Abstract
All genetic markers from phage T2 are partially excluded from the progeny of mixed infections with the related phage T4 (general, or phage exclusion). Several loci, including gene 56 of T2, are more dramatically excluded, being present in only ∼1% of the progeny. This phenomenon is referred to as localized marker exclusion. Gene 69 is adjacent to gene 56 of T4 but is absent in T2, being replaced by completely nonhomologous DNA. We describe SegF, a novel site-specific DNA endonuclease encoded by gene 69, which is similar to GIY–YIG homing endonucleases of group I introns. Interestingly, SegF preferentially cleaves gene 56 of T2, both in vitro and in vivo, compared with that of phage T4. Repair of the double-strand break (DSB) results in the predominance of T4 genes 56 and segF in the progeny, with exclusion of the corresponding T2 sequences. Localized exclusion of T2 gene 56 is dependent on full-length SegF and is likely analogous to group I intron homing, in which repair of a DSB results in coconversion of markers in the flanking DNA. Phage T4 has many optional homing endonuclease genes similar to segF, whereas similar endonuclease genes are relatively rare in other members of the T-even family of bacteriophages. We propose that the general advantage enjoyed by T4 phage, over almost all of its relatives, is a cumulative effect of many of these localized events.
Keywords: Gene conversion, homing endonuclease, phage exclusion, T-even bacteriophage
Homing endonucleases are site-specific DNA endonucleases that are typically found encoded within introns and inteins (in-frame insertions that are removed by protein self-splicing). These endonucleases are required for initiation of a process known as homing (Dujon 1989), a phenomenon that was originally recognized as the unidirectional inheritance of the mitochondrial ω allele of Saccharomyces cerevisiae. Subsequently, it was discovered that the ω allele is an optional intron in the mitochondrial large (21S) subunit rRNA gene and that its pattern of inheritance is a consequence of gene conversion initiated by the endonuclease encoded within the intron (Jacquier and Dujon 1985). Homing is therefore described as an endonuclease-initiated process resulting in transfer of the intron and the endonuclease encoded within it, into the cognate intronless allele of the gene (Lambowitz and Belfort 1993).
Intron-encoded homing endonucleases are phylogenetically widespread, occurring in Archaea, Bacteria, and Eukarya. They have been grouped into protein families based on conserved amino acid motifs: the LAGLIDADG, GIY–YIG, His–Cys box, and H–N–H families. Recently, structural similarities between proteins having the His–Cys box and H–N–H motifs have led to the suggestion that these may be consolidated into a single group (Kühlmann et al. 1999). Structural and biochemical information about representative members of these protein families has recently been reviewed (Chevalier and Stoddard 2001).
Remarkably, despite this diversity, homing endonucleases have similar characteristics. Their DNA targets are the cognate intronless alleles, and they generally recognize long asymmetric sequences spanning the intron-insertion site (IIS), usually making a double-strand break (DSB) close to the IIS. However, the presence of an intron disrupts this recognition sequence, rendering the intron-containing gene resistant to cleavage. This target site specificity, combined with long recognition sequences (ranging from 14 to 40 bp), ensures that host genomes are protected from these endonucleases. Most homing endonucleases leave a 2–5-base 3′ overhang at the cut site, and these ends initiate recombination and repair (Chevalier and Stoddard 2001). Interestingly, with the exception of the GIY–YIG family, representatives of the other homing endonuclease families are also found inserted into inteins, which are in-frame insertions that are removed by protein self-splicing. They are specific for inteinless DNA targets and promote intein mobility in precisely the same manner as for group I introns.
The double-strand break (DSB) repair model (Szostak et al. 1983), initially proposed for gene conversion in yeast, was adapted to explain homing of group I introns (Zinn and Butow 1985). Homing is a localized gene-conversion event surrounding the target site that has sustained a DSB. In the refined model, the DSB in the recipient molecule is extended into a gap by exonucleolytic degradation, leaving 3′ single-stranded overhangs that can invade the homologous region in the intact donor, which is used as a template for DNA synthesis. This recombination-dependent repair copies the intron (or intein), the endonuclease gene encoded within it, and additional flanking markers into the recipient. Analysis in bacteriophage T4 suggests that intron homing may occur via multiple pathways. Alternatives to the DSB repair pathway include synthesis-dependent strand annealing, helicase-mediated reverse branch migration, and topoisomerase-mediated recombination pathways (Mueller et al. 1996a). All models predict that there is coconversion of flanking markers with a frequency inversely proportional to the distance of the markers from the cleavage site (for review, see Lambowitz and Belfort 1993).
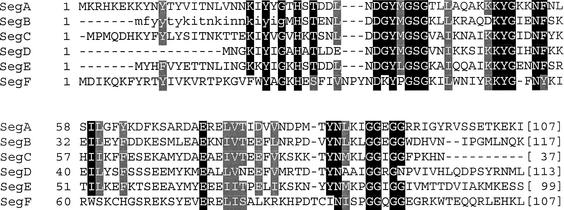
Several free-standing open reading frames (ORFs) carry motifs characteristic of the homing endonucleases, the best studied example being the nuclear-encoded yeast HO mating-type switch endonuclease with the LAGLIDADG motif (Kostriken et al. 1983; Dalgaard et al. 1997a). Sharma et al. (1992) described a family of five genes in bacteriophage T4 that encode proteins that are similar over their N-terminal halves to the GIY–YIG intron endonucleases, and are denoted seg (for similarity to endonucleases encoded by group I introns; Fig. 1). The seg genes are optional among close relatives of T4 (the T-even phages), several of them being absent in phage T2 (Kutter et al. 1996). Two members of this protein family, SegA and SegE, have been shown to possess double-stranded DNA endonuclease activity (Sharma and Hinton 1994; Kadyrov et al. 1996). One could reasonably envisage a scenario similar to homing of group I introns for these free-standing endonuclease genes. Starting from a DSB, a recombination-dependent repair event would result in transfer of the endonuclease and its flanking DNA into the recipient. Consistent with this is the demonstrated plasmid-to-phage mobility of segE (Kadyrov and Kryukov 1996). Indeed, the simple existence of an EcoRI site, surrounded by sequences that are homologous to those around the ecoRI gene, is sufficient to convert EcoRI into a homing endonuclease (Eddy and Gold 1992).
Figure 1.
Amino acid sequence alignment of gp69 (SegF) with members of the Seg family. Alignment was generated with ClustalW 1.8 (Thompson et al. 1994) using amino acid sequences with GenBank accession nos.: SegA (AAD42654), SegB (AAD42655), SegC (AAD42656), SegD (AAD42657), SegE (AAD42658), and SegF/gp69 (AAD42517). The sequence in lower case shows the N terminus of a different database entry for SegB (CAA93270). Conserved residues are shaded. The numbers in brackets indicate the numbers of residues following the aligned sequence.
The phenomenon of partial exclusion (Russell and Huskey 1974) by phage T4 during mixed infection with closely related phages of the T-even family has never been explained. The genomes of T-even phages are colinear, and mutations in almost all the genes of T4 can be complemented by the wild-type (WT) allele from the other phages. Nevertheless, contrary to the expected frequency of 50% from each parent, the frequency of most genetic markers from phage T2 in the progeny of a mixed infection with T4 is 10%–30%. However, a more extensive site-specific exclusion is exerted locally at several genomic sites, among which the most dramatically excluded regions are genes 32 and 56, wherein the T2 alleles were present in <1% of the progeny (Russell and Huskey 1974).
Interestingly, there are polymorphisms between phages T4 and T2 near these sites of local exclusion. They were first detected as nonhomologous loops in the DNA heteroduplex analysis of Kim and Davidson (1974) and later confirmed by sequence analysis. Adjacent to gene 56 is an open reading frame (gene 69) that is absent in T2 (Gary et al. 1998), and next to gene 32 is ORF 32.1, which is also absent in T2 (Repoila et al. 1994).
