Abstract
The JAK/STAT signal transduction pathway controls numerous events in Drosophila melanogaster development. Receptors for the pathway have yet to be identified. Here we have identified a Drosophila gene that shows embryonic mutant phenotypes identical to those in the hopscotch (hop)/JAK kinase and marelle (mrl)/Stat92e mutations. We named this gene master of marelle (mom). Genetic analyses place mom's function between upd (the ligand) and hop. We further show that cultured cells transfected with the mom gene bind UPD and activate the HOP/STAT92E signal transduction pathway. mom encodes a protein distantly related to the mammalian cytokine receptor family. These data show that mom functions as a receptor of the Drosophila JAK/STAT signal transduction pathway.
Keywords: Drosophila, JAK/STAT, signal transduction, cytokine receptor
The JAK/STAT signal transduction pathway was identified through studies of the transcriptional activation response to a variety of cytokines and growth factors (Decker 1999; Levy 1999; Mui 1999; Yeh and Pellegrini 1999; Chatterjee-Kishore et al. 2000; Imada and Leonard 2000). Cytokines, including interferons (IFNs) and interleukins (ILs), are a broad group of mostly soluble factors that exert diverse effects on several biological processes such as immunity, hematopoiesis, and inflammation, as well as neural and embryonic development (Smith et al. 1992; Blalock 1994; Williams 2000).
A Drosophila melanogaster JAK/STAT signal transduction pathway has been identified (Hou and Perrimon 1997; Dearolf 1999; Zeidler et al. 2000). The Drosophila JAK is encoded by the hopscotch (hop) gene (Binari and Perrimon 1994), and the Drosophila STAT is encoded by the marelle/stat92E gene (marelle is the French word for hopscotch; Hou et al. 1996; Yan et al. 1996). An unpaired (upd) secreted glycoprotein identifies an in vivo ligand activating the HOP/STAT92E pathway (Harrison et al. 1998). Mutations in all three genes were first identified by virtue of their distinctive embryonic phenotypes. When the function of any one of the three genes is removed from the embryo, a partial loss of expression of several pair-rule genes occurs; and consequently, some abdominal segments are lost or fused together. In addition to its role in embryonic segmentation, the HOP/STAT92E pathway is also involved in several other developmental events, including sex determination, polarity determination in the eye, and imaginal tissue formation (Hou and Perrimon 1997; Dearolf 1999; Jinks et al. 2000; Sefton et al. 2000; Zeidler and Perrimon 2000; Zeidler et al. 2000). Hyperactivation of the HOP/STAT92E pathway results in melanonic or leukemia-like tumor formation in larvaes and adult flies (Dearolf 1998). Yet the receptor for the HOP/STAT92E pathway is still missing.
In this study we have identified a novel Drosophila gene encoding a protein distantly related to the mammalian cytokine receptor family. Our genetic analysis and results from cultured cells show that this gene functions as a receptor for the HOP/STAT92E pathway.
Results
Mutations in the mom loci suppress the eye phenotype of overexpression of upd
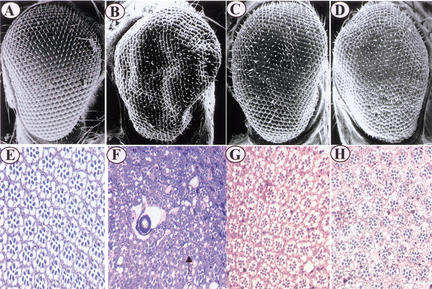
Our recent results show that overexpression of upd using a UAS–upd and GMR–Gal4 driver (Freeman 1996) causes compound eye dramatic overgrowth in the adult eye because of an increase in the number of ommatidia (Fig. 1, cf. B with A). The average number of ommatidia in the compound eye of UAS–upd/GMR–Gal4 female flies is 978 ± 10 compared with 745 ± 7 in wild-type flies. Histological sections through the overgrown eyes revealed that most ommatidia have normal photoreceptor cells and regular cell size (Fig. 1, cf. F with E), indicating that UPD activity mainly regulates cell proliferation in the compound eye. However, the ommatidia look more crowded and have irregular space and arrangement, and several big vacuoles are integrated into the ommatidia lattice (Fig. 1F). The severity of eye morphology appears proportional to the strength of the HOP/STAT92E-mediated signaling, because removing one copy of hop partially suppresses the big eye phenotype (Fig. 1C,G; the average number of ommatidia is 854 ± 9). We took advantage of this sensitized system to conduct a screen for mutations that reduce (suppressors) or increase (enhancers) the degree of eye size. We reasoned that a twofold reduction in the dose of a gene (by mutating one of its two copies) that functions downstream of upd should dominantly alter signaling strength, which, in turn, should visibly modify the eye size. Based on this assumption, we first screened available X-chromosome P-element insertion mutations and identified one complementation group of suppressors with four alleles at the cytological location 18E (Fig. 1D,H; the average number of ommatidia is 813 ± 10). Based on its presumed role in the HOP/STAT92E signal transduction pathway described below, we have named this novel gene master of marelle (mom). The relative strength of four mom alleles in suppressing the UAS–upd/GMR–Gal4 fly big eye phenotype is mom1 > mom2 > mom3 = mom4, and mom1 is the strongest allele.
Figure 1.
mom suppresses UAS–upd/GMR–Gal4 eye phenotype. (A–D) Scanning electron micrographs (SEMs) of adult eyes of the following genotypes: (A) wild type (w1118), (B) UAS–upd/GMR–Gal4, (C) hopC111/+; UAS–upd/GMR–Gal4, (D) mom1/+; UAS–upd/GMR–Gal4. Note that the UAS–upd/GMR–Gal4 deformed eye phenotype is partially suppressed by reducing hop or mom activity by half (cf. C and D with B). (E–H) Tangential sections of eyes of (E) wild-type, (F) UAS–upd/GMR–Gal4, (G) hopC111/+; UAS–upd/GMR–Gal4, (H) mom1/+; UAS–upd/GMR–Gal4. Note that most ommatidia in the deformed eye of UAS–upd/GMR–Gal4 flies have normal photoreceptor cells and regular cell size (cf. F with E), and a few ommatidia have one extra R7 photoreceptor (arrow). However, the ommatidia look more crowded and have irregular space and arrangement; several big vacuoles are integrated into the ommatidia lattice in F. The UAS–upd/GMR–Gal4 eye phenotype is significantly suppressed by hop/+ or mom/+ (cf. G and H with F).
