Abstract
The Notch signaling pathway regulates the commitment and early development of T lymphocytes. We studied Notch-mediated induction of the pre-T cell receptor α (pTa) gene, a T-cell-specific transcriptional target of Notch. The pTa enhancer was activated by Notch signaling and contained binding sites for its nuclear effector, CSL. Mutation of the CSL-binding sites abolished enhancer induction by Notch and delayed the up-regulation of pTa transgene expression during T cell lineage commitment. These results show a direct mechanism of stage- and tissue-specific gene induction by the mammalian Notch/CSL signaling pathway.
Keywords: Notch, CSL, pre-TCRα, enhancer, T lymphocytes
Notch proteins comprise a family of transmembrane receptors conserved throughout evolution and involved in cell fate decisions in many tissues (for review, see Artavanis-Tsakonas et al. 1999; Mumm and Kopan 2000). The mammalian Notch pathway includes at least four Notch receptor genes (Notch1–Notch4) and multiple ligands (Jagged1–Jagged2 and the Delta family). Upon ligand binding, the Notch protein is proteolytically cleaved to release its intracellular domain (NICD), which represents an activated form of the Notch receptor. NICD then translocates into the nucleus and binds through its RAM and ankyrin domains the transcription factor CSL [for CBF1 in humans, Su(H) in Drosophila, and Lag-1 in Caenorhabditis elegans; also known as RBP-Jκ in the mouse]. CSL is a DNA-binding protein that normally represses transcription by virtue of its interaction with several corepressor complexes. The binding of NICD converts CSL into a transcriptional activator and thus induces the expression of target genes, most notably the Hes family of transcriptional repressors. The Hes proteins, in turn, modulate the activity of tissue-specific basic helix–loop–helix (bHLH) transcription factors, further propagating the effects of Notch signaling. In Drosophila, many genes are directly induced by Notch signaling through the binding of the NICD/CSL complex to their regulatory regions (Bray and Furriols 2001). A similar mechanism was shown for the mouse Hes1 gene, a general downstream effector of Notch (Jarriault et al. 1995). However, few tissue-specific Notch targets have been identified in the mammalian system, and the mechanism of their induction by Notch remains obscure.
Several lines of research revealed an essential, nonredundant role of Notch1 receptor signaling in the commitment of lymphoid progenitors to the T cell lineage in the thymus (for review, see Anderson et al. 2001; von Boehmer 2001). For instance, targeted disruption of Notch1 (Radtke et al. 1999) or of its downstream effector Hes1 (Tomita et al. 1999) arrests T cell development at the earliest stages. Conversely, Notch signaling in the bone marrow instructs hematopoetic precursors to adopt a T cell fate (Pui et al. 1999; Jaleco et al. 2001). In addition to its obligatory role in T cell commitment, Notch activity may subsequently favor the specification of the T cell receptor (TCR) αβ-bearing T cells as opposed to the TCRγδ-bearing T cells (Washburn et al. 1997). However, the molecular basis of the regulation of early T cell development by Notch is largely unknown.
Recently, the pre-TCR α chain gene (pTa) was identified as a transcriptional target of Notch signaling in T cells (Deftos et al. 2000). pTa encodes a transmembrane protein that pairs with the newly rearranged TCRβ chain to form the essential pre-TCR signaling complex in the developing T cells (von Boehmer and Fehling 1997). The pTa gene is expressed in immature T cells, and its up-regulation coincides with irreversible T cell lineage commitment in both murine and human thymic precursors (Res and Spits 1999; Rothenberg 2000). Moreover, pTa is required for αβ but not γδ T cell development (Fehling et al. 1995), and may facilitate the αβ lineage commitment by providing an instructive signal from the pre-TCR (Aifantis et al. 1998). The expression and function of pTa are thus consistent with Notch activity, further suggesting that pTa represents a T-cell-specific functional target of Notch. To gain insight into the modulation of stage- and tissue-specific gene expression by Notch signaling, we studied the mechanism of Notch-mediated pTa induction in developing T lymphocytes.
