Abstract
All eukaryotic cells respond to the accumulation of unfolded proteins in the endoplasmic reticulum (ER) by signaling an adaptive pathway termed the unfolded protein response (UPR). In yeast, a type-I ER transmembrane protein kinase, Ire1p, is the proximal sensor of unfolded proteins in the ER lumen that initiates an unconventional splicing reaction on HAC1 mRNA. Hac1p is a transcription factor required for induction of UPR genes. In higher eukaryotic cells, the UPR also induces site-2 protease (S2P)-mediated cleavage of ER-localized ATF6 to generate an N-terminal fragment that activates transcription of UPR genes. To elucidate the requirements for IRE1α and ATF6 for signaling the mammalian UPR, we identified a UPR reporter gene that was defective for induction in IRE1α-null mouse embryonic fibroblasts and S2P-deficient Chinese hamster ovary (CHO) cells. We show that the endoribonuclease activity of IRE1α is required to splice XBP1 (X-box binding protein) mRNA to generate a new C terminus, thereby converting it into a potent UPR transcriptional activator. IRE1α was not required for ATF6 cleavage, nuclear translocation, or transcriptional activation. However, ATF6 cleavage was required for IRE1α-dependent induction of UPR transcription. We propose that nuclear-localized IRE1α and cytoplasmic-localized ATF6 signaling pathways merge through regulation of XBP1 activity to induce downstream gene expression. Whereas ATF6 increases the amount of XBP1 mRNA, IRE1α removes an unconventional 26-nucleotide intron that increases XBP1 transactivation potential. Both processing of ATF6 and IRE1α-mediated splicing of XBP1 mRNA are required for full activation of the UPR.
Keywords: Endoplasmic reticulum, stress response, transcription factors, signal transduction, posttranscriptional regulation
The endoplasmic reticulum (ER) is the compartment where protein folding occurs prior to transport to the extracellular surface or to different intracellular organelles. It is a highly evolved folding factory where proteins attain their final folded conformation without excessive misfolding and/or aggregation. This process depends on molecular chaperones that provide local environments favorable for protein folding. However, under a variety of conditions, these folding reactions are compromised and protein aggregation occurs. As a consequence, the cell activates adaptive signaling pathways that are programmed to enhance the folding capabilities and limit the folding load on the ER. This response is called the unfolded protein response (UPR). The UPR is essential for survival of all eukaryotic cells under conditions of ER stress and is also essential for differentiation and/or survival of eukaryotic cells that secrete high levels of proteins. The UPR is also implicated in the pathogenesis of a number of diseases (Aridor and Balch 1999). Finally, this response may have evolved as part of a nutrient-sensing mechanism to couple energy availability and metabolic processes with differentiation state (Schroder et al. 2000; Scheuner et al. 2001). The UPR culminates in the induction of the ER stress-response genes, including those that encode BiP (GRP78), GRP94, ERP72, calreticulin, CHOP-10 (GADD153), and XBP1 (X-box binding protein) (for review, see Chapman et al. 1998; Kaufman 1999; Mori 2000).
One molecular mechanism for signaling the UPR was elucidated in Saccharomyces cerevisiae. In S. cerevisiae the only proximal sensor of the UPR is the protein kinase and endoribonuclease Ire1p (Nikawa and Yamashita 1992; Cox et al. 1993; Mori et al. 1993). Although the precise mechanism remains to be elucidated, the accumulation of unfolded proteins in the ER lumen promotes Ire1p dimerization, trans-autophosphorylation, and activation of its endoribonuclease (RNase) activity (Shamu and Walter 1996; Welihinda and Kaufman 1996). The only known substrate for the site-specific RNase activity of Ire1p is HAC1 mRNA, encoding a basic leucine zipper (bZIP)-containing transcription factor that binds to the UPR element (UPRE) upstream of responsive genes (Cox and Walter 1996; Mori et al. 1996). Ire1p cleaves HAC1 mRNA at two sites to remove a 252-nt intron. Ire1p endonucleolytic cleavage after guanosine at both the 5′ and 3′ splice-site junctions leaves a 2‘,3′-cyclic phosphate at the 3′ end of the 5′ exon and a free 5′ hydroxyl group at the 5′ end of the 3′ exon. These exons are tethered together by base pairing and subsequently ligated by tRNA ligase Rlg1p (Sidrauski et al. 1996; Gonzalez et al. 1999). In contrast to unspliced HAC1 mRNA that is poorly translated, the spliced form is efficiently translated (Mori et al. 1996; Chapman and Walter 1997; Ruegsegger et al. 2001). As the level of Hac1p rises in the cell, the genes that harbor UPRE within their promoters are induced at the transcriptional level. In yeast there are ∼381 transcriptional targets of the UPR that encode functions ranging from protein folding, protein translocation, and protein transport, to protein degradation within the secretory pathway (Travers et al. 2000).