The maternal effect phenotypes of mom, hop, and stat92E are identical
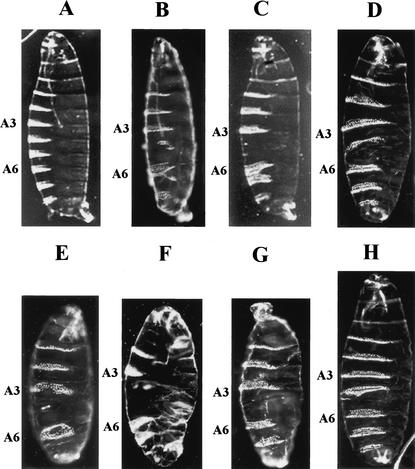
mom is required for zygotic viability because mutant animals homozygous for the mom allele die as early larvae. The dead larvae showed a posterior spiracle defect (see Fig. 3B, below). In addition to its zygotic function, mom is required maternally for normal embryonic segmentation because mom embryos derived from females lacking germ-line mom1 activity die with segmentation defects that resemble the phenotype of hop and stat92E embryos (Fig. 2; Perrimon and Mahowald 1986; Hou et al. 1996). As is the case with hop and stat92E embryos, the severity of the defects observed in mom embryos is dependent on the paternal contribution. Both paternally rescued and unrescued mom embryos show a consistent deletion of the fifth abdominal segment and the posterior midventral portion of the fourth abdominal segment (Fig. 2D,G). Additional defects in the thoracic segments and the head and tail regions are observed in unrescued mom embryos (Fig. 2G). The maternal phenotypes associated with the mom1 mutation are identical to those observed with complete loss of hop and stat92E gene activities (Fig. 2B,C,E,F; Perrimon and Mahowald 1986; Hou et al. 1996).
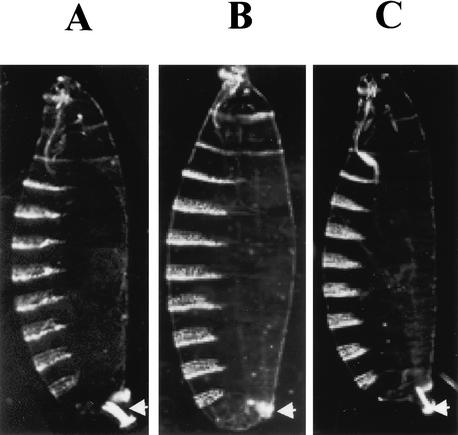
Figure 3.
hopTum-l suppresses the mom posterior spiracle phenotype. Dark-field cuticle pattern of (A) wild-type embryo, (B) mom1/Y embryo, and (C) hopTum-l mom1/Y embryo. Arrows point to the posterior spiracle structures. A gain-of-function mutation of hop (hopTum-l) rescues the mom1 posterior spiracle defect. mom1 mutant embryos show the strongest posterior spiracle defect; mom2 mutant embryos have mild posterior spiracle defect, and mom3 and mom4 mutant embryos have wild-type posterior spiracles.
Figure 2.
Abdominal defects associated with mom, stat92E, and hop embryos. (A) Dark-field micrograph of a wild-type embryo showing eight abdominal dentical belts. Paternally rescued (B) hop, (C) stat92E, and (D) mom GLC embryos are missing abdominal segment 5 and part of segment 4. (E) hop, (F) stat92E, and (G) mom null embryos have additional defects in the thoracic segments and the head and tail regions. (H) UAS–mom-N/matα4–GAL–VP16-Gal4 embryo. Expression of the dominant-negative form of mom generates the similar mutant phenotype to mom (cf. H with G). A3 and A6 indicate the respective position of the abdominal 3 and 6 segments.
mom is epistatic to hop
We further analyzed whether mom operates upstream or downstream of hop by testing genetic interaction between the mom loss-of-function mutation and a hyperactive hop allele. Hemizygous mom1 zygotic mutant larvaes show defects of the posterior spiracle (Fig. 3B), an organ that is connected to the tracheal system and used by the young larvae for gas exchange (Hu and Castelli-Gair 1999). If mom is required to transduce the HOP signal, then a hop gain-of-function mutation should have no effect on the posterior spiracle phenotype of the mom mutation. We used the dominant temperature-sensitive hop allele hopTum-l for this experiment. When grown above 25°C, flies heterozygous for hopTum-l have increased tyrosine kinase activity (Harrison et al. 1995). Hemizygous mom1 and hopTum-l mutant larvae forms wild-type posterior spiracles (Fig. 3C), suggesting that the hop gain-of-function mutation rescues the mom1 mutant phenotype. This result further suggests that mom is a member of the HOP/STAT92E signal transduction pathway and functions upstream of the HOP tyrosine kinase.
UPD, MOM, and HOP regulate STAT92E protein expression
In the mammalian cell culture system, the JAK/STAT pathway becomes activated when a ligand binds to its receptor, inducing tyrosine phosphorylation of the monomeric STAT molecule. Tyrosine phosphorylation causes the STAT protein to dimerize with another STAT molecule via reciprocal SH2 domain–phophotyrosine interactions, and the dimer translocates to the nucleus (Decker and Kovarik 1999).
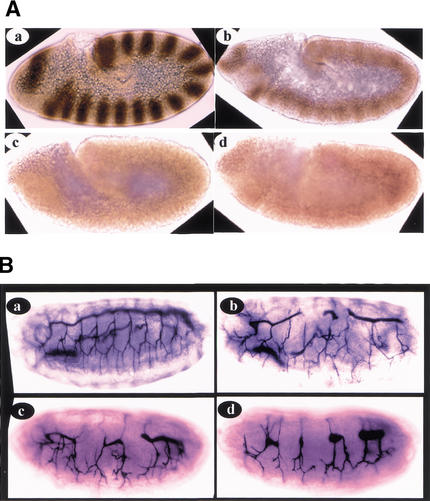
To explore the function of mom in activating the HOP/STAT92E signal transduction pathway, we compared protein levels and distributions of STAT92E in wild-type embryos and mutant embryos of upd, mom, and hop genes. We stained embryos using affinity-purified anti-STAT92E antibodies (Fig. 4A). Although strong STAT92E expressions are detected as 15 clear stripes during stage 9 in the wild-type embryo, STAT92E expression is dramatically reduced in upd, mom, and hop mutant embryos (Fig. 4A). As in wild-type embryos, the remaining STAT92E protein in mutant embryos is localized in both the nucleus and cytoplasm (data not shown). These data suggest that MOM and the UPD/MOM/HOP signaling pathway regulate STAT92E protein expression.
Figure 4.
