the immunoprecipitates within each panel (A–G) were electrophoresed on the same gel. In some of the panels different exposures of the lanes were used to generate the figure. Only the regions where the S2 and S3 cleavage products migrate are shown.
We can only detect S3-cleaved N in anti-N immunoprecipitates of embryonic extracts expressing LNRLexA (Fig. 6A, lane 7), in accord with the increase in Npp114LexA that coimmunoprecipitates with Su(H). In anti-N immunoprecipitates of extracts of embryos expressing NLexA, although we cannot detect S3-cleaved N, there is a band of ∼97 kD that migrates more rapidly than S3-cleaved N (Fig. 6A, lanes 2,3, ▴), that does not associate with Su(H) (Fig. 6A, lane 1) but is dependent on Psn, as it is lacking in Psn embryos (Fig. 6D, cf. lanes 1 and 5 [WT] with lane 4 [Psn]). We will refer to this N cleavage product as Np97 and would like to reiterate that, although its production depends on Psn, it is not “S3” because it does not associate with Su(H). Np97 is also found to varying extents in anti-N immunopreciptates of extracts of embryos expressing LNLexA (see, e.g., Fig. 6F, lane 3). S2-cleaved N can be detected in anti-N immunoprecipitates of embryos expressing each of the three N proteins (Fig. 6A).
Although loss of Delta or Kuzbanian in embryos expressing NLexA causes a reduction in both S2-cleaved N as well as in Np97 (Fig. 6D, cf. lane 1 [WT] with lane 2 [Dl] and lane 3 [kuz]), the ratio of the two cleavage products differs in the two genotypes. In extracts of embryos lacking Dl, the ratio is qualitatively the same as that we observed in wild-type embryos (Fig. 6E, cf. lanes 2 and 5 [WT] with lanes 3 and 6 [Dl]), whereas in extracts of embryos lacking kuz, the level of Np97is reduced to a greater extent (Fig. 6E, lane 7). In kuz extracts there is a protein the size of S2-cleaved N (S2*); however, it is present in much lower levels than is S2-cleaved N in Psn embryos (Fig. 6D), and it is not further processed to produce the Psn-dependent Np97.
As we observed in Psn embryos expressing NLexA, in extracts of Psn embryos expressing LNLexA or LNRLexA, there is an accumulation of S2-cleaved N (Fig. 6F, lane 6 [LNLexA]; Fig. 6G, lane 6 [LNRLexA]). There is also a great reduction in the level of full-length N (data not shown). Expression of LNRLexA does not suppress the neurogenic phenotype of Psn embryos (data not shown), in accord with the lack of S3-cleaved N found in the anti-N immunoprecipitates (Fig. 6G, cf. lanes 3 and 6). Whereas in extracts of Dl embryos expressing LNLexA or LNRLexA, the ratio of cleavage products found in anti-N immunoprecipitates is qualitatively the same as in extracts of wild-type embryos expressing LNLexA or LNRLexA (Fig. 6F, lanes 3 and 4 [LNLexA]; Fig. 6G, lanes 3 and 4 [LNRLexA]), in extracts of kuz embryos the Psn-dependent cleavage products are not present (Fig. 6F, lane 5 [LNLexA]; Fig. 6G, lane 5 [LNRLexA]). Again, as we observed in extracts of kuz embryos expressing NLexA, in kuz embryos expressing LNLexA or LNRLexA, there is a protein the size of S2-cleaved N; however, it is much less abundant than is S2-cleaved N in Psn embryos, and it is not further processed to produce Psn-dependent cleavage products.
The data we have presented in this section indicate that loss of Kuzbanian affects the cleavage of Notch differently than does the loss of Delta. Loss of kuz results in the specific loss of Psn-dependent cleavage products. A protein the size of S2-cleaved N is still present. Although this might appear to indicate that kuz is responsible for S3 cleavage of N, data are presented below showing that kuz is actually responsible for S2 cleavage, and that the protein the size of S2-cleaved N that is found in kuz embryos can be generated by TACE, another ADAM family member. This TACE-generated product is not efficiently processed to produce S3-cleaved N.
Gain-of-function Notch molecules that cannot signal via Delta are dependent on kuzbanian for their activity
That there is a difference in the cleavage products found in anti-N immunoprecipitates of Dl and kuz embryos expressing NLexA, LNLexA, or LNRLexA suggests that kuz is not functioning merely to produce a soluble form of Delta that can then act as a ligand for Notch. If this were, indeed, the case, there should still be a requirement for kuz for the function of Dl-independent gain-of-function N molecules even in the absence of Dl. We tested this in two ways.