Results
The pTa enhancer contains CSL-binding sites
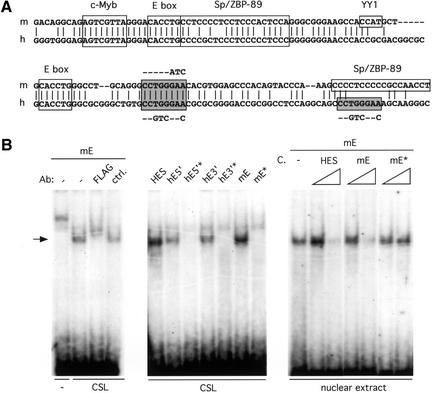
Previously, we identified an upstream pTa enhancer (Reizis and Leder 1999) that is conserved between mice and humans and appears both necessary and sufficient for correct pTa expression in T cells (Reizis and Leder 2001). As illustrated in Figure 1A, a conserved site (CCTGGGAA) homologous to the consensus CSL-binding sequence (CGTGGGAA; Tun et al. 1994) was identified within the core enhancer region. In addition, an identical sequence was found 30 bp downstream in the human but not in the mouse enhancer. To test the ability of these sites to bind CSL, the corresponding short oligonucleotide probes were tested in an electrophoretic mobility shift assay (EMSA) using in vitro translated FLAG-tagged human CSL protein. As shown in the left panel of Figure 1B, the CSL protein and the mouse enhancer site (mE) formed a complex that could be supershifted by anti-FLAG Ab. A similar complex was formed by both human enhancer sites, as well as by a known CSL-binding site from the Hes1 promoter (Fig. 1B, middle panel). In contrast, all three pTa enhancer sites harboring mutations illustrated in Figure 1A failed to bind CSL. These data confirm the specific interaction between the CSL protein and the sites from the pTa enhancer. Furthermore, a complex of similar mobility was formed when the mE site was incubated with a T cell nuclear extract (Fig. 1B, right panel). The formation of this complex was inhibited by the unlabeled Hes1 site and by the wild-type mE site, but not by the mutated mE site. Although a supershift experiment could not be performed owing to insufficient quality of the available anti-CSL antibodies, the size and specificity of the observed complex are consistent with CSL. Moreover, no additional complexes with other nuclear factors were observed. These results suggest, but do not prove, that CSL might be a major nuclear factor interacting with the CSL sites from the pTa enhancer.
Figure 1.
CSL-binding sites in the pTa enhancer. (A) Alignment of the mouse (m) and human (h) enhancer core sequences. Previously identified binding sites are boxed, and the potential CSL-binding sites are highlighted as shaded boxes. The mutations introduced into CSL sites are shown next to the sites. (B) The binding of CSL to sites from the pTa enhancer. EMSA was performed with radio-labeled oligonucleotide probes, and the resulting CSL–DNA complexes are indicated by an arrow. The probes included CSL-binding sites from the Hes1 promoter (HES), the mouse pTa enhancer (mE), and the 5′ and 3′ sites from the human pTa enhancer (hE5′ and hE3′, respectively). The corresponding probes harboring the mutations shown in A are indicated by an asterisk (mE*, hE5′*, and hE3′*). (Left panel) In vitro translated CSL or a control translation reaction (–) were incubated with the mE probe in the presence of anti-FLAG Ab or a control Ab (ctrl.). (Middle panel) In vitro translated CSL was incubated with the indicated probes. (Right panel) Nuclear extract from BW5147 cells was incubated with the mE probe in the presence of 10-fold and 100-fold molar excess of the indicated unlabeled competitor probes (C).