Metazoan cells have evolved a more extensive signaling pathway to mediate more diverse responses upon activation of the UPR. In metazoan cells this response also signals translation attenuation to reduce the amount of proteins that require folding. In addition, if adaptation is not sufficient, apoptosis is activated, possibly mediated by the c-Jun N-terminal protein kinase JNK, and activation of caspases 7 and 12 (Chandler et al. 1998; Nakagawa et al. 2000; Urano et al. 2000a).
There are at least three known proximal sensors of the UPR in metazoan species. Two related homologs of yeast IRE1, referred to as IRE1α and IRE1β, were identified in both the murine and human genomes (Tirasophon et al. 1998; Wang et al. 1998; Iwawaki et al. 2001). Whereas IRE1α is constitutively expressed in all cells and tissues, IRE1β expression is restricted to gut epithelial cells. Overexpression of either IRE1α or IRE1β can activate a reporter gene that harbors an ER stress-response element (ERSE) in a manner that requires the endoribonuclease activity of IRE1 (Tirasophon et al. 2000). These results support the idea that the IRE1 signaling pathway is conserved from yeast to human and is sufficient to activate the UPR.
Translational attenuation is mediated by the protein kinase PERK/PEK, which is activated under conditions identical to those that activate IRE1 (Shi et al. 1998; Harding et al. 1999; Liu et al. 2000; Bertolotti et al. 2001). After trans-autophosphorylation, PERK phosphorylates its only known substrate, the α subunit of eukaryotic translation initiation factor 2 (eIF2α), to limit polypeptide chain initiation, and thereby protects cells under conditions where proteins cannot fold properly. However, disruption of this signaling pathway, by deletion of PERK or mutation of the phosphorylation site in eIF2α, produced unexpected results. Indeed, the cells were very sensitive to activation of the UPR, but they were also defective for transcriptional induction of the UPR (Harding et al. 2000; Scheuner et al. 2001). It was proposed that under conditions of increased eIF2α phosphorylation, AUG-codon recognition by the 40S scanning ribosomal subunit becomes less efficient; therefore, the ribosome scans through upstream open reading frames within the 5′ end of mRNAs to gain access to the authentic AUG codons. At least one potential target for this translational up-regulation upon eIF2α phosphorylation is the transcriptional activator of the CREB/ATF (cyclic AMP response element binding protein/activating transcription factor) family, ATF4 (Harding et al. 2000; Scheuner et al. 2001).
Finally, an alternative pathway for gene activation upon accumulation of unfolded proteins in the ER was discovered by identification of proteins that bind the ERSE through a yeast one-hybrid screen (Haze et al. 1999). Mori and coworkers identified ATF6 as a type-II ER-associated transmembrane bZIP-containing transcription factor. Upon activation of the UPR, ATF6 is proteolyzed to release a cytosolic fragment that migrates to the nucleus to activate transcription of genes through binding to the ERSE (Haze et al. 1999). ATF6 binds to CCACG of the ERSE (CCAAT-N9-CCACG) when the CCAAT is bound to NF-Y (Li et al. 2000; Yoshida et al. 2000, 2001a). BiP, CHOP-10, and XBP1 are genes that are induced by overexpression of ATF6 (Yoshida et al. 1998). Proteolysis of ATF6 is sufficient to activate expression of a BiP-promoter reporter construct. ATF6 is cleaved within the lumenal and transmembrane domains by site-1 and site-2 proteases (S1P and S2P), respectively, which are known to cleave sterol response element binding protein (SREBP) upon sterol deprivation (Ye et al. 2000). Therefore, at least a portion of the UPR signaling has coevolved with the sterol-deprivation response pathway.