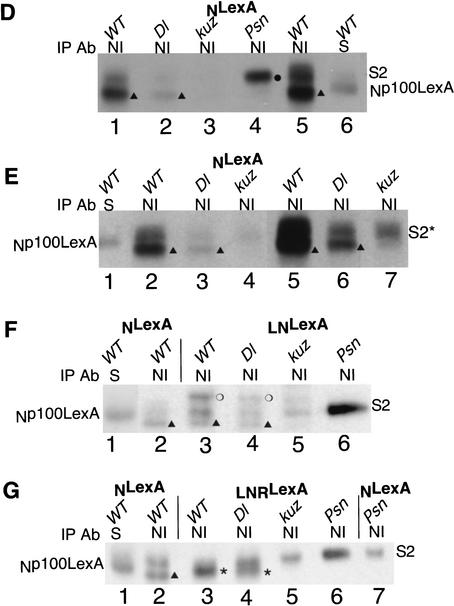
First, we generated transgenic flies that express N protein deleted for EGF repeats 1–18 as well as the LNG repeats (NΔEGF1–18 LNRLexA; see Fig. 1B). We and others have shown that N molecules deleted for EGF repeats 11 and 12 cannot transduce a Dl-dependent signal (Lieber et al. 1993; Lawrence et al. 2000). Expression of NΔEGF1–18 LNRLexA in a wild-type background produces an antineurogenic phenotype (Fig. 7A). Unlike the results observed with LNLexA and LNRLexA, loss of Dl did not significantly reduce the severity of this phenotype (Fig. 7, cf. A with B). Nor did the loss of Dl decrease the level of Npp114LexA derived from NΔEGF1–18 LNRLexA that associates with Su(H) (Fig. 7D, lanes 6,7; two experiments were quantitated). Thus none of the function or processing of NΔEGF1–18 LNRLexA seems to be due to Dl.
Figure 7.
A Dl-independent N protein requires kuz for activity. (A–C) Ventral views of anti-HRP stains of embryos expressing ΔEGF 1–18 LNRLexA under control of daGAL4, in wild-type (A), Dl (B), or kuz (C) backgrounds. (D) Extracts of embryos expressing ΔEGF 1–18 LNRLexA under control of daGAL4 were immunoprecipitated with anti-NI antibody (lanes 2,3) or anti-Su(H) antibody (lanes 6,7). The extracts in lanes 2 and 6 are from wild-type embryos, and the extracts in lanes 3 and 7 are from Dl embryos. (Lane 1) An anti-NI immunoprecipitate of wild-type embryos expressing NLexA; (lanes 4,5) anti-Su(H) immunoprecipitates of embryos expressing NLexA. The immunoprecipitates in lanes 1–4 were treated with phosphatase. (*, lanes 2,3) S3-cleaved N; (●, lane 1) S2-cleaved N. (E) Extracts of embryos expressing ΔEGF 1–18 LNRLexA under control of daGAL4 were immunoprecipitated with anti-NI antibody (lanes 3,4) or anti-Su(H) antibody (lanes 6,7). The extracts in lanes 3 and 6 are from wild-type embryos, and the extracts in lanes 4 and 7 are from kuz embryos. (Lane 2) An anti-NI immunoprecipitate of wild-type embryos expressing NLexA; (lanes 1,5) anti-Su(H) immunoprecipitates of embryos expressing NLexA. The immunoprecipitates in lanes 1–4 were treated with phosphatase. (*, lane 3) S3-cleaved N. The Westerns in both D and E were reacted with anti-LexA antisera.