Induction of the pTa enhancer by Notch
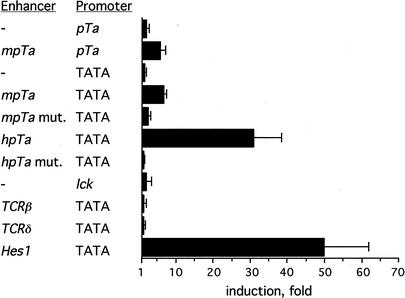
To test whether the pTa enhancer can be activated by Notch signaling, we transiently expressed reporter constructs containing enhancer fragments together with the NICD expression vector. To lower the background activity of the enhancer, we used cells in which T-cell-specific regulatory elements are normally inactive, such as the human embryonic kidney cell line 293. As shown in Figure 2A, the mouse pTa enhancer placed upstream of its cognate promoter or of a heterologous TATA box was weakly but reproducibly induced by NICD. Moreover, the human pTa enhancer was strongly activated by NICD, consistent with the presence of an additional CSL-binding site. No induction was observed with other well-characterized T-cell-specific regulatory elements such as the TCRβ and TCRδ enhancers and the lck proximal promoter, whereas a short Hes1 promoter fragment containing a critical CSL-binding site was strongly induced. Importantly, mutations of the single CSL-binding site in the mouse pTa enhancer or of both CSL-binding sites in the human pTa enhancer completely abolished their induction by NICD. Such specific induction of the pTa enhancer by NICD was observed also in NIH3T3 fibroblasts and in a pTa-negative mature T cell line BW5147 (data not shown). Induction of both the Hes1 promoter and the pTa enhancer was similarly reduced in the absence of the RAM domain of NICD, which mediates the interaction with CSL. Furthermore, the induction of the pTa enhancer by NICD was inhibited by a dominant-negative form of CSL that is unable to bind DNA (data not shown). Altogether, these results show specific induction of the pTa enhancer activity by Notch signaling, which is dependent on the interaction of CSL with its binding sites in the enhancer.
Figure 2.
Induction of the pTa enhancer by NICD. Cell line 293 was transfected with reporter constructs together with the NICD expression vector. Normalized reporter activities in the presence of NICD or of an empty expression vector were determined, and their ratio is presented as fold induction by NICD. The results represent the mean + SD of three independent experiments. Reporter constructs contained the indicated enhancer fragments upstream of the mouse pTa promoter (pTa) or of the SV40 promoter TATA box (TATA). The mouse (mpTa) or human (hpTa) pTa enhancers were either wild-type or harbored the mutations shown in Figure 1A (mpTa mut., hpTa mut.).
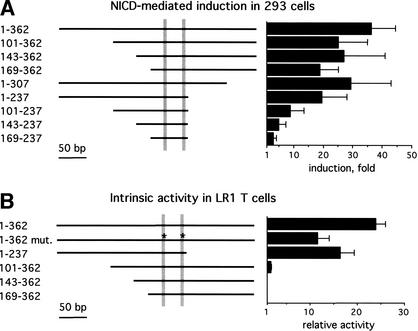
Because of its robust induction by NICD, in subsequent experiments we concentrated on the human pTa enhancer. To further explore the mechanism of Notch-mediated pTa enhancer induction, we tested a panel of truncated enhancer fragments. Figure 3A shows that, in contrast to the full-length enhancer, a short enhancer fragment containing both CSL-binding sites was not activated by Notch. Deletions at either the 5′ or 3′ end only marginally affected the induction. However, 5′ deletions of a fragment lacking the 3′ flanking region gradually decreased the induction to background levels. These data suggest that both the CSL sites and the flanking enhancer regions are required for the induction by Notch, with the 5′ and 3′ regions facilitating the induction in a redundant manner.
Figure 3.
Notch-mediated induction and intrinsic activity of the pTa enhancer fragments. Cells were transfected with reporter constructs containing the indicated human pTa enhancer fragments upstream of the TATA box. The CSL-binding sites (positions 188–95 and 224–31) are shown as shaded bars, and asterisks indicate mutations in these sites as shown in Figure 1A. The results represent the mean + SD of three independent experiments. (A) The induction of the pTa enhancer fragments by NICD in 293 cells was determined as in Figure 2. (B) The relative activity of the pTa enhancer fragments in an immature T cell line, LR1, was determined as the ratio of reporter activity to the activity of an enhancerless reporter vector.
Next, we attempted to define the regions of the pTa enhancer that mediate its Notch responsiveness and its specific activity in immature T cells. Figure 3B illustrates the intrinsic activity of pTa enhancer fragments in a pTa-positive immature T cell line, LR1. The mutations in CSL-binding sites, which drastically reduced the induction by Notch (Fig. 2A), caused only a moderate reduction of the enhancer activity in immature T cells. Conversely, 5′ deletions completely abolished the intrinsic enhancer activity, despite their marginal effect on the induction by Notch (Fig. 3A). This is consistent with the presence of a c-Myb-binding site and additional unidentified sites in the 5′ enhancer region. These data confirm that the enhancer integrates signals from T-cell-specific transcription factors and from the Notch/CSL pathway via separate binding sites.