To identify the transcriptional targets of ATF6, a PCR-amplification technique was applied to identify the consensus ATF6-binding-site motif TGACGTG(G/A) (Wang et al. 2000). Surprisingly, when multiple copies of the ATF6-binding motif (5× ATF6) were placed upstream of luciferase, expression from this promoter element was induced in response to ER stress. Therefore, all the information necessary for UPR transcriptional activation is contained within this short, synthetic ATF6-binding motif. Although the downstream target genes of ATF6 are being elucidated at this time, it is likely that a large percentage of ATF6-responsive genes are targets of the UPR. In this report, we have used the 5× ATF6 reporter as a monitor for UPR signaling in wild-type and mutant cells.
Thus, at present, there is a dilemma as to how UPR pathways signaling through IRE1, PERK, and ATF6 are coordinated to act in concert for transcriptional induction. To address this question, we studied the UPR in IRE1α-null murine embryonic fibroblasts (MEFs). Surprisingly, IRE1α-null MEFs were not defective for induction of the major targets of the UPR; however, they were specifically defective in induction of the 5× ATF6-binding-site reporter gene. By analysis of the defect in these cells, we identified that a novel form of XBP1 is absent in IRE1α-null cells and showed that XBP1 mRNA is a substrate for the endoribonuclease activity of IRE1α. Further analysis using S2P-deficient Chinese hamster ovary (CHO) cells showed that IRE1α-mediated UPR signaling requires coordination of IRE1α-independent ATF6 processing and IRE1α-dependent XBP1 mRNA splicing.
Results
IRE1α-null murine embryonic fibroblasts (MEFs) have an intact UPR
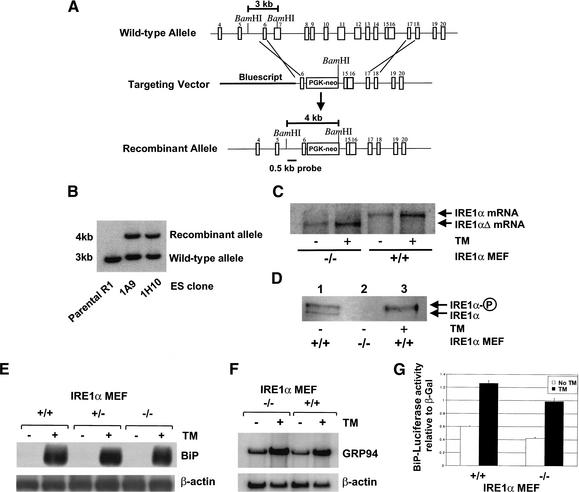
The role for IRE1α in the UPR was studied using IRE1α-null MEFs. Exon 7 to exon 14 from the IRE1α gene was deleted by homologous recombination in R1 embryonic stem cells using a PGK-neo targeting vector (Fig. 1A), and the presence of the deleted IRE1α locus was shown by Southern blot analysis (Fig. 1B). The IRE1α deletion was confirmed by Northern blot and Western blot analysis. As expected from the homologous replacement, the homozygous IRE1α-null MEFs express a smaller IRE1α mRNA transcript compared to that detected in wild-type MEFs (Fig. 1C). The predicted protein product from the deleted IRE1α allele would lack the ER transmembrane domain and would therefore likely be mislocalized to the lumen of the ER. Because the endogenous level of IRE1α expression is very low, expression of IRE1α protein was analyzed by immunoprecipitation using an anti-IRE1α lumenal-domain antibody and Western blot analysis using the same antibody. As a positive control, IRE1α was analyzed in a tunicamycin-treated pancreatic β-cell line known to express IRE1α. Tunicamycin inhibits N-linked glycosylation and activates the UPR. Under these conditions, only the phosphorylated form of IRE1α protein is detected, as previously described (Fig. 1D, lane 3; Tirasophon et al. 1998, 2000). Although nonphosphorylated and phosphorylated species of IRE1α were detected in the wild-type MEFs, anti-IRE1α antibody-reactive protein was not detected in IRE1α-null MEFs (Fig. 1D, lanes 1,2).