Although the function of NΔEGF1–18 LNRLexA is independent of Dl, production of an antineurogenic phenotype by NΔEGF1–18 LNRLexA is completely dependent on the presence of kuz. There is no suppression of the kuz neurogenic phenotype by expression of NΔEGF1–18 LNRLexA (Fig. 7C). This is unlike the result observed when LNLexA or LNRLexA was expressed in a kuz background (Fig. 5C,D). This discrepancy will be discussed in a later section. In accord with the inability of NΔEGF1–18 LNRLexA to suppress the kuz neurogenic phenotype, there is a drastic reduction in the level of Npp114LexA that associates with Su(H), 85%–95% (Fig. 7E, lanes 6,7; two experiments were quantitated). Again, the decrease in the level of Npp114LexA derived from NΔEGF1–18 LNRLexA that associates with Su(H) is much greater than we observed with LNLexA or LNRLexA in the absence of kuz (Fig. 5F). The decrease in processing of NΔEGF1–18 LNRLexA in the absence of kuz is also apparent in anti-N immunoprecipitates of kuz extracts expressing NΔEGF1–18 LNRLexA (Fig. 7E, lanes 3,4).
The data presented in this section show that kuz has a role in the cleavage of N independent of any role it may have in the cleavage of Delta. As was the case with kuz embryos expressing NLexA, LNLexA, or LNRLexA, the loss of kuz is manifested in anti-N immunoprecipitates of extracts expressing NΔEGF1–18 LNRLexA by the loss of S3-cleaved N. In the next section we present data showing that kuz is actually responsible for S2 cleavage. In contrast to kuz embryos expressing NLexA, LNLexA, or LNRLexA, there is very little accumulation of a protein the size of S2-cleaved N in kuz embryos expressing NΔEGF1–18 LNRLexA . Below we present data suggesting that this is because TACE cleaves this substrate less efficiently.
Removal of kuzbanian activity by RNA-mediated interference in S2 cells abolishes cleavage of a transmembrane gain-of-function Notch molecule
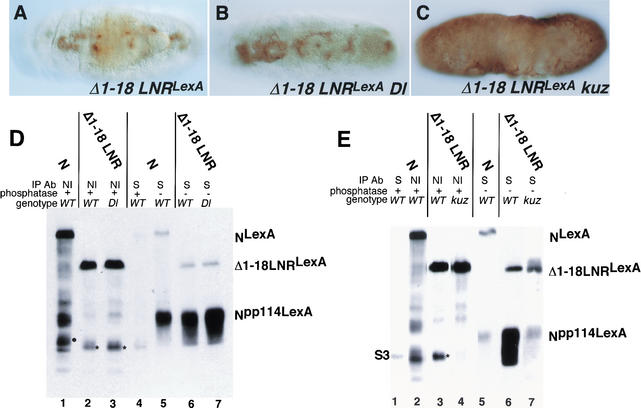
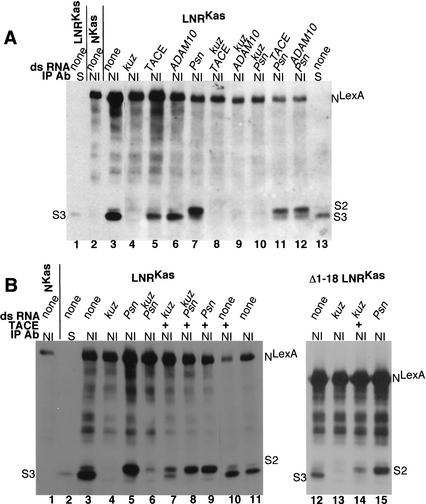
The second way we asked whether kuz has a Dl-independent role in N processing was to use RNA-mediated interference (RNAi) in S2 cells to remove kuz activity. S2 cells do not express any known N ligands (Fehon et al. 1990; Rebay et al. 1991), and RNAi has been shown to be an effective method for removing gene function in S2 cells (Clemens et al. 2000). For these experiments we deleted 381 amino acids from the carboxyl terminus of N, in order to achieve greater separation between S2 and S3 cleavage products. (NKasLexA and LNRKasLexA are shown in Fig. 1B.) As we observed in embryos expressing LNRLexA, anti-N immunoprecipitates of S2 cells expressing LNRKasLexA contain S3-cleaved N that comigrates with Np100KasLexA, the phosphatased soluble domain of LNRKasLexA that associates with Su(H) (Fig. 8A, lanes 1,3). This S3-cleaved N is not visible to any great extent in anti-N immunoprecipitates of S2 cells expressing NKasLexA (Fig. 8A, lane 2). As we observed in anti-N immunoprecipitates of extracts of Psn embryos expressing LNRLexA (Fig. 6G, lane 6), cleavage of LNRKasLexA is blocked at S2 in cells in which RNAi has been used to remove Psn function (Fig. 8A, lane 7). Thus both S2 and S3 cleavages of LNRKasLexA can occur in S2 cells.