(A) upd, mom, and hop regulate STAT92E protein expression. Stage 9 wild-type, updYC43/Y, mom1, and hopC111 null embryos are stained with the affinity-purified anti-STAT92E antibodies. All embryos are oriented anterior left and dorsal up. (a) Whereas strong STAT92E expressions (brown) are detected as 15 stripes in the stage 9 wild-type embryo, STAT92E expression is dramatically reduced in (b) upd, (c) mom, and (d) hop mutant embryos. (B) mom and other components of the HOP/STAT92E pathway regulate tracheal formation. (b) In paternal rescued hopC111 embryos, a defective tracheal system was formed, which generally has several disruptions in the main trunk and several branches. (a) Tracheal structure in a wild-type embryo revealed by mAb2A12 staining. (c) mAb2A12 stainings of paternal rescued stat92E6346 and (d) mom1 embryos revealed tracheal defects similar to that of hopC111 embryos (cf. c and d with b). These data suggest that MOM and the HOP/STAT92E signal transduction pathway play an indispensable role in tracheal formation.
MOM and other components of the HOP/STAT92E pathway regulate tracheal formation
The posterior spiracle defects of the mom mutation lead us to further examine functions of the HOP/STAT92E pathway in tracheal formation.
The tracheal forms from 10 tracheal pits, 1 per hemisegment. The trachealess gene selects the tracheal primordia in the embryonic ectoderm and drives the conversion of these planar epithelial regions into tracheal pits (Metzger and Krasnow 1999). The tracheal pits then sprout successively finer branches and fuse together, forming the tracheal network. The trachea is further connected to the posterior spiracle, forming a functional tracheal system (Hu and Castelli-Gair 1999; Metzger and Krasnow 1999). We examined tracheal formation in mom, hop, and stat92E mutants by using an enhancer trap line in the trachealess gene (1-eve-1; Perrimon et al. 1991) and an antibody [(mAb)2A12] that stains tracheal branches and trunks (Sutherland et al. 1996). In hop null embryos, trachealess expressions are completely abolished and tracheal formation is completely blocked (data not shown). In paternal rescued embryos, a defective tracheal system forms (Fig. 4B, cf. b with a), generally with several disruptions in the main trunk and several branches (Fig. 4Bb). Because all of our mom and stat92E mutants are enhancer trap lines, we cannot directly look at trachealess gene expression by using the 1-eve-1 enhancer trap line. However, in the paternal rescued stat92E and mom embryos, similar to the hop embryos, a defective tracheal system formed (Fig. 4B, cf. c and d with a), generally with several disruptions in the main trunk and several branches (Fig. 4Bc,d). These data suggest that MOM and the HOP/STAT92E signal transduction pathway play an indispensable role in tracheal formation.
Molecular cloning of the mom gene
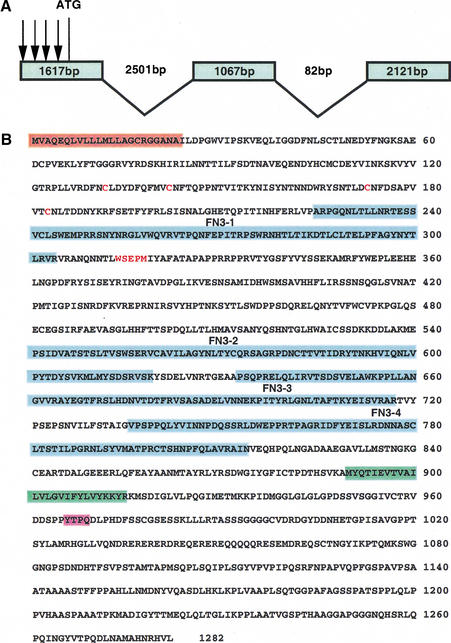
To identify the gene product encoded by mom, we cloned the genomic sequences flanking the P-element insertion associated with mom1, following plasmid rescue. Subfragments of the rescued genomic DNA were then used to screen various cDNAs and search Drosophila genomic databases (for details, see Materials and Methods). The largest cDNA recovered was a full-length 4.8-kb clone, which encodes a conceptual protein of 1282 amino acids (Fig. 5B). The sequence context of the putative initiation codon is in good agreement with the consensus sequence for translation initiation.
Figure 5.
(A) Intron/exon structure of the mom gene. Comparison of a 4.8-kb mom cDNA (flybase gene number CG14226) and genomic DNA from the locus reveals that the mom transcript is made up of 3 exons. The first, a 1617-bp exon, is separated from the second, 1067-bp, exon by 2501 bp. The third, 2121-bp, exon is separated from the second exon by 82 bp. In the four mom alleles, all the P elements (arrows) are inserted in the 5′ untranslated region of the cDNA. (B) Deduced amino acid sequence of the MOM protein. The conserved domains are highlighted using different colors. The probable signal peptide is shaded with orange; the potential transmembrane domain is shaded with green; the four fibronectin type III (FN3) repeats are shaded with blue; the consensus motifs for STAT binding are shaded with pink; and the cysteine residues and WSEPM sequence conserved in the cytokine receptor family are shown as red letters.
The following lines of evidence support the argument that the cDNA we isolated (Fig. 5B) corresponds to mom function. First, high and uniformly distributed levels of maternal mom RNA are detected during the syncytial blastoderm stage, a pattern very similar to the one observed for hop and stat92E transcripts (data not shown). Second, in the four mom alleles, all the P elements are inserted in the 5′ untranslated region of the cDNA (Fig. 5A). Third, expression of the cDNA under the control of a heat-shock promoter in transgenic flies fully rescued the lethality of mom mutant flies. Finally, expression of a dominant-negative form of the cDNA generated the same phenotype as mom mutations (Fig. 2H).
mom encodes a protein distantly related to the mammalian cytokine receptor family
The amino acid sequence analysis identified two hydrophobic segments: the first one near the N terminus (Fig. 5B, shaded with orange) is a probable signal peptide; the second one in the middle (Fig. 5B, shaded with green) may identify the transmembrane domain. In the extracellular domain, the most remarkable feature is the presence of four fibronectin type III (FN3) repeats of ∼90 amino acids each (Fig. 5B, shaded with blue). Fibronectin type III repeats are typically 90–100 amino acids long with the location of aromatic amino acids and nearby hydrophobic residues being conserved (Norton et al. 1990; Patthy 1990). Many cytokine receptors have FN3 repeats in their extracellular domains (Taga and Kishimoto 1997).