The CSL-binding sites are required for the onset of pTa expression during T cell commitment
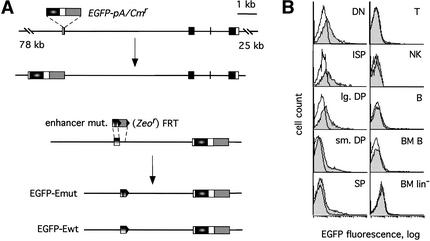
To test the function of CSL-binding sites in the regulation of pTa expression in vivo, we mutated these sites in the context of an entire pTa locus. To this end, we introduced the EGFP reporter into the human pTa gene within a 113-kb genomic BAC clone, and then mutated both CSL-binding sites in the pTa enhancer (Fig. 4A). The resulting BAC clones containing either wild-type or mutated enhancers (constructs EGFP–Ewt and EGFP–Emut, respectively) were used to generate transgenic reporter mice. Multiple transgenic lines of each type were established and analyzed for the expression of EGFP in lymphocytes by flow cytometry.
Figure 4.
Generation of transgenic reporter mice to examine the function of CSL sites in vivo. (A) Strategy for modification of the human pTa locus by homologous recombination in bacteria. The coding regions, UTR, and the upstream enhancer of pTa are shown as black, white, and vertical gradient boxes, respectively. Prokaryotic selection markers (Cmr and Zeor) are indicated by gray boxes. The EGFP gene (elliptical gradient box) and polyA signal (pA, white box) were introduced into the first pTa exon within the genomic BAC clone. Subsequently, mutations of both CSL sites (vertical bars) were introduced into the pTa enhancer. After the recombination and Flp-mediated excision, an Alu repeat 3′ to the enhancer was replaced by a single FRT site (black triangle). The resulting constructs contained either the wild-type (EGFP–Ewt) or a mutated (EGFP–Emut) enhancer sequence. (B) Expression of the modified pTa locus in transgenic mice. Shown are histograms of EGFP fluorescence in the indicated lymphocyte subsets of an EGFP–Ewt transgenic mouse (shaded traces) and a control nontransgenic mouse (empty traces). (Left panel) Thymocyte subsets were defined as follows: DN (CD3−CD4−CD8−), ISP (CD3−CD4−CD8+), large DP (large CD3−CD4+CD8+), small DP (small CD3−/lowCD4+CD8+) and SP (CD3highCD4+CD8− and CD3highCD4−CD8+). (Right panel) Other lymphocyte subsets included splenic T cells (CD3high), NK cells (NK1.1+), splenic B cells (B220+CD3−), bone marrow (BM) B cells (lin+B220+), and bone marrow lineage marker-negative cells (lin−B220−).
The expression of EGFP was generally low and heterogeneous, with only a fraction of cells within each population exhibiting green fluorescence (Figs. 4B, 5). Such weak expression may result from the lower intrinsic activity of the human pTa enhancer (Reizis and Leder 2001); in addition, technical issues arising from our BAC modification scheme cannot be excluded. Nevertheless, the EGFP signal was clearly detected in the majority of transgenic lines (7 out of 9 for the EGFP–Ewt and 4 out of 5 for the EGFP–Emut constructs), and it invariably followed the pattern illustrated in Figure 4B. EGFP was expressed at the highest level in the earliest double-negative (DN, CD4−CD8−) thymocyte precursors, and then decreased in subsequent immature single-positive (ISP, CD4−CD8+), double positive (DP, CD4+CD8+), and mature single-positive (SP, CD4+CD8− or CD4−CD8+) stages of thymocyte development. Peripheral lymphocytes including T, B, and natural killer (NK) cells, as well as immature lineage-negative cells and developing B cells in the bone marrow, were EGFP-negative. This expression pattern is consistent with that of the mouse pTa BAC transgene (Reizis and Leder 2001) and of the pTa gene itself (von Boehmer and Fehling 1997), confirming the proper regulation of the human pTa transgenes. Both the EGFP–Ewt and the EGFP–Emut constructs were expressed in the same manner, revealing no difference in the lineage specificity or in the down-regulation of transgene expression (data not shown).
Figure 5.