Figure 8.
Kuzbanian promotes the cleavage of Notch. (A, lanes 3–12) are anti-NI immunoprecipitates of S2 cells expressing heat-shock-induced LNRKasLexA (see Fig. 1B) that have been treated with the indicated double-stranded (ds) RNAs. (Lane 2) An anti-NI immunoprecipitate of S2 cells expressing heat-shock-induced NKasLexA (see Fig. 1B); (lanes 1,13) anti-Su(H) immunoprecipitates of S2 cells expressing LNRKasLexA. (B, lanes 3–15) anti-NI immunoprecipitates of S2 cells expressing heat-shock-induced LNRKasLexA (lanes 3–11) or ΔEGF 1–18 LNRKasLexA (lanes 12–15; see Fig. 1B) that have been treated with the indicated double-stranded (ds) RNAs. The cells in lanes 7–10 and 14 were cotransfected with an actin-driven TACE construct. (Lane 1) An anti-NI immunoprecipitate of S2 cells expressing heat-shock-induced NKasLexA; (lane 2) an anti-Su(H) immunoprecipitate of S2 cells expressing LNRKasLexA. All the immunoprecipitates were phosphatased prior to electrophoresis, and the Westerns were reacted with anti-LexA antisera. (S2, S3) Locations of S2 and S3 cleavage products.
When kuz double-stranded RNA is used to remove kuz function, no processing of LNRKasLexA is observed (Fig. 8A, lane 4). Neither Drosophila TACE nor Drosophila ADAM 10 double-stranded RNA has any affect on S3 cleavage (Fig. 8A, lanes 5,6). Therefore, in the absence of Dl, the cleavage of LNRKasLexA is dependent on kuz. This kuz-dependent cleavage acts upstream of Psn activity, as we do not observe S2-cleaved N in cells in which the function of both kuz and Psn has been removed by RNAi (Fig. 8A, lane 10). In contrast, removal of TACE or ADAM10 function along with that of Psn results in accumulation of S2-cleaved N (Fig. 8A, lanes 11,12), indicating that in S2 cells neither TACE nor ADAM10 is required for S2 cleavage of LNRKasLexA. In fact, by Northern blot analysis, we could not detect any TACE or ADAM10 RNA in S2 cells, although using the same probes, we were able to detect TACE and ADAM10 in embryo RNA, and we could detect kuz and Psn mRNAs on Northerns of S2 cell RNA (data not shown). We then asked whether kuz RNAi would block cleavage of wild-type N when S2 cells expressing wild-type N were aggregated with S2 cells expressing Dl. We found that double-stranded kuz RNA added to the N-expressing cells blocks the production of Npp114, the phosphorylated soluble cytoplasmic domain that associates with Su(H), but double-stranded TACE RNA has no effect (data not shown).
Brou et al. (2000) have reported that TACE mediates S2 cleavage of mammalian N. We therefore asked whether exogenous TACE could complement the loss of kuz activity generated by RNAi in S2 cells expressing LNRKasLexA. As can be seen in Figure 8B, lane 7, although the production of S3-cleaved N is restored upon cotransfection of an actin TACE construct, the amount of S3 product compared with a product that migrates at the size of S2-cleaved N is substantially lower than in cells with endogenous kuz (Fig. 8B, cf. lanes 7 and 3). This pattern of N cleavage products is similar to the pattern we observe in kuz embryos (see Fig. 6G, lane 5), and suggests that although TACE is able to cleave N at a position close to that cleaved by Kuz, this product is not efficiently cleaved further to produce S3-cleaved N.
In vivo the function of NΔEGF1–18 LNRLexA is completely dependent on kuz activity. As we observed for LNRKasLexA, treatment of S2 cells with double-stranded kuz RNA abolishes the processing of NΔ1–18 LNRKasLexA (Fig. 8B, lane 13), and treatment with double-stranded Psn RNA results in the accumulation of S2-cleaved N (Fig. 8B, lane 15). We therefore asked if exogenous TACE could restore the production of S3-cleaved N derived from NΔEGF1–18 LNRKasLexA in S2 cells that had been treated with double-stranded kuz RNA. We found that hardly any S3 product was generated (Fig. 8B, lane 14), accounting for the in vivo kuz dependence.