In this work we show a site-specific double-stranded DNA endonuclease activity associated with gp69. Further, we show that the preferred target for this endonuclease is in gene 56 of T2 and that DNA repair is associated with localized unidirectional transfer of markers from T4 into T2, around the cut site. Our results provide a molecular basis to explain marker exclusion between closely related T-even phages.
Results
A BLASTP (Altschul et al. 1997) search with the corrected T4 gp69 sequence from Gary et al. (1998) resulted in a best match to another T4 protein, MobD (as already pointed out by Mosig et al. 1998). MobD is a putative mobile element with an H–N–H motif at its N terminus (Kutter et al. 1996), and can be aligned with gp69 over ∼90 amino acids at their C termini. However, four of the next six protein sequences identified in this search are matches between the N terminus of gp69 and the corresponding regions of the SegA, SegB, SegD, and SegE proteins of phage T4. The Seg proteins are a subgroup of the GIY–YIG family of homing endonucleases, lacking some typically conserved residues characteristic of this family (Kowalski et al. 1999) but having other novel positions that are very well conserved within the Seg subgroup. A ClustalW 1.8 (Thompson et al. 1994) alignment of gp69 with the known Seg proteins showed that significant sequence similarity between these proteins extends over approximately their N-terminal halves (Fig. 1). Biochemical and structural analysis of I-TevI, a well-studied member of the GIY–YIG class, has revealed that the N-terminal half of the protein (containing the GIY–YIG sequence module) comprises the catalytic domain, whereas the C terminus serves as the DNA-binding domain (Derbyshire et al. 1997; Kowalski et al. 1999). These observations led us to consider the possibility that gp69 encodes a DNA endonuclease.
Aware of the difficulty in cloning homing endonuclease genes because of their toxicity in Escherichia coli, we used cell-free extracts to synthesize the protein encoded by gene 69 in a test for putative DNA endonuclease activity.
gp69 is a site-specific, double-strand DNA endonuclease with a cleavage site in T2 gene 56
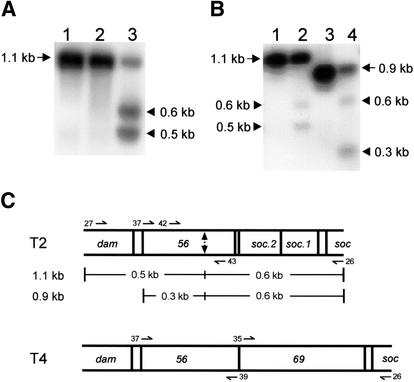
In selecting a substrate for a putative gp69 endonuclease, we were guided by properties of other Seg endonucleases. SegA and SegE, unlike their intron-encoded counterparts, cleave DNA hundreds of base pairs away from their insertion sites (Sharma and Hinton 1994; Kadyrov et al. 1996). Interestingly, like the other seg genes, gene 69is optional in the T-even phages, being absent in T2 and replaced by two small genes of unknown function, soc.1 and soc.2 (Gary et al. 1998). Hence, we decided to test endonuclease activity on a 1.1-kb fragment, amplified by PCR from T2 DNA, that extends from the end of dam (DNA adenine methylase) to the beginning of soc(small outer capsid protein), two genes that are highly similar in T2 and T4. After incubation with the in vitro synthesized gp69, DNA was resolved on an agarose gel, capillary-blotted, and probed with the radiolabeled 1.1-kb PCR product. Two discrete cleavage products of ∼0.6 kb and ∼0.5 kb were detected only with programmed extracts containing gp69 (Fig. 2A). Therefore, gp69 is a site-specific double-strand DNA endonuclease that joins the Seg family of proteins as the sixth member and is called SegF (or F-TevVI according to Belfort and Roberts 1997).
Figure 2.
gp69 is a double-strand DNA endonuclease that cleaves in T2 gene 56. (A) A 1.1-kb DNA fragment was generated by PCR amplification of T2 DNA using primers 27 and 26 (lane 1) and incubated with in vitro synthesized gp69 (lane 3) and with mock transcription/translation extract (lane 2). The reaction products were separated by electrophoresis, blotted, and analyzed by Southern hybridization using radiolabeled substrate DNA as a probe. The arrow shows the DNA substrate with size indicated. Triangles show the cleavage products with inferred sizes indicated. (B) 1.1-kb and 0.9-kb DNA fragments were generated by PCR amplification of T2 DNA using primer pairs 27–26 and 37–26, respectively. Substrate DNAs were incubated alone (lanes 1,3) or with in vitro synthesized gp69 (lanes 2,4), and analyzed by Southern hybridization using radiolabeled substrate DNA (27–26) as a probe. Labeling is as in A. (C) Genomic organization of T2 and T4 in the region around gene 56: A schematic representation of the amplified T2 DNA fragments and the deduced location of the cleavage site (indicated by a double-ended arrow) are shown. The positions of the oligonucleotides used for PCR are shown, with the arrowheads indicating the 3′ ends.
To determine the approximate location of the cleavage site in gene 56, endonuclease assays were repeated on two substrates of different sizes that share a common boundary in T2 DNA (Fig. 2B). Southern blot analysis of the cleavage products confirmed that the 1.1-kb fragment is cleaved into two fragments of 0.6 kb and 0.5 kb, whereas the 0.9-kb fragment gives products of 0.6 kb and 0.3 kb (Fig. 2B). From the sizes of the fragments obtained, we inferred that the DSB is in gene 56 of T2, located ∼60% from the beginning of the gene (Fig. 2C).
Mapping of the SegF cleavage site in T2 gene 56
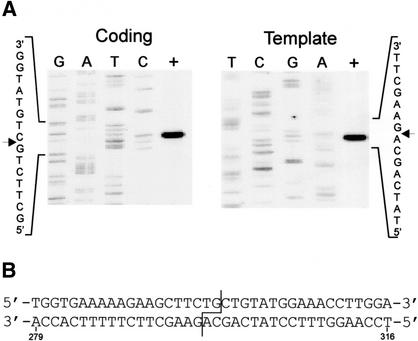
Having narrowed the location of the cleavage site to gene 56, we used a 261-bp fragment amplified from T2 and purified His6-tagged SegF to determine the precise cleavage site. Using end-labeled primers in the PCR, we generated target molecules that were end-labeled on only one strand. After incubation with SegF, the cleavage products were resolved on a denaturing polyacrylamide gel beside sequencing ladders generated from the same end-labeled primers. This permitted us to precisely map the location of the double-strand break (Fig. 3A). SegF cleaves leaving a 2-base 3′ overhang, as is the case for I-TevI and I-TevII (Bell-Pedersen et al. 1990). The cut sites are at residues 298 and 296 from the start of gene 56, on the coding and template strands, respectively. Figure 3B summarizes these results and shows the location of the DSB on the sequence of T2 gene 56.
Figure 3.
Mapping the cleavage site of SegF. (A) Substrate DNAs were synthesized by PCR amplification of pBST2ab4243 DNA using primers differentially end-labeled on either the coding (top) or template (bottom) strand. After incubation with a 1:5 dilution of purified SegFHis6, the cleavage products were separated on a denaturing 6% polyacrylamide gel (+) alongside sequencing ladders that were generated from pBST2ab4243 by extension with the respective end-labeled primers. Sequencing lanes are labeled with the appropriate dideoxynucleotides. The sequence around the cleavage site (indicated by an arrow) on each strand is shown. (B) The sequence surrounding the cleavage site in T2 gene 56. The numbering below is with respect to the start codon of gene 56. The staggered line indicates cleavage sites.