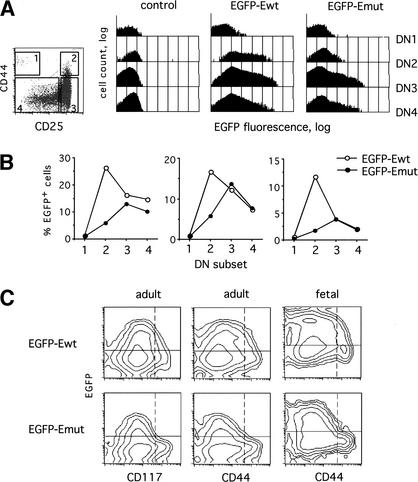
EGFP reporter expression in the earliest thymocyte subpopulations. (A) Thymocytes were stained with a cocktail of mAb to lineage markers, and the lineage-negative thymocytes (1%–2% of the total) were gated on the basis of CD44 and CD25 expression as shown on the density plot at the left. Shown are the histograms of the EGFP fluorescence in the gated DN thymocyte subsets (DN1–DN4) from a control nontransgenic mouse or from representative transgenic mice. The logarithmic scale was used on the Y axis to facilitate comparison between the different subsets. (B) Mice from three independent transgenic lines of each type were stained as above, and the percentage of EGFP-positive cells was determined in each DN subset. Control wild-type thymocytes contained <0.5% positive cells in each subset. The representative staining results are shown as pairwise comparisons between the EGFP–Ewt (open circles) and EGFP–Emut (filled circles) lines with similar levels of overall EGFP expression. (C) Adult or 16.5 days postcoitum (dpc) fetal thymocytes from the same transgenic lines as in A were stained with mAb to the lineage markers, CD25 and either CD44 or CD117. Shown are contour plots of EGFP versus CD117 or CD44 fluorescence in lineage-negative CD25+ cells encompassing subsets DN2 and DN3. Horizontal lines show the baseline EGFP fluorescence in nontransgenic control mice, and vertical dashed lines define the CD44/CD117high (DN2) subset.
Next, we compared the expression of EGFP–Ewt and EGFP–Emut constructs in DN thymocytes, which include the earliest T cell precursors undergoing T cell commitment, αβ/γδ cell fate choice, and pre-TCR signaling. The DN population can be subdivided into at least four subsets based on the expression of cell surface markers CD25 and CD44, as well as of the CD117 (c-kit) receptor (Shortman and Wu 1996). The DN1 (CD25−CD44+CD117+) subset includes the earliest pluripotent thymic precursors, which up-regulate CD25 and become committed to T cell lineage at the DN2 (CD25+CD44+CD117+) stage. These cells subsequently down-regulate both CD44 and CD117 to become DN3 (CD25+CD44−CD117low) cells undergoing TCR rearrangement and pre-TCR signaling. The latter event induces the loss of CD25 and rapid proliferation to the DN4 (CD25−CD44−CD117−) and then to the ISP and DP stages. The major up-regulation of pTa expression occurs at the DN2 stage, coinciding with T cell commitment (von Boehmer and Fehling 1997; Rothenberg 2000).
Figure 5A shows the expression of EGFP in the DN thymocyte subsets from representative EGFP–Ewt and EGFP–Emut transgenic lines. As expected, the EGFP–Ewt transgene was scarcely detectable in DN1 cells and was strongly induced in the DN2 subset. However, the EGFP–Emut transgene was only marginally up-regulated in DN2 cells, instead reaching its peak of expression at the DN3 stage. These expression patterns are illustrated in Figure 5B, which shows pairwise comparison of EGFP–Ewt and EGFP–Emut transgenic lines manifesting similar overall EGFP levels. Indeed, each construct exhibited the same expression profile in multiple lines, independent of the level of expression. The delayed onset of the EGFP–Emut transgene expression is further evident in the gated CD25+ DN cells that include DN2 and DN3 subsets (Fig. 5C). When plotted against CD44 or CD117, EGFP was distributed uniformly within the CD25+ population from the EGFP–Ewt transgenic mice (convex plots). In contrast, EGFP was detected only in the more mature CD44low or CD117lowCD25+ cells from the EGFP–Emut transgenic mice (concave plots). These differences in EGFP expression were observed in both adult and fetal transgenic DN thymocytes. Altogether, these data suggest that the CSL-binding sites are required for the stage-specific induction of pTa expression upon T cell commitment.