Discussion
Pan and Rubin (1997) originally proposed that Kuz cleaves Notch. This proposal is in accord with the cell-autonomous requirement for kuz both in Drosophila and in C. elegans (Rooke et al. 1996; Sotillos et al. 1997; Wen et al. 1997). More recently it has been suggested that the phenotypes resulting from loss of kuz are attributable to its role in the processing of Dl (Qi et al. 1999), although in that case the requirement for kuz would be non-cell-autonomous. We have shown that Kuz can associate with N (Fig. 2), and that removal of kuz activity from S2 cells, which do not express Dl, blocks the processing of ligand-independent gain-of-function N molecules (Fig. 8). In vivo, a gain-of-function N molecule that is completely Dl-independent displays an absolute requirement for kuz (Fig. 7). Our results show that kuz can mediate the cleavage of N, and are therefore in agreement with Pan and Rubin's original proposal. However, whereas Pan and Rubin proposed that kuz mediates S1 cleavage, our data suggest that kuz is responsible for S2 cleavage.
The role of kuz/ADAM10 in the N pathway in vertebrates is uncertain. It has been shown that expression of a dominant-negative form of mouse Kuz causes the overproduction of neurons in Xenopus (Pan and Rubin 1997) and inhibits Delta-1-like-induced transactivation of a HES-1 reporter in HeLa cells expressing Notch-1 (Jarriault et al. 1998). Yet both S2 and S3 cleavages of a ligand-independent N protein occur in cells derived from kuz mice (Mumm et al. 2000). Whereas mammals have a single kuz/ADAM10 gene, Drosophila has two, kuz, the focus of this work, and another ADAM10 homolog. Perhaps Drosophila kuz has evolved a function distinct from that of ADAM10.
Although both LNLexA and LNRLexA function in the absence of the ligand Dl, the activity of LNRLexA is greater than that of LNLexA. This difference is not caused by an enhanced affinity of Kuz for LNRLexA, as both associate equally with Kuz in S2 cells (data not shown). This suggests that the difference in activity of these gain-of-function N molecules results from an enhanced ability of LNRLexA to be cleaved by Kuz. In fact, the interaction of LN and LNR with Kuz is no greater than that of wild-type N, and the association of Kuz with any of the N molecules occurs in the absence of Dl (Fig. 2; data not shown). In this regard, the association of Kuz with N is like that of Kuz with ephrin-A2, which forms a stable complex with Kuz prior to Eph receptor binding. The binding of clustered Eph receptors to the Kuz–ephrin-A2 complex activates Kuz and triggers ephrin-A2 cleavage (Hattori et al. 2000). Likewise, the binding of Dl to the Kuz–N complex could activate Kuz and trigger N cleavage.
A curious result is the difference in Kuz dependence of LNRLexA and ΔEGF1–18 LNRLexA (for NΔEGF1–18 LNRLexA) in vivo. We offer two explanations for this observation. The first takes into account the difference in Dl responsiveness between Δ1–18 LNRLexA and LNRLexA.LNRLexA, which has reduced activity in the absence of Dl, still retains some function in the absence of Kuz (Figs. 4 and 5), whereas Δ1–18 LNRLexA, which is completely Dl-independent, cannot function in the absence of kuz (Fig. 7). One possible explanation for the discrepancy is that there are two pathways that mediate N cleavage and function in embryos. One pathway requires Dl but is independent of Kuz, and the other pathway requires Kuz but is independent of Dl. LNRLexA can operate in both pathways, so that upon removal of either Dl or Kuz, LNRLexA still functions via the alternative remaining pathway. Δ1–18 LNRLexA can only operate in the kuz pathway, so that upon removal of kuz it is nonfunctional. There is, however, no strong evidence pointing to a Dl-independent N pathway involving cleavage in embryos, and given the requirement for Dl in the germ line for the differentiation of follicle cells (López-Schier and St. Johnston 2001), the generation of embryos that are maternally Dl null is not straightforward.