SegF cleaves gene 56 of T4 but prefers T2 DNA in vitro
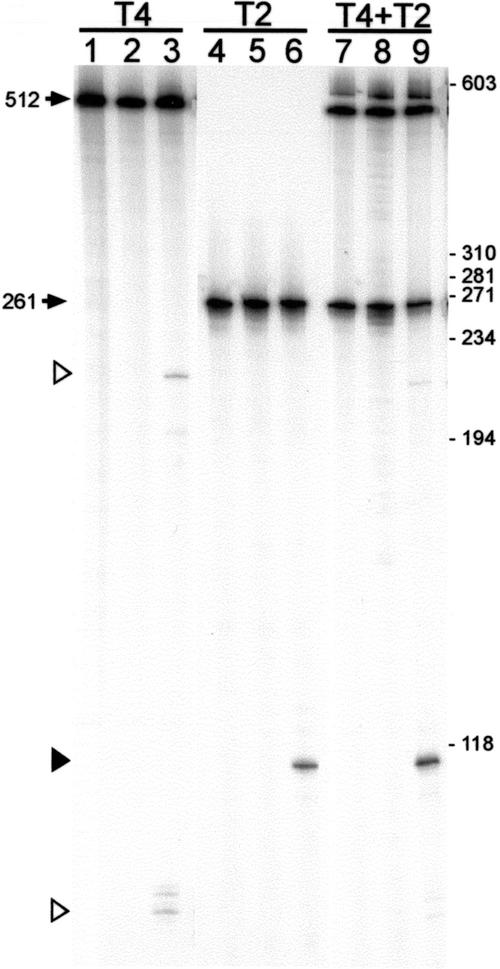
A similar experiment showed that SegF cleaves T4 DNA at several sites on both strands (data not shown). To more accurately compare the activity of SegF against both phage sequences, substrates of different sizes, isotopically labeled on only one strand, were prepared. PCR amplification of T4 gene 56 yields a 512-bp product that is cleaved at several locations on the template strand, yielding a fragment of ∼200 bp (as expected for cleavage at the same site as in T2), and several fragments of ∼90 bp, further downstream in gene 56 (Fig. 4, lanes 1–3). Incubation of the same amount of SegF with a 261-bp PCR product containing the T2 cleavage site showed a single cleavage product of the expected size on the template strand (Fig. 4, lanes 4–6). To discover if the higher accumulation of the T2 product is due to a preference for this substrate, the T2 and T4 DNAs were mixed and incubated with SegF (Fig. 4, lanes 7–9). Although T4 cleavage products can still be detected, the clear bias toward T2 is maintained in this mixed cleavage assay. SegF has a preference in vitro for the T2 cleavage site compared with any of the sites in T4 gene 56.
Figure 4.
SegF prefers T2 DNA in vitro. Substrate DNAs that were end-labeled on their template (bottom) strands were generated by amplification of T4 DNA (primers 37 and 39) and of T2 DNA (primers 42 and 43). The positions of primers are shown in Figure 2C. Substrates were incubated singly (lanes 1–6) and together (lanes 7–9), with addition of in vitro synthesized SegF (lanes 3,6,9), a mock transcription/translation extract (lanes 2,5,8), or reaction buffer only (lanes 1,4,7). Cleavage products were resolved on a denaturing 6% polyacrylamide gel. Arrows indicate DNA substrates with the sizes indicated. Open triangles indicate the major cleavage products from a substrate with the T4 sequence; a solid triangle indicates the product from a substrate with the T2 sequence. Positions of the ΦX174/HaeIII DNA size standard are indicated on the right.
SegF preferentially cleaves T2 DNA in vivo
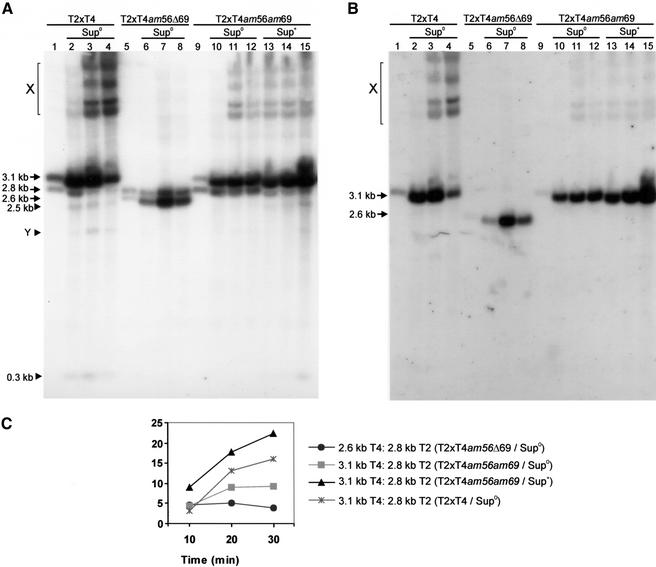
The in vitro experiments described above used unmodified PCR-generated DNA substrates. However, T4 and T2 phage DNAs are highly modified: cytosines are completely replaced by 5-hydroxymethyl cytosine residues, that are then further modified by glycosylation. To determine if T2 DNA is also preferred in vivo, we examined the accumulation of double-strand breaks in DNA isolated from mixed infections with T4 am56 (amE51) that were either wild type or carried a deletion (Δ69) or an amber mutation (am69) in gene 69. The Δ69 mutation is a 513-bp in-frame deletion lacking amino acids 22–192 of gp69, whereas am69 changes the tyrosine codon (TAC) at residue 24 to an amber (TAG) Stop codon. DNA isolated at various times after infection from E. coli B40 (strr) or B40 (supF, strr), was digested with restriction endonuclease PacI (which is active on fully modified DNA), separated on an agarose gel, and analyzed by Southern hybridization. Preliminary experiments (data not shown) confirmed the expected size (3.1 kb) of the restriction fragment containing T4 gene 56 (Kutter et al. 1994), and showed that the equivalent T2 restriction fragment is 2.8 kb. One of the PacI sites is between genes dam and 56, 360 bp upstream of the SegF cleavage site (Kutter et al. 1994). The Δ69 mutation reduces the size of the T4 restriction fragment to 2.6 kb.
DNA was isolated from cells that were coinfected with wild-type phages, and analyzed with mixed probes that hybridize to target site restriction fragments from both phages. Products of ∼2.5 kb and ∼0.3 kb accumulate (precisely those expected from SegF cleavage of the 2.8-kb T2 PacI fragment), along with another band of about 1.5 kb (labeled Y) that appears at later times (Fig. 5A, lanes 1–4). In addition to these smaller products, slower-migrating hybridizing species (labeled X) also accumulate at later times. These slower-migrating products could represent recombination intermediates. Similar slow-migrating bands, shown to be homing-specific recombination intermediates, have been observed during in vivo homing assays with I-TevI. However, in this case the intermediates could only be observed under conditions where T4 resolvase was limiting (Mueller et al. 1996b).
Figure 5.
SegF preferentially cleaves T2 DNA in vivo. (A) E. coli B40 strr (Sup0) or B40 supF strr (Sup+) cells were infected with phage T2 in combination with phage T4 or T4am56Δ69 or T4am56am69. Total DNA was prepared from the mixture of phages before addition to cells (lanes 1,5,9) and from phage-infected cells 10 min (lanes 2,6,10,13), 20 min (lanes 3,7,11,14), and 30 min (lanes 4,8,12,15) after infection. The DNA was digested with restriction endonuclease PacI, separated by electrophoresis, and analyzed by Southern hybridization. The probe was a mixture of random-primer-labeled DNA that was generated by amplification of T2 and T4 DNAs with primers 37 and 26 (see Fig. 2C). Arrows indicate PacI restriction fragments containing the T2 and T4 target DNAs, and solid triangles indicate putative cleavage products from T2. The sizes shown to the left were approximated using a DNA ladder mix as size standards. Slower-migrating DNA species are indicated by X. (B) Blot A was stripped and reprobed with end-labeled T4-specific oligonucleotide 35 (see Fig. 2C). (C) Hybridization signals from the T4 target DNA (3.1-kb PacI fragment in wild type and T4am56am69 or 2.6-kb fragment from T4am56Δ69) and T2 target DNA (2.8-kb PacI fragment) from blot A were determined using a phosphorimager. The ratios of intensities of the T4 to T2 bands were determined and normalized with respect to the input ratio.