Computer-assisted homology searches in the GenBank and EMBL databases identified several receptor-linked protein tyrosine phosphotases (PTPs; Streuli et al. 1990) and cytokine receptors (Taga and Kishimoto 1997). However, the homology between MOM and PTPs is only limited to the FN3 repeats; MOM does not have the protein tyrosine phosphatase (PTPase) domains. MOM is distantly related to the cytokine receptor family, particularly the gp130-subfamily proteins (Taga and Kishimoto 1997). All these receptors contain, in their extracellular region, a domain of ∼200 amino acid residues, which is conserved in the family. This domain is characterized by four positionally conserved cysteine residues in its N-terminal half and a WSXWS motif in the C-terminal end. It is suggested that this domain consists of seven β-strands positioned antiparallel to form a barrel-like shape so that a trough formed between two barrel-like modules functions as a ligand-binding pocket. The predicted primary sequence of MOM includes a cytokine receptor family domain containing four conserved cysteine residues (red letters in Fig. 5B) and a WSEPM (red letters in Fig. 5B) sequence that resembles the WSXWS motif in the cytokine receptor domain of the gp130-subfamily receptors, followed by four fibronectin type III (FN3) repeats. The MOM cytoplasmic domain is more diversified from the gp130-subfamily receptors. The potential box 1 and 2 motifs are less visible in the MOM sequence. Nevertheless, MOM has one tyrosine residue (Y966) fitting a YXXQ consensus motif (shaded with pink in Fig. 5B) for STAT binding in the gp130 protein and may bind STAT92E protein.
In conclusion, MOM and the gp130-subfamily receptors have certain homology in domain structures rather than in primary amino acid sequence. MOM may be a distantly related member of the gp130-subfamily receptors.
MOM binds UPD through its extracellular domain
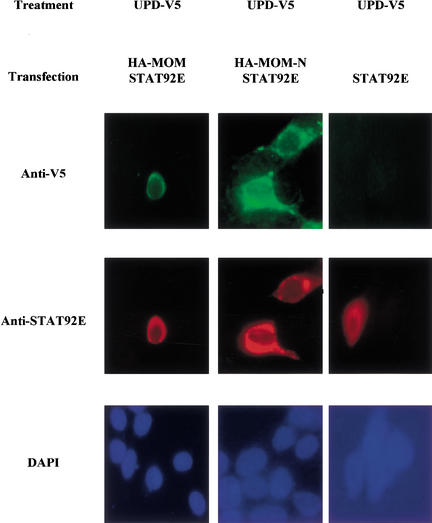
The results described above suggest that MOM is a receptor for UPD. To further study the interaction between these molecules without specific antibodies, we subcloned their cDNAs in epitope-tagged mammalian expression vectors. Then we cotransfected 293T cells with V5-tagged UPD and HA-tagged MOM and detected the expression by immunofluorescence and Western blot with specific mouse monoclonal anti-tag antibodies (data not shown). To examine the direct binding of UPD to MOM, we transfected 293T cells with UPD–V5. The ligand was released to the medium by treating the cells with heparin as described in Materials and Methods. Subsequently, the concentrated conditioned medium was applied to 293T cells nontransfected and transfected with HA–MOM and with a truncated form containing the N-terminal domain, MOM-N. Of note, given that we do not have specific antibodies for MOM and both anti-HA and anti-V5 antibodies have the same animal origin, prohibiting double staining, we used an indirect approach to evidence their presences in transfected cells. As it is known that cells transfected with two DNAs will incorporate both at the same time, we transfected 293T cells with HA–MOM along with STAT92E. Therefore, cells stained with rabbit anti-STAT antibody should be those also expressing MOM. As expected, we detected V5 staining in cells containing STAT92E and transfected with MOM or MOM-N (Fig. 6, left and middle panels), whereas we did not observed the presence of the ligand in control cells transfected only with STAT92E (Fig. 6, right panel). These data show that UPD can be detected in 293T cells only when MOM is present, which indicates a physical interaction between these two molecules.
Figure 6.
UPD protein binds MOM. Transfected 293T cells were incubated with concentrated conditioned medium from 293T cells, producing UPD–V5. Following incubation with conditioned medium, the cells were washed, fixed in paraformaldehyde/PBS, and incubated with mouse anti-V5 and rabbit anti-STAT92E antibodies. Only 293T cells expressing STAT92E/HA–MOM and STAT92E/HA–MOM-N show strong staining when incubated with UPD and anti-V5 antibody (left and middle panels). Cells transfected with HA–STAT92E alone show a background staining when incubated with UPD-containing conditioned medium (right panel). These results suggest that UPD specifically binds to 293T cells expressing MOM.
Activated MOM binds STAT92E
The gp130-subfamily of receptors has no intrinsic tyrosine kinase domain, but is constitutively associated with tyrosine kinase JAKs. A tyrosine residue fitting a YXXQ consensus motif at the C terminus of the receptors provides a binding site for STAT (Hirano et al. 1997). A C-terminal YXXQ sequence was found in MOM, suggesting that MOM may bind STAT92E.
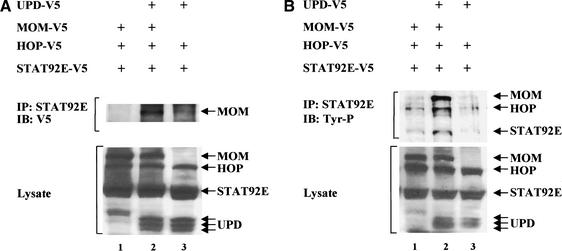
We directly examined the physical interactions between MOM and STAT92E in cotransfection experiments. We prepared cell lysates from S2 cell lines expressing V5-epitope-tagged MOM, HOP, and STAT92E with either UPD–V5 or vector alone. The lysates were immunoprecipitated using anti-STAT92E antibodies and then probed using anti-V5 antibodies. MOM only coimmunoprecipitated with STAT92E in the upd–V5 and mom–V5 transfected cells (Fig. 7A, lane 2). These data suggest that STAT92E binds to the activated MOM receptor.
Figure 7.
(A) MOM binds STAT92E. S2 cells were transfected with V5-epitope-tagged upd, mom, hop, and stat92E in different combinations as indicated. Cellular lysates were immunoprecipitated (IP) with anti-STAT92E antibodies and blotted (IB) with an anti-V5 antibody (top panel). MOM only coimmunoprecipitated with STAT92E in the upd–V5-transfected cells (lane 2). These data suggest that STAT92E binds to the activated MOM receptor. Expression of UPD, MOM, HOP, and STAT92E was assessed in whole-cell extracts by immunoblot analysis (lower panel). UPD appears as three bands because of partially glycosylated intermediates (Harrison et al. 1998). (B) MOM mediates activation of the HOP/STAT92E signal transduction pathway. We cotransfected S2 cells with V5-epitope-tagged upd, mom, hop, and stat92E in different combinations as indicated. Anti-STAT92E immunoprecipitates were prepared and tested for reactivity with the anti-phosphotyrosine antibody 4G10. Whereas UPD, MOM, HOP, and STAT92E proteins are detectable in the transfected samples (lower panel), increased tyrosine phosphorylation of MOM, HOP, and STAT92E is detected in immunoprecipitates prepared from mom–V5- and upd–V5-transfected cells (top panel).