We therefore favor the hypothesis that the difference in kuz-dependence of LNRLexA and Δ1–18 LNRLexA in vivo is owing to their differing abilities to be cleaved by TACE. Brou et al. (2000) have shown that TACE, another member of the ADAM family of metalloproteases, mediates S2 cleavage of mammalian N in vitro. Drosophila S2 cells do not contain any detectable TACE RNA (data not shown), and exogenous TACE only poorly complemented the RNAi-mediated loss of kuz activity (Fig. 8B, lane 7). The restoration of some S3 product upon expression of TACE is in accord with the residual activity of LNRLexA in kuz embryos (Fig. 5), and in vitro TACE and N do interact, albeit less well than do Kuz and N (data not shown). In the absence of a TACE mutant, we are unable to say for certain whether kuz and TACE have redundant functions, if another member of the ADAM family is responsible for the residual S3 cleavage, or if the residual in vivo activity is caused by the expression of LNRLexA from a heterologous promoter. Hardly any S3 product was generated from Δ1–18 LNRLexA by exogenous TACE in S2 cells that had been treated with kuz double-stranded RNA (Fig. 8B, lane 14), accounting for the in vivo Kuz-dependence. It is not clear why the ability of exogenous TACE to produce S3-cleaved N differs between LNRLexA and Δ1–18 LNRLexA; however, in S2 cells, even in the presence of endogenous Kuz, Δ1–18 LNRLexA is not cleaved as well as is LNRLexA (Fig. 8B; data not shown), suggesting that perhaps differences in the secondary structure of the molecules account for their differing responses to TACE.
The pattern of cleavage products generated by expression of TACE in kuz− S2 cells also provides an explanation for the in vivo biochemical data we present in Figure 6, which had seemed to suggest that kuz is responsible for S3 cleavage. It is intriguing that both in kuz embryos (Fig. 6) and in TACE-complemented kuz− S2 cells (Fig. 8) there is an accumulation of a protein the size of S2-cleaved N, which is not efficiently cleaved further to produce S3-cleaved N. We propose that although TACE can cleave N at juxtamembrane sites, a large fraction of this cleavage is occurring at a site close to but distinct from the S2 site that allows for efficient S3 cleavage. This suggests that cleavage of N at any juxtamembrane site is not immediately followed by efficient S3 cleavage.
A surprising result was the suppression of the kuz neurogenic phenotype by expression of NLexA (Fig. 5B). In fact, the size of the nervous system in NLexA kuz embryos is smaller than in LNRLexA kuz embryos (Fig. 5, cf. B [NLexA kuz] with D [LNRLexA kuz]). As kuz embryos are neurogenic, the suppression must result from the overexpression of N. This, along with the fact that the suppression does not involve the association of the cytoplasmic domain of N with Su(H) (Fig. 5F), suggests that in the absence of kuz, overexpressed N is competitively interacting with a protein required for neurogenesis. Wild-type N accumulates to a higher steady-state level on the cell surface than does LN or LNR (Fig. 3G–I; data not shown). It has been shown that expression of the extracellular domain of N can disrupt the establishment of proneural clusters in the developing wing disc (Brennan et al. 1999).
In summary, we have shown that in flies S2 cleavage can be mediated by kuz. This contrasts with mammalian data that suggest S2 cleavage occurs via TACE. The discrepancy might be owing to mechanistic differences between flies and mammals as has also been shown for S1 cleavage (Kidd et al., in prep.).
Materials and methods
Constructs
All constructs were generated by standard methods. kuz and kuzDN DNAs used for the S2 cell experiments were from D.J. Pan (Pan and Rubin 1997). The kuz DNA used for in vitro translation was from EST clone ID SD03071. The hIR DNA was from C.R. Kahn. The Psn DNA was from I. Livne-Bar (Boulianne et al. 1997). RT-PCR, using primers based on the then preliminary Drosophila genome project, was used to amplify a segment of Drosophila TACE DNA (FlyBase accession no. CG7908), which was then used to screen a Drosophila cDNA library. RT-PCR, using primers based on the preliminary Drosophila genome project, was used to amplify a segment of Drosophila ADAM10 DNA (FlyBase accession no. CG1964).
Antibodies
All the anti-N antibodies and the Su(H) antibody have been described previously (Kidd et al. 1989, 1998; Lieber et al. 1993). Anti-myc antibody was from Calbiochem or Sigma. The anti-LexA antibody was generated in mice against amino acids 1–87 of LexA.
Flies
daGAL4 (daG32) flies were from E. Knust (Wodarz et al. 1995). arm GAL4 files were from J.P. Vincent (Sanson et al. 1996). The Dl allele used, Dlx, is described by FlyBase (1998). kuze29-4 FRT flies (Rooke et al. 1996) were from D.J. Pan. The Psn allele used was PSC1, and hs FLP; PSC1 FRT flies were from G. Struhl (Struhl and Greenwald 1999).