The two major products (2.7 kb and 2.8 kb) predicted from SegF cleavage of the T4 restriction fragment could have been obscured by the uncleaved T2 restriction fragment. To investigate this, the membrane was stripped and reprobed with a 5′-end-labeled oligonucleotide (ab35) that hybridizes only to T4 DNA. Figure 5B shows that only the uncleaved T4 restriction fragment is labeled by the probe; there is no indication of lower-molecular-mass products, even after longer exposure (data not shown). Interestingly, the slower-migrating products hybridize to the T4-specific probe, indicating that these carry T4-specific sequences.
T2-specific cleavage products do not accumulate during coinfection with T4am56Δ69 (Fig. 5A,C, lanes 5–8), where the endonuclease activity of SegF should be absent. Although it is possible that the 2.6-kb T4 restriction fragment might obscure the 2.5-kb T2 product band, absence of a 0.3-kb product confirms the absence of SegF activity. Interestingly, the slower-migrating bands are also absent under these conditions.
Another indication that the faster-migrating DNA products are caused by cleavage by SegF is provided by analysis of DNA from mixed infections of T2 and T4am56am69. These cleavage products cannot be detected during infection of a Sup0 bacterial host, but appear (at reduced levels) in a tyrosine-inserting suppressor strain (Fig. 5A,C, lanes 9–15). The slower-migrating bands are present in infections of Sup+ as well as (with a delay) of Sup0 cells. This suggests that the nonsense mutation am69 may be leaky, or partially suppressed, even in the translationally stringent (strr) Sup0 cells.
Taken together, the experiments presented in Figure 5 show that gp69 (SegF) is a site-specific endonuclease in vivo, exhibiting a marked preference for T2 DNA, and may also be required for production of the slower-migrating DNA products that appear in mixed infections with phage T2.
Figure 5A also presents evidence consistent with a SegF-dependent conversion of the T2 PacI restriction fragment into the T4 form. In the complete absence of SegF (Fig. 5A, lanes 6–8) the ratios of the T2 and T4 bands do not change during the course of infection, whereas in the presence of wild-type SegF (Fig. 5A, lanes 2–4), the ratio of the T4-specific form to that of T2 increases steadily. This change is most likely the result of recombinational insertion, via the slower-migrating DNA species, into the T4 genome. This apparent conversion is reduced when segF contains an amber mutation (Fig. 5A, lanes 10–12), but is restored when in an amber-suppressing host that inserts the wild-type amino acid (Fig. 5A, lanes 13–15). These observations are presented graphically in Figure 5C.
SegF is required for marker exclusion of T2 gene 56
The properties of SegF provide an explanation for the strong exclusion of the T2 allele of gene 56 from the progeny of mixed infections with T4 (Pees and De Groot 1970; Russell and Huskey 1974). A preferential DSB in T2 gene 56 might initiate recombination-dependent repair using intact T4 DNA as a template, thus resulting in gene conversion originating from the cut site. This would effectively transfer T4 markers in and around gene 56 into the T2 phage genome.
To determine if SegF is involved in marker exclusion, we measured the inheritance of the E51 amber mutation in T4 gene 56 in mixed infections with T2. T4am56 or T4am56Δ69 were coinfected with T2 in B40 (supF, strr). Progeny from each cross were plated on B40 (supF, strr) to allow growth of all phages. Progeny with wild-type (T2) gene 56 were determined by replica-plating individual plaques on Sup+ and Sup0 hosts. Table 1 summarizes results from three independent experiments. The strong exclusion of the T2 gene 56 marker (Table 1, top) is similar to that reported by Russell and Huskey (1974) and Pees and De Groot (1970). Even in experiment 1, where 70% of the input phage were T2, only 3% of the progeny contained the T2 allele of gene 56, consistent with the number of cells that were not coinfected with T4. In contrast, ∼20% of the progeny of coinfection with T4am56Δ69 (with an in-frame deletion in gene 69 retaining only 37 of 224 amino acids) carry the wild-type marker from T2 gene 56 (Table 1, bottom). This is in the range of transmission (10%–40%) of T2 markers that are not located in zones of high-level exclusion (see Figs. 1 and 5 in Russell and Huskey 1974). This shows that an intact SegF is required for the local exclusion of T2 gene 56.
Table 1.
Role of SegF in exclusion of T2 allele of gene 56
| Phages
|
Exp. no.
|
Input
WTa
|
Progeny WTa
|
|---|---|---|---|
| T2L × T4am56 | 1 | 0.71 | 0.03 |
| 2 | 0.53 | 0.00 | |
| 3 | 0.48 | 0.01 | |
| T2L × T4am56Δ69 | 1 | 0.79 | 0.17 |
| 2 | 0.54 | 0.20 | |
| 3 | 0.51 | 0.22 |
Crosses were performed as described in Material and Methods. The host used for the coinfections was B40 supF strr (Sup+).
Fraction with wild-type marker (in gene 56) from T2 is indicated.
When a similar experiment was done comparing wild-type gene 69 with am69 T4 phage, there was little difference in transmission of the T2 gene 56 allele when mixed infections were performed on the nonpermissive host (data not shown). This is, however, consistent with the data in Figure 5, which show that limited expression of the endonuclease activity of gene 69 is sufficient for substantial accumulation of presumed recombination intermediates and for selective replication of T4 DNA in the vicinity of gene 56.
Discussion
The product of T4 gene 69 is a site-specific DNA endonuclease, SegF
The function of gene 69, originally described by Macdonald and Mosig (1984), has been elusive (Macdonald and Mosig 1984; Mosig and Macdonald 1986; for review, see Kreuzer and Morrical 1994). We show here that gp69 is a sequence-dependent DNA endonuclease, and that its amino acid sequence aligns with the group of five previously described Seg proteins in phage T4 (Fig. 1), which constitute a subgroup of the GIY–YIG family of homing endonucleases (Sharma et al. 1992; Kowalski et al. 1999). Therefore, we propose that gene 69 should be renamed segF.
Homing endonucleases are, by definition, intron-encoded and typically make double-strand breaks in intronless alleles of their cognate genes, 2–5 bp from the IIS. However, there are some exceptions, such as the three intron-encoded endonucleases of the T-even family of bacteriophages that cleave 13–25 bp from the IIS (Bell-Pedersen et al. 1990; Eddy and Gold 1991) and the Bacillus phage SP82 intron endonuclease (I-HmuII), which introduces a single-strand break 52 bp downstream of the IIS (Goodrich-Blair and Shub 1996). In contrast, the Seg proteins are not intron-encoded, but are free-standing ORFs inserted intergenically in bacteriophage T4, and (for the two cases examined, segA and segE) cleave DNA hundreds of base pairs from their insertion sites (Sharma and Hinton 1994; Kriukov et al. 1996). SegF is similar, with its gene inserted between conserved genes dam and 56, and it creates a staggered double-strand cut at its preferred cleavage site in gene 56 of T2, 210 and 212 bp upstream from its insertion site (Figs. 3, 6).
Figure 6.