Figure 1.
Generation and characterization of IRE1α-null MEFs. (A) Schematic representation of the predicted recombination of targeting vector and the mIRE1α locus. The bar indicates the position of a 0.5-kb BamHI–XhoI fragment used as a probe for Southern hybridization. (B) Southern analysis of ES recombinant clones (1A9 and 1H10) compared to the parental R1 cells. (C) Northern blot analysis of IRE1α-null MEFs. Wild-type and IRE1α-null MEFs were treated with or without 10 μg/mL tunicamycin for 6 h prior to harvesting total RNA for Northern blot analysis. The blot was probed with an [α-32P]-labeled 3.6-kb EcoRI–XbaI fragment from pED–hIRE1α cDNA. (D) Western blot analysis of wild-type and IRE1α-null MEFs. Proteins were prepared from wild-type and IRE1α-null MEFs (lanes 1,2) and from the pancreatic β-cell line HIT-T15 (lane 3). HIT-T15 was treated with 10 μg/mL tunicamycin for 6 h prior to protein harvest. The proteins were subjected to SDS-PAGE and Western blot analysis using anti-IRE1α lumenal-domain antibody. Phosphorylated and nonphosphorylated forms of IRE1α are indicated. (E,F) Northern blot analysis of wild-type and IRE1α-null MEFs. Wild-type, heterozygous, or homozygous IRE1α-null MEFs were treated with or without 10 μg/mL tunicamycin for 6 h prior to harvesting total RNA for Northern blot analysis. One blot was probed with [α-32P]-labeled hamster BiP cDNA and β-actin cDNA (E), and another blot was probed with [α-32P]-labeled mouse GRP94 DNA and β-actin cDNA (F). Quantification of the results showed that tunicamycin induced GRP94 mRNA 6.7-fold and 5.4-fold in wild-type and IRE1α-null MEFs, respectively. (G) BiP reporter gene expression in IRE1α-null MEFs. The reporter plasmids containing the luciferase gene under control of the rat BiP promoter and β-galactosidase under control of the CMV promoter were cotransfected into wild-type and IRE1α-null MEFs. The transfected cells were treated with 2 μg/mL tunicamycin for 16 h prior to harvest. The luciferase activities are presented relative to CMV β-galactosidase activities. Similar results were obtained from three independent experiments.
To test the requirement for IRE1α in UPR-transcriptional induction, wild-type and IRE1α-null MEFs were treated with tunicamycin for 6 h, and RNA was prepared for Northern blot analysis. Both wild-type and heterozygous IRE1α+/− cells showed comparable BiP mRNA induction upon tunicamycin treatment. However, BiP mRNA induction was also observed in homozygous IRE1α-null MEFs, although quantification of the results suggested a slightly reduced induction (10%) in the IRE1α-null MEFs (Fig. 1E). Induction of GRP94 (Fig. 1F) and CHOP-10 (data not shown) mRNAs was also comparable in the wild-type and IRE1α-null MEFs. To determine whether the increase in BiP mRNA observed reflected transcriptional activity of the BiP promoter, the induction of a BiP-promoter–luciferase-reporter plasmid was studied. Tunicamycin treatment induced luciferase expression from the BiP promoter to similar degrees in wild-type and in IRE1α-null MEFs (Fig. 1G). These results support the hypothesis that IRE1α is not essential for the transcriptional induction of several well-characterized UPR target genes and suggest that at least one additional mechanism for UPR transcriptional induction is intact in IRE1α-null MEFs.