Discussion
Our results indicate that Notch signaling may induce its T-cell-specific target gene pTa through the CSL-binding sites in the pTa enhancer. Regulatory elements of several tissue-specific mammalian genes have been shown to contain CSL-binding sites (Shirakata et al. 1996; Chen et al. 1997; Kannabiran et al. 1997; Lam and Bresnick 1998; Oswald et al. 1998), and in some cases to be induced by NICD (Chen et al. 1997; Oswald et al. 1998). However, it is not clear whether these genes represent authentic targets of Notch signaling, and the role of the CSL sites in vivo has not been studied. In contrast, pTa is up-regulated by NICD in cell lines and in transgenic thymocytes (Deftos et al. 2000). Furthermore, we show that the CSL-binding sites in the pTa enhancer are required for its induction by NICD in vitro and for stage-specific pTa expression in vivo. Thus, pTa appears to represent a model tissue-specific mammalian Notch target that can be directly induced by NICD/CSL binding to its distal regulatory element.
A Notch-responsive enhancer may be regulated through distinct binding sites by the Notch/CSL pathway as well as by other signaling pathways and transcription factors (Flores et al. 2000). Similarly, we could separate the regions of the pTa enhancer mediating its intrinsic activity in immature T cells and its Notch responsiveness. In addition, we show that the flanking regions of the enhancer may facilitate the activation by NICD. This effect is unlikely to involve any specific sites cooperating with the CSL sites, because both 5′ and 3′ flanking fragments of different sizes were effective. Again, no correlation with the intrinsic enhancer activity could be observed: for example, the 3′ flanking region facilitated the induction by NICD (cf. fragments 169–237 and 169–362 in Fig. 3A) yet was dispensable for the enhancer activity in T cells (Fig. 3B). It is possible that a distinct base composition of the flanking regions might render the enhancer more accessible to the NICD/CSL complex. Indeed, both the human and the mouse pTa enhancers are 65% G/C-rich, and the 200-bp core of the human enhancer is 75% G/C-rich. In contrast, only a minimal 60-bp fragment of the Hes1 promoter containing the CSL-binding sites was efficiently activated by NICD in our experiments. These data highlight an important difference between a tissue-specific enhancer integrating multiple signals including Notch, and a specialized Notch-responsive element such as the Hes1 promoter.
Signaling from the Notch1 receptor appears essential for the specification of T cell lineage. In the mouse thymus, T cell commitment is thought to occur at the DN2 stage, characterized by acquisition of the CD25 marker and by up-regulation of pTa expression. Accordingly, the development of DN2 subset is abrogated in the absence of Notch1 (Radtke et al. 1999) or Hes1 (Tomita et al. 1999), and therefore is likely to result from Notch activity. Indeed, we found that Notch1 mRNA was expressed at the highest levels in the DN1 subset, whereas Hes1 was abundantly expressed in the DN1 and DN2 subsets and decreased afterward (B. Reizis, P. Leder, F. Gounari, and H. von Boehmer, unpubl.). In agreement with this, we find that mutation of the CSL-binding sites abolished induction of the pTa transgene at DN2, but had no obvious effects on its subsequent expression. These data highlight a pulse of Notch activity that occurs during T cell commitment and induces a T-cell-specific gene expression program. This induction is likely to involve multiple indirect mechanisms, including the activity of Hes1 (Tomita et al. 1999) and Deltex (Deftos et al. 1998), and the inhibition of bHLH proteins such as E2A (Pui et al. 1999). In addition, our data suggest that at least some T-cell-specific genes might be induced directly by the NICD/CSL transactivation complex.
The conservation of Notch inducibility between the mouse and human pTa enhancers implies a functional significance for this effect. It can be hypothesized that a timely Notch-mediated induction of pTa upon T cell commitment is required to provide optimal levels of the pTa protein for the assembly of the pre-TCR complex at the subsequent stage. In such a scenario, defective Notch signaling would delay the accumulation of pTa and the formation of pre-TCR, thereby impairing αβ T cell development. In contrast, γδ T cells do not use pre-TCR and would not be affected by differences in Notch signaling. Thus, Notch-induced pTa expression during T cell commitment might contribute to the selective promotion of αβ T cell development by activated Notch (Washburn et al. 1997).