UPD activates the HOP/STAT92E pathway through MOM
In the mammalian system, JAK proteins are bound to monomeric cytokine receptors through the membrane-proximal domain (Hirano et al. 1997; Taga and Kishimoto 1997). Signaling is triggered when cytokine binding induces receptor dimerization. This brings the receptor-associated JAK kinases into apposition, enabling them to transphosphorylate each other. The JAK kinases, now activated, phosphorylate a distal tyrosine on the receptor. This receptor phosphotyrosyl residue is subsequently recognized by the SH2 domain of the STAT proteins, drawing them into the receptor complex, where they are activated through phosphorylation on the tyrosine residue by JAKs. (Schindler and Darnell 1995; Wen et al. 1995).
To show MOM-dependent activation of the HOP/STAT92E pathway, we analyzed the tyrosine phosphorylation of MOM, HOP, and STAT92E. We cotransfected S2 cells with V5-epitope-tagged MOM, HOP, and STAT92E with either UPD–V5 or vector alone. Anti-STAT92E immunoprecipitates were prepared and tested for reactivity with the anti-phosphotyrosine antibody 4G10. As shown in Figure 7B, whereas UPD, MOM, HOP, and STAT92E proteins are detectable in the transfected samples (Fig. 7B, lower panel), increased tyrosine phosphorylation of MOM, HOP, and STAT92E is detected in immunoprecipitates prepared from UPD–V5- and MOM–V5-transfected cells (Fig. 7B, top panel). These data are consistent with our hypothesis that MOM is a receptor of UPD to activate the HOP/STAT92E signal transduction pathway.
Discussion
In this report we identified a novel Drosophila gene, mom, that encodes a protein distantly related to the mammalian gp130 subfamily and likely functions as a receptor for the HOP/STAT92E signal transduction pathway. mom shows embryonic mutant phenotypes identical to those in the hopscotch (hop)/JAK kinase and marelle (mrl)/STAT92E mutations: mom, hop, and stat92E have identical embryo mutant cuticle phenotypes; they also show similar tracheal defects. Like upd and hop, mom controls STAT92E protein expression. Genetic analyses place mom's function between upd (the ligand) and hop. We further show that cultured cells transfected with the mom gene bind UPD and activate the HOP/STAT92E signal transduction pathway. The experiments reported here provide a new point of entry for examining the biochemistry of the HOP/STAT92E signal transduction pathway.
Is MOM the only receptor of the HOP/STAT92E signal transduction pathway?
It is important to note that our data do not rule out the possibility that additional molecules in the conditioned medium might associate with UPD and participate in its binding to the receptor. They also do not rule out the possibility that MOM is part of a larger complex at the cell surface; in such a complex MOM would be necessary but might not be sufficient for binding and/or signal transduction, especially because MOM belongs to the gp130 subfamily of receptors.
In mammals, the gp130 subfamily of receptors includes gp130, leukemia inhibitory factor receptor (LIFR), oncostatin M receptor (OSMR), ciliary neurotrophic factor receptor (CTNFR), cardiotrophin-1 receptor (CT-1R), granulocyte colony-stimulating factor receptor (G-CSFR), and the obesity/leptin gene receptor (OBR) (Hirano et al. 1997; Taga and Kishimoto 1997; Grant and Begley 1999). The functional gp130 subfamily of receptors generally comprises multiple components of membrane proteins, and these multichain receptor complexes often share the common signal gp130 as a component critical for signal transduction (Hirano et al. 1997; Taga and Kishimoto 1997; Grant and Begley 1999). Cytokines in the IL-6 family initiate intracellular signaling by binding to a specific ligand-binding subunit and then subsequently form a complex with the signal-transducing receptor component, gp130. IL-6 binds to IL-6R, and the IL-6/IL-6R complex then associates with gp130, allowing it to homodimerize. However, OSMR and G-CSFR are unique among the gp130 subfamily of receptors. In the case of OSMR, OSM binds directly to gp130 with low affinity. Upon binding by OSM, gp130 generates a high-affinity receptor complex by forming a heterodimer with either the LIFR- or the OSM-specific receptor subunit (LIFRβ and OSMRβ, respectively). The G-CSFR shows high homology with gp130; G-CSFR forms homodimers by itself in response to G-CSF.
UPD has a very limited similarity to the IL-6 family of cytokines. It is difficult to predict whether UPD signaling is more like IL-6 or G-CSF. If it is more like IL-6, there should be another receptor subunit that would be functionally similar to IL-6R. UPD may first bind to the missing receptor and then form a functional complex with MOM. On the other hand, if UPD is more like G-CSF, MOM may be the only receptor. UPD binding should promote MOM to form a homodimer and start the signal transduction process. However, UPD may interact with its receptor in a different way. UPD structure is less consistent with a cytokine-type molecule. The UPD protein is extremely basic, with a predicted pI of nearly 12. In contrast to many soluble cytokines, UPD is associated with ECM (extracellular matrix), which may help it bind to the receptor and limit the range of activity of the ligand.
MOM-mediated signal transduction
The gp130 subfamily of receptors has no intrinsic tyrosine kinase domain, but constitutively associates with tyrosine kinase JAKs. The family members possess conserved motifs in the cytoplasmic region, such as box 1, box 2, and box 3 in the membrane proximal-to-distal order. These conserved boxes are important for receptor-mediated signal transduction (Hirano et al. 1997; Taga and Kishimoto 1997; Grant and Begley 1999). The box 1 motif in the cytoplasmic region of gp130 is important for the association with JAK kinases; the box 3 motif provides a docking site for the SH2 domain of STAT3 protein, which recruited to gp130 now serves as a substrate for JAKs. The activation of STAT3 is dependent on the phosphorylation of any one of the four tyrosines (Y767, Y814, Y905, Y915) in the C terminus of the gp130 that have a glutamine residue at the third position behind tyrosine (Y-X-X-Q). MOM has no visible box 1 and 2 motifs but has one tyrosine residue (Y966) fitting a YXXQ consensus motif. MOM binds HOP and STAT92E in biochemical experiments (Fig. 7B).
In addition to the JAK/STAT pathway, multiple signaling molecules are tyrosine-phosphorylated in response to the IL-6 family of cytokines. CNTF, LIF, OSM, and IL-6 induce tyrosine phosphorylation of phospholipase Cγ and SHP-2 (a phosphotyrosine phosphatase, also called PTP1-D, SHPTP-2, PTP2C, and Syp), which is a mammalian homolog of Drosophila corkscrew (csw). The Ras-MAPK pathway is activated by the IL-6 cytokine family. The activation of the Ras-MAPK pathway is possibly mediated by SHP-2 and/or Shc, which bind a Grb2–SOS complex (Hirano et al. 1997; Taga and Kishimoto 1997). Tyrosine 759 of gp130 is required for the tyrosine phosphorylation of SHP-2 and its binding to gp130, the association between SHP-2 and Grb2, and the full activation of MAPK.