5× ATF6 reporter activation is defective in IRE1α-null MEFs
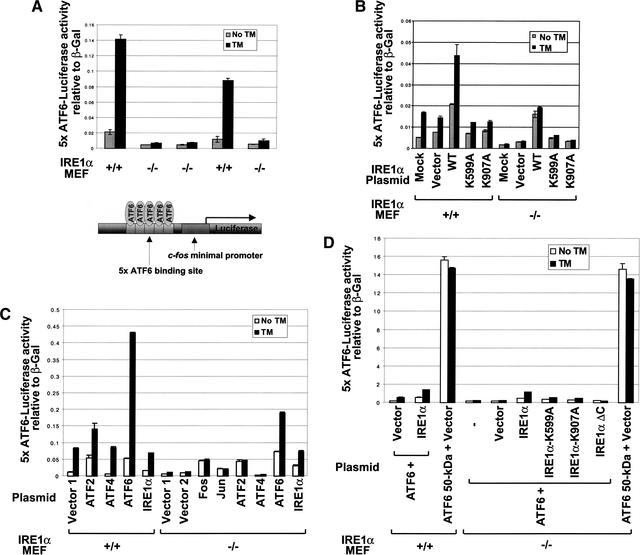
Previous studies indicate that ATF6 cleavage is required for UPR transcriptional induction (Ye et al. 2000). To test whether IRE1α is required for ATF6 cleavage and function, we used a luciferase reporter plasmid under transcriptional control of a multimerized ATF6-binding site (Fig. 2A, bottom). This multimerized ATF6-binding site is sufficient to direct ER stress-induced expression of luciferase (Wang et al. 2000). Previously, we showed that overexpression of wild-type IRE1α activates this 5× ATF6 reporter, whereas overexpression of a kinase and RNase domain-deleted mutant IRE1α (IRE1ΔC) acts in a trans-dominant negative manner to prevent the ER stress-induced expression of the 5× ATF6 reporter (Wang et al. 2000). Surprisingly, compared to wild-type MEFs, tunicamycin-induced expression of the 5× ATF6 reporter gene was completely defective in IRE1α-null MEFs (Fig. 2A). Upon transfection of IRE1α-null MEFs with the 5× ATF6 reporter in the presence of wild-type (WT) IRE1α, kinase-defective K599A mutant IRE1α, or RNase-defective K907A mutant IRE1α, only the wild-type IRE1α complemented the defect in 5× ATF6 reporter expression (Fig. 2B). Therefore, the IRE1α kinase and endoribonuclease activities are required for 5× ATF6 reporter activation. We then tested whether overexpression of several known bZIP/ATF family members could activate 5× ATF6 reporter expression in the IRE1α-null MEFs. Although overexpression of c-Jun, c-Fos, and ATF2 slightly increased the basal level of 5× ATF6 reporter gene expression in the IRE1α-null MEFs, no further increase occurred upon tunicamycin treatment. In contrast, overexpression of intact ATF6 elevated both the basal and the tunicamycin-induced 5× ATF6 reporter gene expression in the IRE1α-null MEFs (Fig. 2C). Tunicamycin-induced expression of the 5× ATF6 reporter gene in IRE1α-null MEFs transfected with wild-type IRE1α was variable, dependent on the tunicamycin concentration and duration of treatment (Fig. 2B,C). Expression of the 50-kD processed form of ATF6 dramatically increased 5× ATF6 reporter activation in both cell types (Fig. 2D). Therefore, overexpression of the 50-kD ATF6 bypassed the IRE1α requirement for 5× ATF6 reporter activation. Because ER stress induction of the 5× ATF6 reporter was completely defective in IRE1α-null MEFs, but could be complemented by overexpression of the 50-kD processed form of ATF6, it was possible that IRE1α was required for ATF6 processing and/or function. Therefore, we studied the requirement for IRE1α in ATF6 cleavage and function.