Materials and methods
Electrophoretic mobility shift assay (EMSA)
A construct containing a FLAG-tagged human CSL (RBP3) cDNA downstream of the T7 promoter was translated in vitro using the TnT T7 system (Promega). Double-stranded oligonucleotide probes contained 8-bp CSL-binding sites shown in Figure 1A, or the CSL-binding site A from the Hes1 promoter (Jarriault et al. 1995), flanked by 7 bp of 5′ sequence and 5 bp of 3′ sequence. Where indicated, the same probes containing the corresponding mutations shown in Figure 1A were used. The 32P-labeled probes were incubated with translated CSL protein or with crude nuclear extract from BW5147 cells, and the resulting complexes were visualized by EMSA as described (Reizis and Leder 1999).
Transfection and reporter assays
The intracellular region of mouse Notch1 (amino acids 1751–2443; Deftos et al. 1998) containing an initiation codon was subcloned into a pCAGGS expression vector. Promoter fragments were subcloned into the β-galactosidase reporter vector pβGal-Basic (Clontech), and enhancer fragments were subcloned in the same vector containing an SV40 promoter-derived TATA box. The following fragments were used, with positions of the corresponding GenBank entries indicated: mouse pTa promoter (1–507 of U27268), mouse lck proximal promoter (841–1117 of M23191), mouse pTa enhancer (1370–1627 of AF132612), human pTa enhancer (39728–39367 of HS475N16) and deletions thereof, mouse TCRβ (494–913 of X07177) and TCRδ (241–568 of X53336) enhancers, and mouse Hes1 promoter fragment (103–165 of D16464). Enhancer mutations were generated by overlap extension PCR. Cells were transfected using Fugene 6 reagent (Roche Molecular Biochemicals), and the activities of β-galactosidase and luciferase in cell lysates were measured 24 h later using chemiluminescent assays. Cell line 293 was transfected with 0.75 μg of reporter vector, 0.25 μg of expression vector, and 0.1 μg of pGL3-Control luciferase expression vector, used for normalization. Cell line LR1 was transfected with 1 μg of reporter vector only.
Transgenic mice
A 113-kb BAC clone RP3–475N16 (GenBank accession no. HS475N16) containing the human pTa locus was modified using ET recombination as described (Muyrers et al. 1999). The initiation codon and the coding region of pTa exon I (positions 35334–35281) were replaced by a fragment containing EGFP (Clontech), BGH polyA signal, and a prokaryotic chloramphenicol-resistance cassette (Cmr). The targeting cassette for the second recombination step contained the pTa enhancer fragment with both CSL sites mutated as shown in Figure 1A, followed by a prokaryotic zeocin-resistance (Zeor) cassette flanked by FRT sites. Upon ET recombination, the Zeor cassette replaced an Alu repeat immediately 3′ to the enhancer (positions 39366–39051). Depending on whether the homologous recombination occurred 5′ or 3′ to the mutated CSL sites, the resulting clones contained either a wild-type or a mutated pTa enhancer (constructs EGFP–Ewt and EGFP–Emut, respectively). The Zeor cassette was removed using FLP-expressing bacterial strain 294-FLP, leaving a single FRT site 3′ to the enhancer. All targeting events were confirmed by PCR amplification and sequencing, and the integrity of the BAC clones was verified by conventional and pulsed-field gel electrophoresis.
The linearized BAC constructs were microinjected into fertilized oocytes of FVB mice, and transgenic offspring were bred with wild-type FVB mice. Hemizygous F1 mice were analyzed at 4–8 weeks of age unless indicated otherwise. Lymphoid cells were stained with direct mAb conjugates and analyzed using a FACSCalibur flow cytometer and CellQuest software (BD Pharmingen). Thymocytes were stained with mAb to CD3 (PE), CD8 (PerCP), and CD4 (APC). For the analysis of EGFP expression in DN thymocyte subsets, cells were stained with mAb to lineage markers CD3, CD4, CD8, B220 and Mac-1 (APC), CD44 or CD117 (PE), and CD25 (biotin), followed by streptavidin-PerCP. Splenocytes and lymph node cells were stained with mAb to CD3 or NK1.1 (PE) and B220 (APC). Bone marrow cells were stained with a PE lineage cocktail (CD3, Ter119, CD19, and Gr1) and B220 (APC).