In flies, the phenotypes caused by loss of function of the HOP/STAT92E pathway and of the Ras-MAPK pathway are very distinct. In the embryo, loss-of-function mutations in the HOP/STAT92E pathway cause segmentation defects. The absence of additional phenotypes, such as terminal defects or poorly differentiated cuticle in germ-line clone-derived embryos, indicates that this pathway does not cross-talk with the Ras-MAPK pathway in either the TORSO or DER/EGFR RTK signaling pathways. In the eye, the Ras-MAPK pathway regulates photoreceptor formation. Changing the activity of the HOP/STAT92E pathway does not affect photoreceptor fate rather than affect cell proliferation.
However, there are some similarities in phenotypes between the HOP/STAT92E pathway and the Drosophila Ras-MAPK pathway. For example, overexpression of hop in the wing disc results in vein phenotypes similar to those seen in animals that express activated forms of D-raf (Harrison et al. 1995). Loss-of-function D-raf mutations result in larval/pupal lethality with underproliferation of diploid tissues (Perrimon et al. 1985), similar to hop mutations (Perrimon and Mahowald 1986). However, these similarities are based on overexpression phenotypes as well as most of the mammalian evidence for cross-talk. These results should be taken with caution just in case an artifactual phenomenon was created by these overexpressions.
Other potential functions of MOM and the HOP/STAT92E signal transduction pathway
The mammalian gp130 subfamily of receptors interacts with the receptors' respective ligands and has pleiotropic functions. G-CSF is the principal hematopoietic growth factor regulating the production of neutrophils, and it is widely used to treat neutropenia in a variety of clinical settings (Welte et al. 1996). The mouse obesity gene encodes a soluble protein (OB or leptin; Zhang et al. 1994) that produces weight-reducing effects in mice when administrated in vivo. The structure of the receptor for this factor (OBR or leptin-R) is highly homologous to that of gp130. The IL-6 family of cytokines shares gp130 as a component critical for signal transduction in the cytokine receptor complexes and has redundant functions (Taga and Kishimoto 1997). Some of these biological activities of IL-6 are also often exerted by other cytokines, namely, IL-11, LIF, OSM, CNTF, and CT-1. The biological activity by which each cytokine was initially identified was growth promotion of myeloma and plasmacytoma for IL-11, growth inhibition of a mouse myeloid leukemia cell line for LIF, growth inhibition of a human melanoma cell line for OSM, promotion of survival of rat-cultured ciliary neurons for CNTF, and induction of cardiac hypertrophy in vitro for CT-1. These cytokines show structural similarity and, more importantly, have biological functions that overlap with those of IL-6.
MOM is the only member of this family of receptors so far identified in Drosophila. The fly has also only one JAK (HOP) and one STAT (STAT92E). It remains to be seen whether this simple MOM/HOP/STAT92E pathway has the pleiotropic functions that are accomplished by a great complex of a homologous receptor family. This simple genetic model system will greatly enhance our understanding of the gp130 subfamily of receptors' biological functions and mediated signal transductions.
Materials and methods
Drosophila stocks
mom1, 1(1)G0441; mom2, l(1)G0264; mom3, l(1)G0367; and mom4, l(1)G0405 were identified from the collection of X-chromosome lethal mutations generated by Ulrich Schaefer in the laboratory of Herbert Jackle (http://flybase.bio.indiana.edu).
Two hop alleles were used in this study: C111, a null allele (Perrimon and Mahowald 1986); and Tum-l, a dominant temperature-sensitive allele (Corwin and Hanratty 1976). Stat92E6346 was described in Hou et al. (1996). updYC43 was described in Harrison et al. (1998).
Transgenic lines of HS–mom, UAS–mom, and UAS–upd were produced by inserting corresponding cDNA into the pCasper-hs and pUAST vectors (Thummel et al. 1988; Brand and Perrimon 1993) and injecting the constructs into flies. UAS–mom-Nwas constructed by inserting a C-terminal truncated mom cDNA (amino acids 1–671) into a pUAST vector. The matα4–GAL-VP16 line contains a construct consisting of the DNA-binding domain (amino acids 1–144) of GAL4 fused to the VP16 transcriptional activation domain expressed from the α4-tubulin promoter and was a gift of D. St. Johnston through the N. Perrimon lab. GMR–Gal4 was from the Bloomington stock center.
Flies were raised on standard Drosophila media at 25°C unless otherwise indicated. Chromosomes and mutations that are not described in the text can be found in Lindsley and Zimm (1990).
Germ-line clones (GLCs)
Females carrying GLCs of mom were induced according to the dominant female sterile technique that uses the mutation Fs(1)K1237 (or OvoD1; Perrimon et al. 1989). Flies were irradiated with a dose of 1000 rads using a Torrex 120D X-ray machine at the end of the first instar larval stage. Such conditions generate ∼7% mosaic females.
At emergence, irradiated mom/OvoD1 females were distributed in lots of 10 per vial with 5–10 w1118 males and checked each day for the presence of eggs. When eggs were found, the clone-containing female was then isolated and studied individually. The females that laid eggs with segmentation defects were pooled together and used for studying mom functions. Ovarioles containing the germ-line clone (+/+) are easily distinguished from the others (+/OvoD1) by the presence of vitellogenic egg chambers.
Rescue of mom lethality
A 2-chromosome HS–mom transgene line was used for rescuing mom lethality. Heat shock at 37°C was administered for 30 min to mom1/FM7;+/+ X +/Y; HS–mom/+ flies every 12 h from the second instar larval stage; more than 20 FM7− male flies were obtained from each vial.
Genetic interaction between hopTum-land mom
To test for interaction between hop and mom, the cuticle phenotype of hemizygous embryos derived from heterozygous females for mom1 and hopTum-l was compared with that of hemizygous embryos derived from heterozygous females for mom1 only.
Molecular cloning
Genomic DNA flanking the l(1)G0441 P element was recovered by plasmid rescue and used to screen a 0–4-h embryonic cDNA library (Brown and Kafatos 1988) and to search the Drosophila EST database. One full-length 4.8-kb clone and several shorter cDNA clones corresponding to one open reading frame were identified. The longest, 4.8-kb clone was selected for further analysis. DNA sequencing was carried out on both strands on an ABI model 310 DNA sequencer using consecutive oligonucleotide primers synthesized to extend the sequences. The mom genomic organization and the P-element insertion site were deduced from a BDGP database (http://www.fruitfly.org). Resequencing confirmed the P-element insertion point and the exon–intron joints. The mom gene is encoded by three exons, and the four P elements are inserted into a 5′ untranslated sequence in the first exon (Fig. 5A).