Figure 2.
5× ATF6 reporter activation is defective in IRE1α-null MEFs. (A) 5× ATF6 reporter gene expression in wild-type and IRE1α-null MEFs. The reporter plasmids containing the luciferase gene under control of 5× ATF6 binding sites and β-galactosidase under control of the CMV promoter were cotransfected into wild-type and IRE1α-null MEFs. The transfected cells were treated with 2 μg/mL tunicamycin for 16 h prior to harvest. The luciferase activities are presented relative to CMV β-galactosidase activities. Similar results were obtained from two independent experiment. (B–D) Wild-type and IRE1α-null MEFs were transfected as in A in the presence of vector alone or vector encoding wild-type IRE1α, kinase-defective (K599A) IRE1α, RNase defective (K907A) IRE1α, C-terminal-deleted IRE1α (IRE1αΔC), ATF2, ATF4, ATF6, processed form of ATF6 (ATF6 50-kD), c-Jun, or c-Fos as indicated. The vector used for IRE1α expression was pEDΔC. The empty vectors used as controls were pEDΔC (B,D), pcDNA3 (Vector 1), and pCMV-HA (Vector 2) (C). MEFs were transfected by either Effectine (B,D) or FuGENE6 (C) according to the manufacturers' recommended procedures. The transfected cells were treated with 10 μg/mL tunicamycin for 6 h (B,D) or 2 μg/mL tunicamycin for 16 h (C) prior to harvest. Similar results were obtained from four independent experiments.
IRE1α is not required for ATF6 cleavage, nuclear translocation, or transcriptional activation
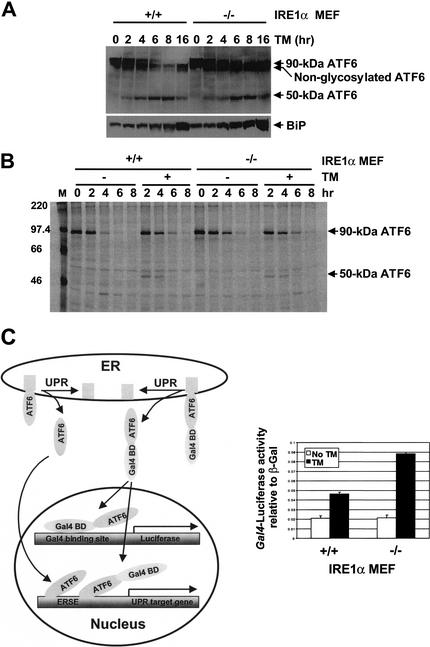
Initial studies showed that IRE1α overexpression in COS-1 cells did not generate the processed form of ATF6 (data not shown). To further analyze the requirement for IRE1α in ATF6 function, we examined ATF6 cleavage by Western blot and radiolabel pulse-chase experiments. Cells were treated with tunicamycin for increasing amounts of time, and ATF6 was monitored by Western blot analysis. The 50-kD processed form of ATF6 was generated at the same rate in both wild-type and IRE1α-null MEFs and accumulated up to 8 h (Fig. 3A, top). BiP protein levels also increased with similar kinetics in the wild-type and IRE1α-null MEFs (Fig. 3A, bottom). To more closely monitor the kinetics of 90-kD ATF6 cleavage and stability, pulse-labeling with [35S]methionine and [35S]cysteine was performed with a chase in the presence or absence of tunicamycin. The labeled ATF6 proteins were immunoprecipitated with anti-ATF6 antibody and subjected to SDS-PAGE and autoradiography (Fig. 3B). The 50-kD processed form of ATF6 was detected in both wild-type and IRE1α-null MEFs after 2 h of tunicamycin treatment. No significant difference in the cleavage and/or stability of ATF6 was detected between wild-type and IRE1α-null MEFs (Fig. 3B). Interestingly, both the intact and processed forms of ATF6 showed a short half-life of ∼2 h.