For the biochemical experiments, to ensure that the protein being characterized is derived from Dl embryos, both the N transgenes and daGAL4 were recombined onto Dl chromosomes. To ensure that the protein being characterized is derived from kuz embryos, the N transgenes were recombined onto kuz chromosomes, and to ensure that the protein being characterized is derived from Psn embryos, the N transgenes were recombined onto Psn chromosomes.
Immunocytyochemistry and immunofluorescence
Immunocytochemistry and immunofluorescence were carried out as described previously (Kidd et al. 1998). For the immunofluorescence experiments on S2 cells shown in Figure 3G–I, Cy5-coupled anti-rabbit IgG was used to detect the outside antibody and Texas Red-coupled anti-mouse IgG was used to detect the inside antibody. For the immunofluorescence experiments on embryos shown in Figure 3, J and K, biotinylated anti-mouse IgG and FITC-coupled streptavidin were used to detect the anti-LexA antibody. All secondary antibodies were from Jackson.
Immunoprecipitations and pull-down assays
With the exception of the experiments presented in Figure 6, B and C, detergent lysis used to prepare the embryonic and S2 cell extracts and immunoprecipitations were carried out as described previously (Kidd et al. 1998). The fractionation presented in Figure 6B was carried out on extracts prepared by isotonic lysis as described (Kidd et al. 1998), with the addition of 5 mM phenanthroline and the removal of molybadate. The fractionation presented in Figure 6C was carried out on extracts prepared by hypotonic lysis, as used to generate extracts for gel shifts (Kidd et al. 1998), with the addition of 5 mM phenanthroline. The nuclei were isolated by pelleting through a sucrose cushion as described by Rio et al. (1986). The preparation of extracts by hypotonic lysis tends to result in the production of some additional cleavage products that are not seen when extracts are prepared by detergent lysis.
GST fusion proteins were produced in the bacterial strain pLysS. After induction and growth for 3 h, proteins were extracted, bound to glutathione beads, and extensively washed in 20 mM Tris (pH 7.6), 1 M NaCl, 1 mM EDTA, 1% Triton X-100, and 2% sarkosyl. After the final wash the beads were equilibrated and stored in 10 mM HEPES (pH 7.6), 150 mM NaCl, 0.1% Triton X-100, 1 mg/mL BSA, and protease inhibitors. Su(H), kuz, and hIR were transcribed and translated in vitro using the Promega TNT system. Subsequent steps were all at 4°C or on ice. In vitro translation products were first precleared for 2 to 3 h in 50 mM HEPES (pH 7.6), 150 mM NaCl, 1% Triton X-100, 5 mM CaCl2, and 1 mg/mL BSA with GST coupled beads. The supernatants were incubated overnight with either GST or GST–N fusion beads, which were then washed 3× in 50 mM HEPES (pH 7.6), 150 mM NaCl, 1% Triton X-100; 2× with 50 mM HEPES (pH 7.6), 250 mM NaCl, 0.1% Triton X-100; and 1× with 10 mM HEPES (pH 7.6), 150 mM NaCl, 0.1% Triton X-100. Proteins were eluted by boiling in Laemmli gel loading buffer.
Scanned autoradiographs were quantitated on a Macintosh computer with the public domain NIH image program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image).
RNAi experiments
PCR using T7 primers was used to amplify kuz, TACE, ADAM10, and Psn DNAs. The following primers were used: kuz, for Figure 8A, 5′ primer, CATTGTATTCGTATCGAT; 3′ primer, GAATGTTGTTGTCGACGA; for Figure 8B to avoid any TACE homologous sequences, 5′ primer, ATGCAACGT CATCCCAAT; 3′ primer, ATAAACGATATTTCGGCG; TACE, 5′ primer, CAAGGACGATGTGGTGCAC; 3′ primer, GTAGATCTTGTGCACCCGATC; ADAM10, 5′ primer, CTC CAGGTCCTGTTCCTC; 3′ primer, CGAAAATCATCGTTG TAC; Psn, 5′ primer, ATGGCTGCTGTCAATCTC; 3′ primer, CAGAGGTCCCTGCCAATG. Double-stranded RNA was generated using a T7 Megascript Kit (Ambion). RNAi was carried out as described by Clemens et al. (2000). Cells were transfected using calcium phosphate 1 d after addition of the double-stranded RNA. The following day the cells were washed and double-stranded RNA was added again. The cells were heat-shocked and lysed the next day.