In situ hybridization and antibody staining
In situ hybridizations to whole-mount embryos by using digoxigenin-labeled antisense mom DNA probe were performed as described (Hou et al. 1996).
Peptides corresponding to the C terminals of STAT92E were used to produce antibodies in rabbits. Antiserum was purified using the peptides as affinity reagents. STAT92E staining was performed using the purified antisera at 1:1000 dilution. Additional antibodies used for immunohistochemistry include anti-tracheal lumen mAb2A12 (Developmental Study Hybridoma Bank). Biotin-conjugated secondary antibodies and the vectastain Elite kit were from Vector.
DNA constructs and cell transfection
mom cDNA was amplified by the PCR technique using its cDNA as the template. The amplified DNA was subcloned into pCEFL, a modified pcDNA3 expression vector containing the elongation factor 1 promoter driving the expression of an in-frame N-terminal tag of nine amino acids derived from the influenza virus hemagglutinin HA1 protein (HA; Wilson et al. 1984). To obtain a C-terminal tagged upd–V5 construct, the PCR-amplified upd cDNA was subcloned into the pcDNA3.1/V5-His A vector (Invitrogen). For S2 cell expression, upd–V5, mom–V5, hop–V5, and stat92E–V5 in pMT/V5-His A vectors (Invitrogen) were constructed.
Transient transfections of S2 cells were performed by using a FuGENE 6 transfection reagent (Boehringer Mannheim No. 1814443) as described in Wei et al. (2000). After 24 h, copper sulfate was added to the medium to a final concentration of 500 μM to modulate expression of transfected genes from the metallothionein promoter according to the manufacturer's instructions (Invitrogen). After another 24 h, cells were collected and the lysates were used for coimmunoprecipitation experiments.
Extracts from cells transfected with epitope-tagged HOP, STAT92E, or MOM were immunoprecipitated with anti-STAT92E antibodies, and the immunocomplexes were recovered with Protein G-sepharose (Pharmacia). Beads were washed three times with PBS containing 1% NP-40 and 2 mM vanadate and loaded in 10% polyacrylamide gels. The presence of MOM in the complexes was analyzed by Western blot using an anti-V5 specific antibody (1:5000). Proteins were visualized by enhanced chemiluminescence detection (Amersham) using goat anti-mouse and anti-rabbit IgGs coupled to horseradish peroxidase as the secondary antibody (Cappel).
To detect tyrosine phosphorylation of MOM, HOP, and STAT92E, extracts from S2 cells transfected with V5-tagged mom, hop, and stat92E with upd–V5 or vector alone were immunoprecipitated with anti-STAT92E antibodies; the tyrosine phosphorylation of MOM, HOP, and STAT92E in the complexes was detected with monoclonal antiphosphotyrosine antibody 4G10 (UBI; 1:10,000).
UPD protein binds to cells transfected with MOM and MOM-N
293T cells were transfected with pcDNA3–upd–V5 by use of lipofectamine (Life Technologies) according to the manufacture's instructions. After transfection, 50 μg/mL of heparin (Sigma H-9399) was added to cells to release UPD to the medium from the extracellular matrix. A day later, the medium was collected and concentrated in Centriprep 10,000 MWCO ultrafiltration devices (Amicon).
Separately, 293T cells were seeded on glass coverslips and transfected with expression vectors for HA–MOM or HA–MOM-N with HA–STAT92E by Lipofectamine Plus Reagents (Life Technologies) as described (Marinissen 2001). After 24 h, the transfected 293T cells were washed twice in PBS and incubated with 1.5 mL of concentrated conditioned medium at 37°C for 3 h. After three 10-min washes with PBS, the cells were fixed and permeabilized with 4% formaldehyde and 0.5% Triton X-100 in 1× PBS for 20 min. After washing with PBS, cells were blocked with 1% bovine serum albumin and incubated with the indicated primary antibodies for 1 h. Mouse anti-V5 and rabbit anti-STAT92E antibodies (1:200) were used to detect V5-tagged or STAT92E molecules. Following incubation, cells were washed three times with 1× PBS and then incubated with the corresponding secondary antibodies (1:200) conjugated with either Fluorescein (FITC) or tetramethylrhodamine B isothiocyanate (TRITC; Jackson ImmunoResearch Laboratories). Coverslips were washed three times and mounted in Vectashield mounting medium with DAPI (Vector Laboratories) and viewed using a Zeiss Axiophot photomicroscope equipped with epifluorescence. Immunofluorescence was photographed using Kodak TMAX 3200 film.
Acknowledgments
We thank D. St. Johnston for matα4–GAL-VP16-Gal4 stock; B. Zbar for allowing us to use his tissue culture hood and incubator; H. Shin for generating transgene lines; and G. Zheng for help with antibody staining of embryos. This work is supported in part by a grant from the U.S. Army (Award no. DAMD17-00-1-0356) to S.X.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL shou@mail.ncifcrf.gov; FAX (301) 846-6145.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.955202.