Figure 3.
IRE1α is not required for ATF6 cleavage, nuclear translocation, or transcriptional activation. (A) Western blot analysis of ATF6. Wild-type and IRE1α-null MEFs were treated with tunicamycin (10 μg/mL) for increasing times, and protein extracts were prepared for Western blot analysis. ATF6 proteins were detected using anti-ATF6 antibody and anti-rabbit immunoglobulin conjugated with horseradish peroxidase and enhanced chemiluminescence. (B) Pulse-chase analysis of ATF6. Wild-type and IRE1α-null MEFs were pulse-labeled with [35S]methionine and [35S]cysteine (0.5 mCi/100-mm dish) for 40 min, and then chase was performed with or without 10 μg/mL tunicamycin for the periods indicated. Proteins were extracted and immunoprecipitated using anti-ATF6 antibody. Immunoprecipitates were subjected to SDS-PAGE, and radiolabeled proteins were visualized using PhosphorImager (Molecular Dynamics). (C) ATF6 cleavage-dependent GAL4 reporter gene expression in wild-type and IRE1α-null MEFs. The reporter plasmids containing the luciferase gene under control of the GAL4 promoter and β-galactosidase under control of the CMV promoter were cotransfected with the GAL4 DNA-binding domain–ATF6 fusion protein expression vector into wild-type and IRE1α-null MEFs. The transfected cells were treated with 2 μg/mL tunicamycin for 16 h prior to harvest. The luciferase activities are presented relative to CMV β-galactosidase activities. Similar results were obtained from two independent experiments. The diagram on the left depicts ATF6 cleavage-dependent GAL4 reporter gene expression that is independent from the transcriptional activity of endogenous ATF6.
To test whether ATF6 nuclear translocation and activation require IRE1α, a GAL4 transactivation assay was used. The GAL4 DNA-binding domain was fused to the N terminus of full-length ATF6. This expression vector was transfected into wild-type and IRE1α-null MEFs with a luciferase reporter construct under transcriptional control of five GAL4 DNA-binding sites. Under these conditions, the expression of luciferase is dependent on binding of the Gal4–ATF6 fusion protein liberated from the ER membrane (Fig. 3C, diagram). After cotransfection the cells were treated with tunicamycin. Tunicamycin induced luciferase expression in both wild-type and mutant MEFs to a similar degree, suggesting that cleavage, nuclear translocation, and transcriptional activation of ATF6 are independent of IRE1α function (Fig. 3C). Therefore, by all these analyses, ATF6 processing and function were not defective in the IRE1α-null MEFs. These results led us to study whether another factor is defective in the IRE1α-null MEFs that is required for transcriptional activation of the 5× ATF6 reporter.
5× ATF6 reporter induction requires IRE1α-dependent splicing of XBP1 mRNA
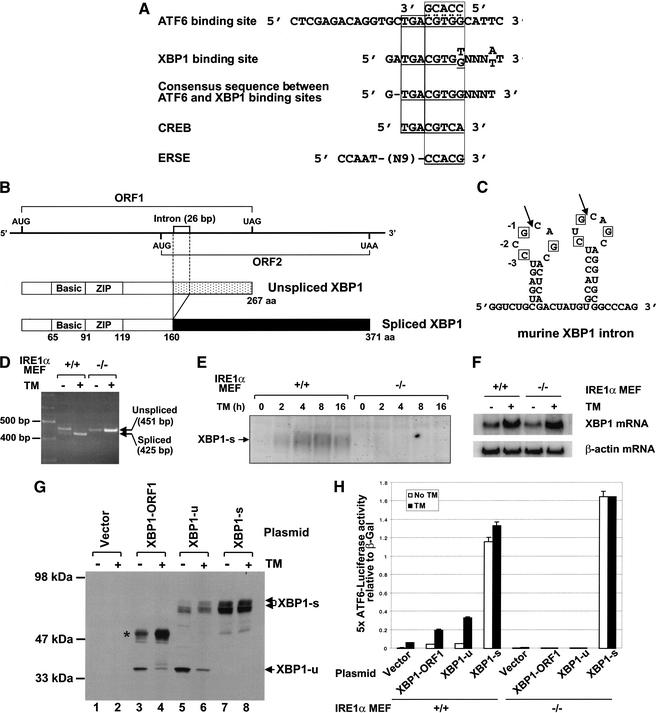