Nucleotide sequence alignment of gene 56 from T4 and T2. Nucleotide sequence alignment of gene 56 from phages T4 (accession no. AF158101) and T2 (Gary et al. 1998), based on the amino acid sequence alignment. Nucleotide gaps are indicated by dashes. Identical nucleotides between T4 and T2 are shaded. SegF cleavage on the coding strand of T2 gene 56 is indicated by an upward-pointing arrow. An asterisk indicates the position of a G → A change in T4am56E51 (Gary et al. 1998). A right-pointing arrow indicates the start of gene 69 in the +1 frame of the T4 gene 56 sequence.
SegF mediates localized exclusion of T2 gene 56 by a process analogous to intron homing
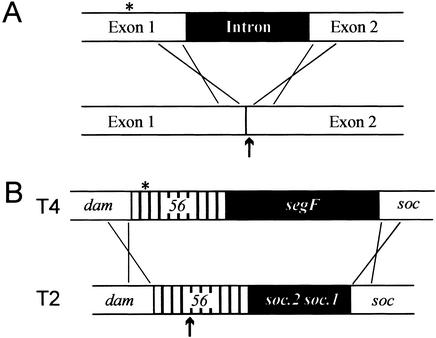
Intron homing is a consequence of a localized gene conversion event initiated by repair of a DSB induced in intronless DNA by the intron-encoded homing endonuclease. Coconversion of flanking markers accompanies intron insertion, but falls off rapidly with increasing distance from the cleavage site (Fig. 7A; Bell-Pedersen et al. 1989). We propose that SegF is a mobile element that practices homing-like mobility into gene 56 sequences of T-even bacteriophages, bringing with it the gene 56 alleles of the donor phage. This provides an explanation for the localized marker exclusion that has been described in genetic crosses (Pees and De Groot 1970; Russell and Huskey 1974).
Figure 7.
Intron versus intronless homing. Filled boxes represent regions of DNA nonidentity. (A) Group I intron homing: An endonuclease encoded within the intron cleaves (↑) the intronless homolog close to the intron insertion site. DSB repair, which includes limited exonuclease digestion of cut DNA, incorporates the intron into the recipient, with frequency of coconversion of flanking markers (*) inversely proportional to their distance from the cleavage site. Insertion of the intron destroys the endonuclease recognition site. (B) Intronless homing by segF. SegF preferentially cleaves T2 gene 56, within a region of partial conservation with the T4 homolog (↑). Because of patchy conservation within gene 56, DSB repair initiates recombination in dam and soc, the closest regions of extensive sequence conservation. segF and the entire T4 gene 56 (with its nonpreferred SegF cleavage site) are transferred to recipients. The coconversion of markers in gene 56 (*) is close to 100%.
Although analogous to intron- or intein-homing, intronless homing of a free-standing gene presents certain unique problems. Homing sites of mobile introns or inteins are generally found in highly conserved regions of essential genes, maximizing the availability of targets (Dalgaard et al. 1997b; Edgell et al. 2000). Generally, the endonuclease cleavage site is immediately adjacent to the intron insertion site, guaranteeing that insertion of the intron (and the endonuclease gene within it) will accompany DSB repair. Furthermore, insertion of the intron disrupts the endonuclease recognition site, rendering the intron-containing gene resistant to cleavage (Fig. 7A). Because the inserted sequence will be removed by RNA- or protein-splicing, there is no harm to the host in disrupting an essential gene. In contrast, intronless ORFs must be intergenic to avoid disrupting gene function. However, intergenic sequences are poor candidates for sequence-specific cleavage/insertion sites. If an intergenic target sequence contains an essential regulatory element, insertion will be deleterious to the host, whereas if the intergenic sequence has no function, it is subject to rapid mutational change and will not persist in the population. segF has adapted to homing, without the need to associate with a splicing element, by using a different strategy.
Gene 56 (dCTP/dUTPase) is essential in the T-even phages, being required to prevent incorporation of cytosine into phage DNA, thereby protecting themselves from the action of phage-encoded endonucleases that are directed against the unmodified cytosine-containing DNA of the bacterial host. Nevertheless, gene 56 has diverged extensively in T2 and T4, being only 71% identical at the nucleotide level and 70% identical at the amino acid level (Gary et al. 1998). Moreover, the regions of identity are distributed in patches throughout the gene (see Fig. 6; Gary et al. 1998). The minimum length of uninterrupted identity needed to support efficient recombination in T4 (or homing of the td intron mediated by I-TevI) is ∼50 bp (Singer et al. 1982; Parker et al. 1999). However, there is no single continuously conserved sequence of 50 bp in gene 56 (Fig. 6).
segF is inserted downstream of, and partially overlapping, gene 56 of T4, whereas the same region of T2 is occupied by two entirely unrelated small ORFs, soc.1 and soc.2 (Gary et al. 1998). The entire region is flanked by two genes, dam and soc, that are well conserved in both phages (Figs. 5D, 7B). Although SegF cleaves T2 DNA within the longest conserved sequence in gene 56, the closest opportunities to initiate DSB repair are in dam and soc. This assures that, even though segF is inserted at a distance from its cleavage site, segF is substituted for soc.1 and soc.2, and all of T4 gene 56 replaces its T2 homolog (removing the preferred substrate for SegF) in the same recombinational event (Fig. 7). Even though the SegF cleavage site is within a 31-bp region of identity between T2 and T4 genes 56 (Fig. 6), the observed preference of SegF for the T2 sequence could be determined by the divergent sequences at either end. For example, the recognition/cleavage site of the related homing enzyme I-TevI covers ∼40 bp.
Alternatively, if the SegF cleavage site were in a gene with long tracts of sequence identity in phages T2 and T4, repair of the DSB would likely produce many events where recombination occurred between the cleavage site and the segF gene (Mueller et al. 1996b). This would result in inefficient transfer of the segF gene to its new home.
In this context, it is not surprising that virtually 100% of the progeny of a mixed infection contain the T4 allele of gene 56, and that this localized marker exclusion is dependent on an intact segF gene. Gary et al. (1998) offered an explanation for exclusion of T2 alleles of gene 56 based on the fact that patchy homology between genes 56 of T2 and T4 would be a barrier to recombination, and that many hybrid genes that would be formed by recombination might not be functional. However, neither of these observations accounts for the directionality of the exclusion. On the other hand, selective cleavage of the T2 gene by SegF assures propagation of the T4 allele by DSB repair (Fig. 7).
Our model for localized marker exclusion requires, in addition to the presence of segF in T4, the T2 sequence at the SegF cleavage site, ∼300 bp from the beginning of gene 56. Consistent with this conclusion, Gary et al. (1998) determined that localized exclusion of genetic markers in gene 56 occurred normally if the T2 phage in the cross was a hybrid containing the first 160 bp of the T4 gene 56. However, localized exclusion did not occur if the T2 hybrid had its gene 56 entirely replaced by the wild-type T4 gene. Interestingly, Okker et al. (1981) isolated a mutant of T2 that was resistant to exclusion of gene 56 markers in crosses with T4, and whose mutation mapped in gene 56, consistent with a change at the SegF cleavage site. Unfortunately, this strain has been lost (R. Okker, pers. comm.).
The modular nature of mobile group I introns
It has long been assumed that group I introns are composite selfish DNA elements, because the introns and their associated endonuclease genes often have different evolutionary histories (Mota and Collins 1988; Lambowitz and Belfort 1993). Phage T4 presents a particularly graphic representation of the modular nature of these genetic elements. Each of the three T4 introns is inhabited by a homing endonuclease, two of which belong to the GIY–YIG gene family and one to the H–N–H family. (N.B., whereas the endonuclease gene in the T4 nrdBintron has suffered a deletion of its proximal half, a complete, active H–N–H endonuclease gene is found in the corresponding intron in the T-even phage isolate RB3 [Eddy and Gold 1991; Shub et al. 1994].) At the same time, 12 additional ORFs, belonging to these two gene families, are inserted intercistronically at various places in the genome (Kutter et al. 1996; this work). It is likely that T4 has been a natural laboratory for trafficking of endonuclease genes between introns and intercistronic sites.