References
- Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative JAK family tyrosine kinase in Drosophila. Genes & Dev. 1994;8:300–312. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- Blalock JE. The syntax of immune-neuroendocrine communication. Immunol Today. 1994;15:504–511. doi: 10.1016/0167-5699(94)90205-4. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a mean of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Chatterjee-Kishore M, van den Akker F, Stark GR. Association of STATs with relatives and friends. Trends Cell Biol. 2000;10:106–111. doi: 10.1016/s0962-8924(99)01709-2. [DOI] [PubMed] [Google Scholar]
- Corwin HO, Hanratty WP. Characterization of a unique lethal tumorous mutation in Drosophila. Mol Gen Genet. 1976;144:345–347. doi: 10.1007/BF00341734. [DOI] [PubMed] [Google Scholar]
- Dearolf CR. Fruit fly “leukemia.”. Biochim Biophys Acta. 1998;1377:M13–M23. doi: 10.1016/s0304-419x(97)00031-0. [DOI] [PubMed] [Google Scholar]
- ————— JAKs and STATs in invertebrate model organisms. Cell Mol Life Sci. 1999;55:1578–1584. doi: 10.1007/s000180050397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T. Introduction: STATs as essential intracellular mediators of cytokine responses. Cell Mol Life Sci. 1999;55:1505–1508. doi: 10.1007/s000180050390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T, Kovarik P. Transcription factor activity of STAT proteins: Structural requirements and regulation by phosphorylation and interacting proteins. Cell Mol Life Sci. 1999;55:1535–1546. doi: 10.1007/s000180050393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of aa cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Grant S, Begley CG. The oncostatin M signalling pathway: Reversing the neoplastic phenotype? Mol Med Today. 1999;5:406–412. doi: 10.1016/s1357-4310(99)01540-3. [DOI] [PubMed] [Google Scholar]
- Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes & Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Nakajima K, Hibi M. Signaling mechanisms through gp130: A model of the cytokine system. Cytokine & Growth Factor Rev. 1997;8:241–252. doi: 10.1016/s1359-6101(98)80005-1. [DOI] [PubMed] [Google Scholar]
- Hou XS, Perrimon N. The JAK-STAT pathway in Drosophila. Trends Genet. 1997;13:105–110. doi: 10.1016/s0168-9525(97)01006-8. [DOI] [PubMed] [Google Scholar]
- Hou SX, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Hu N, Castelli-Gair J. Study of the posterior spiracles of Drosophila as a model to understand the genetic and cellular mechanisms controlling morphogenesis. Dev Biol. 1999;214:197–210. doi: 10.1006/dbio.1999.9391. [DOI] [PubMed] [Google Scholar]
- Imada K, Leonard WJ. The Jak-STAT pathway. Mol Immunol. 2000;37:1–11. doi: 10.1016/s0161-5890(00)00018-3. [DOI] [PubMed] [Google Scholar]
- Jinks TM, Polydorides AD, Calhoun G, Schedl P. The JAK/STAT signaling pathway is required for the initial choice of sexual identity in Drosophila melanogaster. Mol Cell. 2000;5:581–587. doi: 10.1016/s1097-2765(00)80451-7. [DOI] [PubMed] [Google Scholar]
- Levy DE. Physiological significance of STAT proteins: Investigations through gene disruption in vivo. Cell Mol Life Sci. 1999;55:1559–1567. doi: 10.1007/s000180050395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. and Zimm, G. 1990. The genome of Drosophila melanogaster part 4: Genes L-Z, balancers, transposable elements. Dros. Inf. Serv. 68.
- Marinissen MJ, Chiariello M, Gutkind JS. Regulation of gene expression by the small GTPase Rho through the ERK6 (p38γ) MAP kinase pathway. Genes & Dev. 2001;15:535–553. doi: 10.1101/gad.855801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger RJ, Krasnow MA. Genetic control of branching morphogenesis. Science. 1999;284:1635–1639. doi: 10.1126/science.284.5420.1635. [DOI] [PubMed] [Google Scholar]
- Mui ALF. The role of STATs in proliferation, differentiation, and apoptosis. Cell Mol Life Sci. 1999;55:1547–1558. doi: 10.1007/s000180050394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton PA, Hynes RO, Rees DJG. Sevenless: Seven found? Cell. 1990;61:15–16. doi: 10.1016/0092-8674(90)90209-w. [DOI] [PubMed] [Google Scholar]
- Patthy L. Homology of a domain of the growth hormone/prolactin receptor family with type III modules of fibronectin. Cell. 1990;61:13–14. doi: 10.1016/0092-8674(90)90208-v. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Mahowald AP. l(1)hopscotch, a larval-pupal zygotic lethal with a specific maternal effect on segmentation in Drosophila. Dev Biol. 1986;118:28–41. doi: 10.1016/0012-1606(86)90070-9. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Engstrom L, Mahowald AP. A pupal lethal mutation with a paternally influenced maternal effect on embryonic development in Drosophila melanogaster. Dev Biol. 1985;110:480–491. doi: 10.1016/0012-1606(85)90105-8. [DOI] [PubMed] [Google Scholar]
- ————— Zygotic lethals with specific maternal effect phenotypes in Drosophila melanogaster. I. Loci on X chromosome. Genetics. 1989;121:333–352. doi: 10.1093/genetics/121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Noll E, McCall K, Brand A. Generating lineage-specific markers to study Drosophila development. Dev Genet. 1991;12:238–252. doi: 10.1002/dvg.1020120309. [DOI] [PubMed] [Google Scholar]
- Schindler C, Darnell JE., Jr Transcriptional responses to polypepetide ligands: The JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- Sefton L, Timmer JR, Zhang Y, Beranger F, Cline TW. An extracellular activator of the Drosophila JAK/STAT pathway is a sex-determination signal element. Nature. 2000;405:970–973. doi: 10.1038/35016119. [DOI] [PubMed] [Google Scholar]
- Smith MA, Nichols J, Robertson M, Rathjen PD. Differentiation inhibiting activity (DIA/LIF) and mouse development. Dev Biol. 1992;151:339–351. doi: 10.1016/0012-1606(92)90174-f. [DOI] [PubMed] [Google Scholar]
- Streuli M, Krueger NX, Tsai AYM, Saito H. A family of receptor-linked protein tyrosine phosphatases in humans and Drosophila. Proc Natl Acad Sci. 1990;86:8698–8702. doi: 10.1073/pnas.86.22.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland D, Samakovlis C, Krasnow M. Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- Thummel CS, Boulet AM, Lipshitz HD. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- Wei Q, Marchler G, Edington K, Karsch-Mizrachi I, Peterson BM. RNA interference demonstrates a role for nautilus in the myogenic conversion of Schneider cells by daughterless. Dev Biol. 2000;228:239–255. doi: 10.1006/dbio.2000.9938. [DOI] [PubMed] [Google Scholar]
- Welte K, Gabrilove J, Bronchud MH, Platzer E, Morstyn G. Filgrastim (r-metHuG-CSF): The first 10 years. Blood. 1996;88:1907–1929. [PubMed] [Google Scholar]
- Wen Z, Zhong Z, Darnell JE. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- Williams JG. STAT signalling in cell proliferation and in development. Curr Opin Genet Dev. 2000;10:503–507. doi: 10.1016/s0959-437x(00)00119-2. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Yan R, Small S, Desplan C, Dearolf CR, Darnell JJ. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- Yeh TC, Pellegrini S. The Janus kinase family of protein tyrosine kinases and their role in signaling. Cell Mol Life Sci. 1999;55:1523–1534. doi: 10.1007/s000180050392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler MP, Perrimon N. Sex determination: Co-opted signals determine gender. Curr Biol. 2000;10:R682–R684. doi: 10.1016/s0960-9822(00)00686-2. [DOI] [PubMed] [Google Scholar]
- Zeidler MP, Bach EA, Perrimon N. The roles of the Drosophila JAK/STAT pathway. Oncogene. 2000;19:2598–2606. doi: 10.1038/sj.onc.1203482. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]