Acknowledgments
We thank I. Livne-Bar, C.R. Kahn, and D.J. Pan for DNAs and E. Knust, D.J. Pan, G. Struhl, and J.P. Vincent for fly stocks and C. Wesley for advice on S2 cell agregations. This work was supported by a National Institutes of Health grant GM25103 to M.W.Y.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL young@mail.rockefeller.edu; FAX (212) 327-8695.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.942302.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- Boulianne G, Livne-Bar I, Humphreys J, Liang Y, Lin C, Rogaev E, St George-Hyslop P. Cloning and characterization of the Drosophila presenilin homologue. Neuroreport. 1997;8:1025–1029. doi: 10.1097/00001756-199703030-00041. [DOI] [PubMed] [Google Scholar]
- Brennan K, Tateson R, Lieber T, Couso JP, Zecchini V, Martinez Arias A. The Abruptex mutations of Notch disrupt the establishment of proneural clusters in Drosophila. Dev Biol. 1999;216:230–242. doi: 10.1006/dbio.1999.9501. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israël A. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci. 2000;97:6499–503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MAT, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- FlyBase. FlyBase—A Drosophila database. Nucleic Acid Res. 1998;26:85–88. doi: 10.1093/nar/26.1.85. http://flybase.bio.indiana.edu/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. LIN-12/Notch signaling: Lessons from worms and flies. Genes & Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- Hattori M, Osterfield M, Flanagan JG. Regulated cleavage of a contact-mediated axon repellent. Science. 2000;289:1360–1365. doi: 10.1126/science.289.5483.1360. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Le Bail O, Hirsinger E, Pourquie O, Logeat F, Strong CF, Brou C, Seidah NG, Israël A. Delta-1 activation of Notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S, Baylies MK, Gasic GP, Young MW. Structure and distribution of the Notch protein in developing Drosophila. Genes & Dev. 1989;3:1113–1129. doi: 10.1101/gad.3.8.1113. [DOI] [PubMed] [Google Scholar]
- Kidd S, Lieber T, Young MW. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes & Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence N, Klein T, Brennan K, Martinez Arias A. Structural requirements for Notch signalling with delta and serrate during the development and patterning of the wing disc of Drosophila. Development. 2000;127:3185–3195. doi: 10.1242/dev.127.14.3185. [DOI] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. Indirect evidence for Delta-dependent intracellular processing of Notch in Drosophila embryos. Curr Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes & Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israël A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Schier H, St. Johnston D. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes & Dev. 2001;15:1393–1405. doi: 10.1101/gad.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Kopan R. Notch signaling: From the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- Primakoff P, Myles DG. The ADAM gene family: Surface proteins with adhesion and protease activity. Trends Genet. 2000;16:83–87. doi: 10.1016/s0168-9525(99)01926-5. [DOI] [PubMed] [Google Scholar]
- Qi H, Rand MD, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Processing of the Notch ligand Delta by the metalloprotease Kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: Implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Rio DC, Laski FA, Rubin GM. Identification and immunochemical analysis of biologically active Drosophila P element transposase. Cell. 1986;44:21–32. doi: 10.1016/0092-8674(86)90481-2. [DOI] [PubMed] [Google Scholar]
- Rooke J, Pan D, Xu T, Rubin GM. KUZ, a conserved metalloprotease-disintegrin protein with two roles in Drosophila neurogenesis. Science. 1996;273:1227–1231. doi: 10.1126/science.273.5279.1227. [DOI] [PubMed] [Google Scholar]
- Sanson B, White P, Vincent JP. Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature. 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- Schlondorff J, Blobel CP. Metalloprotease-disintegrins: Modular proteins capable of promoting cell–cell interactions and triggering signals by protein-ectodomain shedding. J Cell Sci. 1999;112:3603–3617. doi: 10.1242/jcs.112.21.3603. [DOI] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Sotillos S, Roch F, Campuzano S. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development. 1997;124:4769–4779. doi: 10.1242/dev.124.23.4769. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Nuclear access and action of Notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- Wen C, Metzstein MM, Greenwald I. SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN-12/NOTCH signalling. Development. 1997;124:4759–4767. doi: 10.1242/dev.124.23.4759. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Ye Y, Lukinova N, Fortini ME. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature. 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]