Furthermore, the endonucleases themselves are modular. In particular, the GIY–YIG enzyme I-TevI has an endonuclease domain in its N-terminal half, whereas the C-terminal half comprises the DNA-binding domain. Although the N-terminal half of SegF aligns with the GIY–YIG catalytic domain (Fig. 1), its C-terminal half aligns with the corresponding portion of the H–N–H family gene mobD (Gary et al. 1998). The endonuclease genes, themselves, are clearly shuffling their domains of protein structure.
From selfish element to symbiont? Abundance of free-standing endouclease genes in bacteriophage T4
The frequent occurrence of homing endonuclease genes in T4 stands in sharp contrast to the situation encountered in all of its nearest relatives. The distribution of introns in these phages has been the subject of a careful study involving ∼30 independent T-even isolates. In cases where the corresponding genes could be substantiated, only four other occurrences of the td intron, one of the nrdB intron, and no additional examples of the sunY/nrdD intron were found (Eddy 1992; Edgell et al. 2000). Similarly, segA, which is present in T4, absent in T2, and encodes a T2-specific endonuclease, has been shown to be absent in 25 additional T-even isolates, all of which retain a SegA cleavage site (A. Bell, D. Hinton, and D.A. Shub, in prep.). segF/gene 69 does not appear to be an exception. Although only a small number of phages were examined, two of them, LZ5 and T6, are T2-like (with soc.1 and soc.2), whereas a third phage (RB15) lacks any additional genes between 56 and soc (Mosig et al. 2001).
This distribution of intron endonucleases in the natural population of T-even phages is inconsistent with the situation one would expect based on laboratory experiments. Virtually 100% of the progeny of a coinfection between T4 and one of its relatives contain the homing endonuclease from T4 (Bell-Pedersen et al. 1990; Kadyrov et al. 1997). However, many homing endonucleases have a relaxed specificity, being able to cleave variants of the preferred sequence (Fig. 4; for review, see Jurica and Stoddard 1999). We assume that acquisition of such a gene carries an associated fitness cost, which will eventually purge the endonuclease gene from the progeny of a recipient. This would explain the sporadic distribution of these genes in natural populations. The striking exception of phage T4 to this distribution implies that occasional double-strand breaks are not as harmful to T4 as they are to its relatives. Assuming that phage T4 is specially adapted to repair its DNA, it would rapidly become a haven for homing endonucleases and associated introns, either from other genomes or by transposition within its own DNA.
Whereas one homing endonuclease can ensure the efficient coconversion of flanking genetic markers in genetic crosses, accounting for localized marker exclusion, possession of a suite of similar nucleases, distributed around the phage chromosome, might fragment related T-even genomes into small pieces when they are encountered in a mixed infection. In this way, the additive effect of multiple selfish endonuclease genes may provide an explanation for the preferential inheritance of all T4 genes (general, or phage exclusion) that is displayed in genetic crosses with almost all of its close relatives (Russell and Huskey 1974).
Materials and methods
Media and standard phage techniques are as described in Russell and Huskey (1974).
Recombinant plasmids
pBST2ab4243 was constructed by amplification of T2 DNA using primers 42 and 43 and ligating the polymerase chain reaction (PCR) product to HincII-digested pBluescript II SK(−) (Stratagene). The SegFHis6 expression vector pAA1 was generated by amplifying T4 DNA using primers 35 and 36. The product was digested with XhoI and ligated into pET24a (Novagen), which had been digested with NdeI, filled in with the Klenow fragment of DNA polymerase, and digested with XhoI. pBST4Δ69 was constructed as follows. Two PCR products were generated from T4 DNA using primer pairs 59–60 and 61–62, digested with BamHI and BglII, respectively, and joined with T4 ligase. The resulting 713-bp DNA fragment, containing a 513-bp in-frame deletion extending from codon 22 to 192 of gene 69, was ligated into the HincII site of pBluescript II SK(−). pBST4ab3334 was generated by ligating a PCR product derived from T4 DNA, using primers 33 and 34, into HincII-digested pBluescriptSK(−). pBST4am69 was constructed by PCR of pBSTab3334 using primers 40 and 41. Primer 40 changes the twenty-fourth codon of gene 69 from tyrosine (TAC) to amber (TAG) and the twenty-fifth codon from GCT to GCA. The mutations were confirmed by sequencing.
Bacteria and bacteriophage strains
E. coli strain ER2266 (NEB) was used for the expression of SegFHis6. E. coli XL-1 blue (Stratagene) was the recipient strain for electroporations. E. coli B40 strr (Sup0) and B40 supF strr (Sup+) were from J.D. Karam (Tulane University Medical Center, New Orleans). supF is a tyrosine-inserting amber suppressor. B. Singer (University of Colorado, Boulder) contributed Bacteriophage T4D (Doermann) and T4amE51 (carrying a G → A change at position 67 that results in a change from tryptophan to amber at codon 20 of gp56; Gary et al. 1998). T2L (Luria) was a gift from M. Belfort (Wadsworth Center, Albany, NY).
To construct phage mutant T4am56Δ69, E. coliharboring plasmid pBST4Δ69 was infected with T4amE51. Progeny were screened for recombinant phage that carried the deletion by plaque hybridization using oligonucleotide 63 that spans the deletion site. The presence of the deletion was confirmed by sequencing. In addition, the presence of the amber mutation in gene 56 was confirmed by plating on Sup0 versus Sup+ hosts. Similarly, T4am56am69 was constructed via recombination between plasmid pBST4am69 and phage T4amE51, and progeny were screened by plaque hybridization with oligonucleotide 40 that distinguishes between wild-type and am69 alleles. The presence of the amber mutation in gene 56 was confirmed by plating the recombinants on Sup0 versus Sup+ hosts.
Phage crosses
E. coli B40 strr or B40 supF strr cells were grown at 37°C to an A590 of 0.3 and coinfected with phages T2L and T4am56am69 or T4am56Δ69, each at a multiplicity of infection of 4. After 10 min of incubation without shaking at 30°C, the infected cultures were diluted 10−4 into fresh, prewarmed medium and grown with vigorous shaking at 30°C for 2.5 h. Chloroform was added to ensure complete lysis, and progeny of the mixed infections were titrated on B40 supFstrr. Wild-type progeny were determined by replica-plating on Sup0 versus Sup+ hosts.
Phage DNA isolation
Phages were isolated by CsCl step gradient, and subsequently DNA was isolated as described in Kricker and Carlson (1994).
Molecular biology
PCR, cloning, and Southern analysis were all performed essentially as described in Sambrook et al. (1989). Capillary blotting was used to transfer separated nucleic acid fragments from agarose gels to Hybond-N membrane (Amersham). Plaque hybridizations involved plaque lifts onto Hybond-N membranes as described in the supplier's protocol. Hybridization was carried out overnight in 1× Denhardt's solution, 3× SSC, and 0.1% SDS, using end-labeled oligonucleotide at 50°C, or with denatured random primer labeled probe (∼107 cpm) at 60°C. The blots were washed for 15 min in 2× SSC and 0.1% SDS at room temperature followed by a 10-min wash at a stringent temperature for each probe.
Protein expression
Templates for in vitro protein synthesis were obtained by amplification of T4 gene 69 using primers 25 and 26. Primer 25 incorporates a T7 gene 10 promoter and a Shine–Dalgarno sequence fused to phage sequences upstream of the ORF. Transcription/translation was done using the Modulis Systems (MBI fermentas) as described in the supplier's protocol.