Acknowledgments
We thank A. Harrington and K. Cozine for oocyte injections, E. Manet for the RBP3 plasmid, C. Bassing for TCR enhancer clones, M. Deftos and M. Bevan for the NICD cDNA and for helpful discussions, and J. Michaelson and R. Weiss for critical reading of the manuscript. B.R. was supported in part by a postdoctoral fellowship from the Cancer Research Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL leder@rascal.med.harvard.edu; FAX: (617) 432-7944.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.960702.
References
- Aifantis I, Azogui O, Feinberg J, Saint-Ruf C, Buer J, von Boehmer H. On the role of the pre-T cell receptor in αβ versus γδ T lineage commitment. Immunity. 1998;9:649–655. doi: 10.1016/s1074-7613(00)80662-7. [DOI] [PubMed] [Google Scholar]
- Anderson AC, Robey EA, Huang YH. Notch signaling in lymphocyte development. Curr Opin Genet Dev. 2001;11:554–560. doi: 10.1016/s0959-437x(00)00232-x. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bray S, Furriols M. Notch pathway: Making sense of Suppressor of Hairless. Curr Biol. 2001;11:R217–R221. doi: 10.1016/s0960-9822(01)00109-9. [DOI] [PubMed] [Google Scholar]
- Chen Y, Fischer WH, Gill GN. Regulation of the ERBB-2 promoter by RBPJκ and NOTCH. J Biol Chem. 1997;272:14110–14114. doi: 10.1074/jbc.272.22.14110. [DOI] [PubMed] [Google Scholar]
- Deftos ML, He YW, Ojala EW, Bevan MJ. Correlating Notch signaling with thymocyte maturation. Immunity. 1998;9:777–786. doi: 10.1016/s1074-7613(00)80643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor α gene in development of α β but not γ δ T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Jaleco AC, Neves H, Hooijberg E, Gameiro P, Clode N, Haury M, Henrique D, Parreira L. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J Exp Med. 2001;194:991–1002. doi: 10.1084/jem.194.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kannabiran C, Zeng X, Vales LD. The mammalian transcriptional repressor RBP (CBF1) regulates interleukin-6 gene expression. Mol Cell Biol. 1997;17:1–9. doi: 10.1128/mcb.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT, Bresnick EH. Identity of the β-globin locus control region binding protein HS2NF5 as the mammalian homolog of the Notch-regulated transcription factor Suppressor of Hairless. J Biol Chem. 1998;273:24223–24231. doi: 10.1074/jbc.273.37.24223. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Kopan R. Notch signaling: From the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Muyrers JPP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucl Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Liptay S, Adler G, Schmid RM. NF-κB2 is a putative target gene of activated Notch-1 via RBP-Jκ. Mol Cell Biol. 1998;18:2077–2088. doi: 10.1128/mcb.18.4.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Reizis B, Leder P. Expression of the mouse pre-T cell receptor α gene is controlled by an upstream region containing a transcriptional enhancer. J Exp Med. 1999;189:1669–1678. doi: 10.1084/jem.189.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The upstream enhancer is necessary and sufficient for the expression of the pre-T cell receptor α gene in immature T lymphocytes. J Exp Med. 2001;194:979–990. doi: 10.1084/jem.194.7.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Res P, Spits H. Developmental stages in the human thymus. Semin Immunol. 1999;11:39–46. doi: 10.1006/smim.1998.0152. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. Stepwise specification of lymphocyte developmental lineages. Curr Opin Genet Dev. 2000;10:370–379. doi: 10.1016/s0959-437x(00)00098-8. [DOI] [PubMed] [Google Scholar]
- Shirakata Y, Shuman JD, Coligan JR. Purification of a novel MHC class I element binding activity from thymus nuclear extracts reveals that thymic RBP-Jκ/CBF1 binds to NF-κB-like elements. J Immunol. 1996;156:4672–4679. [PubMed] [Google Scholar]
- Shortman K, Wu L. Early T lymphocyte progenitors. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- Tomita K, Hattori M, Nakamura E, Nakanishi S, Minato N, Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes & Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J κ. Nucl Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H. Coming to grips with Notch. J Exp Med. 2001;194:F43–F46. doi: 10.1084/jem.194.7.f43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H, Fehling HJ. Structure and function of the pre-T cell receptor. Ann Rev Immunol. 1997;15:433–452. doi: 10.1146/annurev.immunol.15.1.433. [DOI] [PubMed] [Google Scholar]
- Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, Robey E. Notch activity influences the αβ versus γδ T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]