ER2566 (pAA1) was grown in LB supplemented with kanamycin (30 μg/mL) at 37°C to an A600 of 0.6. Cultures were induced at 25°C with IPTG (0.5 mM). After 3.0 h of induction, cells were harvested at 6000g, resuspended at 10 mL/g cells of lysis cocktail (20 mM Tris/HCl at pH 8.0, 0.5 M NaCl, 2 mM imidazole, 1 mM PMSF, 2 μg/mL leupeptin) and at stored at −80°C. Frozen cells were thawed and sonicated to complete lysis. Insoluble material was removed by two rounds of centrifugation at 12,000g for 30 min. PEI (5%) was added to the supernatant to a final concentration of 0.1%, and the precipitate was removed by centrifugation at 10,000g. After filtration, proteins were applied to a chelating column (HiTrap, Pharmacia), charged with 50 mM NiSO4, and equilibrated in 20 mM Tris (pH 8.0), 500 mM NaCl, and 2 mM imidazole. The column was washed with 20 mM Tris/HCl (pH 8.0), 500 mM NaCl, and 60 mM imidazole. Proteins were eluted with 20 mM Tris/HCl (pH 8.0) and 500 mM NaCl over a linear gradient of imidazole from 0.06 M to 0.7 M. One-milliliter fractions were collected into 1 mL of 20 mM Tris/HCl (pH 8.0) and 0.5 M NaCl to prevent precipitation. Fractions containing SegF were pooled and applied to a 1-mL Heparin column (HiTrap, Pharmacia), equilibrated in 20 mM Tris/HCl (pH 8.0) and 0.5 M NaCl. The column was washed and protein was eluted in 20 mM Tris/HCl (pH 8.0) over a 15-mL linear gradient of 0.5 to 1.55 M NaCl.
Endonuclease assays
Radiolabeled or unlabeled PCR-generated substrates were incubated with SegF in the presence of 20 mM Tris/HCl (pH 8.0), 5 mM MgCl2, and 2 mM ATP at 37°C. Reactions were terminated with 10 mM EDTA and freezing on dry ice.
Detection of double-stranded breaks in vivo
E. coli strain B40 supF strr (Sup+) or B40 strr (Sup0) cells were grown at 37°C to an A590 of 0.3 and coinfected with phages T2 and T4, T4am56am69, or T4am56Δ69, each at a multiplicity of infection of 4. After a 10-min incubation without shaking, the infected cultures were incubated with vigorous shaking at 37°C. DNA was prepared from 1 mL of infected culture by pelleting cells for 5 min. Released phage particles in the supernatant were pelleted by centrifugation for 1 h. The combined pellets were frozen in dry-ice ethanol, followed by thawing and resuspension in 300 μL of lysis buffer (100 mM NaCl, 50 mM Tris-HCl at pH 7.5, 0.2% SDS, 10 mM EDTA, 0.3 mg/mL proteinase K) and incubated at 65°C for 1 h. After extraction with phenol, DNA was precipitated and resuspended in H2O. Total DNA was digested with PacI, resolved on a 1% agarose gel, and analyzed by Southern hybridization.
Oligonucleotides
In the following, coordinates in brackets refer in the case of T2 to Figure 1A in Gary et al. (1998) and in the case of T4 to GenBank accession no. AF158101, or as otherwise noted. Bold letters indicate mismatches to the wild-type sequence. Incorporated restriction sites are shown in italics.
T2-specific
42: 5′-CCAGAAACAAATCGTCATCCTG [191–212]. 43: 5′-GTGAAACTGGTCAATCAGTTC [complement: 431–451]
T4-specific
25: 5′-TAATACGACTCACTATAGGGCAATTT TAGGAGG/CAAGATAATGGATATTAAAC [T7 promoter/complement: 16268–16287]. 33: 5′-GTTGGCCTTGGAATGA ATGC [complement: 16340–16359]. 34: 5′-CTAGAACCATG GCATTTTGACC [16075–16096]. 35: 5′-TGGATATTAAACA AAAATTTTATAG [complement: 16255–16279]. 36: 5′-TTA CATGATCTCGAGTACATCTTCTTTTCTG [15594–15624]. 39: 5′-TATCCATTATCTTGACGTTC [16274–16293]. 40: 5′-CG TCTTTTGGTAGGCAGGGAAACATGA [complement: 16194–16221]. 41: 5′-CCTTTCGGCGTCCTAAC [16222–16238]. 59: 5′-CCGCGAACTTCTGACTTCTC [complement: 16527–16546]. 60: 5′-CGCGGATCCGACACCTTTCGGCGTCCTAAC [16218–16238]. 61: 5′-GGAAGATCTTCAGCAACAGGCCCGGTTG [complement: 15689–16006]. 62: 5′-CCAGTTACTTTCCACAA ATCTTC [15334–15357]. 63: 5′-GGTGTC/GGATCTT/CAG CAAC-3′ [complement: 16018–16023/GGATCTT/complement: 15700–15706].
Common (T4 and T2)
26: 5′-CGCGAGTACTAGCCATG [T4: 15558–15574]. 27: 5′-CGGAAAGGAAAACACTCTTC [1286–1307 {T2 sequence (accession: M22342)}]. 37: 5′-ATGGCT CACTTTAATGAATG [T4: complement: 16766–16785].
Acknowledgments
We thank Marlene Belfort and Richard Zitomer for useful discussions, Richard Bonocora and David Edgell for advice and a critical reading of the manuscript, and National Institute of Health grant GM37746 for support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL shub@albany.edu; FAX (518) 442-4354.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.960302.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M, Roberts RJ. Homing endonucleases: Keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D, Quirk SM, Aubrey M, Belfort M. A site-specific endonuclease and co-conversion of flanking exons associated with the mobile td intron of phage T4. Gene. 1989;82:119–126. doi: 10.1016/0378-1119(89)90036-x. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Quirk S, Clyman J, Belfort M. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: Mechanistic and evolutionary implications. Nucleic Acids Res. 1990;18:3763–3770. doi: 10.1093/nar/18.13.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier BS, Stoddard BL. Homing endonucleases: Structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ, Moser MJ, Holley WR, Chatterjee A, Mian IS. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonuclease and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acid Res. 1997a;15:4626–4638. doi: 10.1093/nar/25.22.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard JZ, Moser MJ, Hughey R, Mian IS. Statistical modeling, phylogenetic analysis and structure prediction of a protein splicing domain common to inteins and hedgehog proteins. J Comput Biol. 1997b;4:193–214. doi: 10.1089/cmb.1997.4.193. [DOI] [PubMed] [Google Scholar]
- Derbyshire V, Kowalski JC, Dansereau JT, Hauer CR, Belfort M. Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J Mol Biol. 1997;265:494–506. doi: 10.1006/jmbi.1996.0754. [DOI] [PubMed] [Google Scholar]
- Dujon B. Group I introns as mobile genetic elements: Facts and mechanistic speculations—A review. Gene. 1989;82:91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- Eddy SR. “Introns in the T-even bacteriophages.” Ph.D. dissertation. Boulder, CO: University of Colorado; 1992. [Google Scholar]
- Eddy SR, Gold L. The phage T4 nrdBintron: A deletion mutant of a version found in the wild. Genes & Dev. 1991;5:1032–1041. doi: 10.1101/gad.5.6.1032. [DOI] [PubMed] [Google Scholar]
- ————— Artificial mobile DNA element constructed from the EcoRI endonuclease gene. Proc Natl Acad Sci. 1992;89:1544–1547. doi: 10.1073/pnas.89.5.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgell DR, Belfort M, Shub DA. Barriers to intron promiscuity in bacteria. J Bacteriol. 2000;182:5281–5289. doi: 10.1128/jb.182.19.5281-5289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary TP, Colowick EN, Mosig G. A species barrier between bacteriophages T2 and T4: Exclusion, join-copy and join-cut-copy recombination and mutagenesis in the dCTPase gene. Genetics. 1998;148:1461–1473. doi: 10.1093/genetics/148.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich-Blair H, Shub DA. Beyond homing: Competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell. 1996;84:211–221. doi: 10.1016/s0092-8674(00)80976-9. [DOI] [PubMed] [Google Scholar]
- Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Stoddard BL. Homing endonucleases: Structure, function and evolution. Cell Mol Life Sci. 1999;55:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Kryukov VM. Mobility of the T4 phage segE gene: The high-frequency transfer of segEfrom plasmid into phage genome depends on the intactness of the gene. Dokl Akad Nauk. 1996;346:86–89. [PubMed] [Google Scholar]
- Kadyrov FA, Kaliman AV, Kryukov VM. The phage T4 SegE endonuclease. I. Cloning, expression and biochemical characterization of endonuclease activity. Mol Biol (Moscow) 1996;30:654–660. [PubMed] [Google Scholar]
- Kadyrov FA, Shlyapnikov MG, Kryukov VM. A phage T4 site-specific endonuclease, SegE, is responsible for a non-reciprocal genetic exchange between T-even-related phages. FEBS Lett. 1997;415:75–80. doi: 10.1016/s0014-5793(97)01098-3. [DOI] [PubMed] [Google Scholar]
- Kim J-S, Davidson N. Electron microscope heteroduplex study of sequence relations of T2, T4, and T6 bacteriophage DNAs. Virology. 1974;57:93–111. doi: 10.1016/0042-6822(74)90111-1. [DOI] [PubMed] [Google Scholar]
- Kostriken R, Strathern JN, Klar AJ, Hicks JB, Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983;35:167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Kowalski JC, Belfort M, Stapleton MA, Holpert M, Dansereau JT, Pietrokovski S, Baxter SM, Derbyshire V. Configuration of the catalytic GIY–YIG domain of intron endonucleases I-Tev I: Coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer KN, Morrical SC. Initiation of DNA replication. In: Karam JD, et al., editors. Molecular biology of bacteriophage T4. Washington, DC.: ASM press; 1994. pp. 28–53. [Google Scholar]
- Kricker M, Carlson K. Isolation of T4 phage DNA. In: Karam JD, et al., editors. Molecular biology of bacteriophage T4. Washington, DC.: ASM press; 1994. pp. 455–456. [Google Scholar]
- Kriukov VM, Shlyapikov MG, Kadyrov FA. Endonuclease SegE of phage T4. II. Recognition site. Mol Biol (Moscow) 1996;30:786–791. [Google Scholar]
- Kühlmann UC, James GR, Hemmings AM. Structural parismony in endonuclease active sites: Should the number of homing endonuclease families be refined? FEBS Lett. 1999;463:1–2. doi: 10.1016/s0014-5793(99)01499-4. [DOI] [PubMed] [Google Scholar]
- Kutter E, Stidham T, Guttman B, Kutter E, Batts D, Peterson S, Djavakhishvili T, Arisaka F, Mesyanzhinov V, Ruger W, et al. Genomic map of Bacteriophage T4. In: Karam JD, et al., editors. Molecular biology of bacteriophage T4. Washington, DC.: ASM press; 1994. pp. 502–508. [Google Scholar]
- Kutter E, Gachechiladze K, Poglazov A, Marusich E, Shneider M, Aronsson P, Napuli A, Porter D, Anzhinov VM. Evolution of T4 related phages. Virus Genes. 1996;11:285–297. doi: 10.1007/BF01728666. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Belfort M. Introns as mobile genetic elements. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Mosig G. Regulation of a new bacteriophage T4 gene69, that spans an origin of DNA replication. EMBO J. 1984;12:2863–2871. doi: 10.1002/j.1460-2075.1984.tb02221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosig G, Macdonald PM. A new membrane-associated DNA replication protein, the gene 69 product of bacteriophage T4, shares a patch of homology with Escherichia colidnaA protein. J Mol Biol. 1986;189:243–248. doi: 10.1016/0022-2836(86)90395-5. [DOI] [PubMed] [Google Scholar]
- Mosig G, Colowick NE, Pietz BC. Several new bacteriophage T4 genes, mapped by sequencing deletion endpoints between genes 56 (dCTPase) and dda(a DNA-dependent ATPase-helicase) modulate transcription. Gene. 1998;223:143–155. doi: 10.1016/s0378-1119(98)00238-8. [DOI] [PubMed] [Google Scholar]
- Mosig G, Gewin J, Luder A, Colowick N, Vo D. Two recombination-dependent DNA replication pathways of bacteriophage T4, and their roles in mutagenesis and horizontal gene transfer. Proc Natl Acad Sci. 2001;98:8306–8311. doi: 10.1073/pnas.131007398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota EM, Collins RA. Independent evolution of structural and coding regions in a Neurosporamitochondrial intron. Nature. 1988;332:654–656. doi: 10.1038/332654a0. [DOI] [PubMed] [Google Scholar]
- Mueller JE, Clyman J, Huang Y, Parker MM, Belfort M. Intron mobility in phage T4 occurs in the context of recombination-dependent DNA replication by way of multiple pathways. Genes & Dev. 1996a;10:351–364. doi: 10.1101/gad.10.3.351. [DOI] [PubMed] [Google Scholar]
- Mueller JE, Smith D, Belfort M. Exon coconversion biases accompanying intron homing: Battle of the nucleases. Genes & Dev. 1996b;10:2158–2166. doi: 10.1101/gad.10.17.2158. [DOI] [PubMed] [Google Scholar]
- Okker RJH, Pees E, Bom V. Partial exclusion of bacteriophage T2 by bacteriophage T4: An exclusion-resistant mutation in gene56 of T2. J Gen Virol. 1981;53:13–19. doi: 10.1099/0022-1317-53-1-13. [DOI] [PubMed] [Google Scholar]
- Parker MM, Belisle M, Belfort M. Intron homing with limited exon homology: Illegitimate double-strand-break repair in intron acquisition by phage T4. Genetics. 1999;153:1513–1523. doi: 10.1093/genetics/153.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pees E, De Groot B. Partial exclusion of bacteriophage T2 with T4-glucosylated DNA in crosses with bacteriophage T4. Genetica. 1970;41:541–550. doi: 10.1007/BF00958933. [DOI] [PubMed] [Google Scholar]
- Repoila F, Tetart F, Bouet J-Y, Krisch HM. Genomic polymorphism in the T-even bacteriophages. EMBO J. 1994;13:4181–4192. doi: 10.1002/j.1460-2075.1994.tb06736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RL, Huskey RJ. Partial exclusion between T-even bacteriophages: An incipient genetic isolation mechanism. Genetics. 1974;78:989–1014. doi: 10.1093/genetics/78.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sharma M, Hinton DM. Purification and characterization of the SegA protein of Bacteriophage T4, an endonuclease related to proteins encoded by group I introns. J Bacteriol. 1994;176:6439–6448. doi: 10.1128/jb.176.21.6439-6448.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Ellis RL, Hinton DM. Identification of a family of bacteriophage T4 genes encoding proteins similar to those present in group I introns of fungi and phage. Proc Natl Acad Sci. 1992;89:6658–6662. doi: 10.1073/pnas.89.14.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shub DA, Goodrich-Blair H, Eddy SR. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem Sci. 1994;19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- Singer BS, Gold L, Gaus P, Doherty DH. Determination of the amount of homology required for recombination in bacteriophage T4. Cell. 1982;31:25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn AR, Butow RA. Nonreciprocal exchange between alleles of the yeast mitochondrial 21S rRNA gene: Kinetics and the involvement of a double-strand break. Cell. 1985;40:887–895. doi: 10.1016/0092-8674(85)90348-4. [DOI] [PubMed] [Google Scholar]