XBP1 is a bZIP transcription factor of the CREB/ATF protein family that binds to an identical sequence motif as ATF6 (Fig. 4A; Clauss et al. 1996). Indeed, XBP1 was also isolated as an ERSE-binding factor in the same yeast one-hybrid screen used to identify ATF6 (Haze et al. 1999). During the course of our studies, we discovered that two protein products are derived from the Caenorhabditis elegans and human XBP1 mRNAs, where the larger product is translated from a spliced form of XBP1 mRNA that is generated upon ER stress (Shen et al. 2001; Yoshida et al 2001b). Therefore, we used the sequence information to clone the full-length cDNA for murine XBP1. The murine XBP1 gene structure is very similar to the human XBP1, having conserved two open reading frames, an intron, and a bZIP domain in the N terminus (Fig. 4B,C). The translation products from the first and second open reading frames (ORFs) consist of 267 and 222 amino acids in the mouse and 261 and 212 amino acids in the human, respectively. Splicing of the 26-nt intron would generate a frame-shift and a fusion of the first ORF to the second ORF, to yield a larger protein product of 371 and 376 amino acids in the mouse and human, respectively. Only one base differs between the human and murine 26-base intron. RT–PCR analysis of RNA isolated from tunicamycin-treated wild-type and IRE1α-null MEFs using PCR primers designed to amplify the region encompassing the overlap between ORF1 and ORF2 showed that XBP1 mRNA splicing is induced by ER stress and requires IRE1α (Fig. 4D). DNA sequence analysis confirmed the removal of 26 nucleotides from the shorter RT–PCR product. The 425-nt fragment from spliced XBP1 mRNA was detected in wild-type MEFs after tunicamycin treatment. In contrast, this spliced form of XBP1 mRNA was not detected in IRE1α-null MEFs before or after tunicamycin treatment. Western blot analysis using an antibody that reacts with only the longer XBP1 product derived from the spliced XBP1 mRNA showed that a 55-kD heterogeneous-sized species appeared with time after tunicamycin treatment in wild-type MEFs (Fig. 4E). This polypeptide was not detected before tunicamycin treatment (Fig. 4E). Although a small amount of the spliced XBP1 mRNA was detected by RT–PCR prior to tunicamycin treatment, this analysis was not quantitative. Therefore, we cannot correlate the presence of the spliced mRNA with protein expression. This polypeptide was not detected in the IRE1α-null MEFs (Fig. 4E). We propose that this 55-kD protein is translated from XBP1 mRNA that is spliced in an IRE1α-dependent reaction. As expected from the presence of ERSE in the XBP1 promoter and correct ATF6 processing in IRE1α-null MEFs, XBP1 mRNA was induced with tunicamycin treatment in IRE1α-null MEFs (Fig. 4F).
Figure 4.
5× ATF6 reporter induction requires IRE1α-dependent splicing of XBP1 mRNA. (A) Alignment of ATF6, XBP1, CREB, and ERSE DNA sequence motifs. The entire oligonucleotide sequence used to construct the 5× ATF6 reporter is shown. The 5′ sequence located outside of the boxed region is the fixed flanking sequence used to generate random oligonucleotides (Wang et al. 2000). (B) Schematic representation of unspliced and spliced forms of the murine XBP1 mRNA and protein coding regions. The translated portion of the two open reading frames, the 26-bp intron, and the bZIP domains are depicted. (C) The predicted mRNA secondary structure at the splice-site junctions in XBP1 mRNA. The 3 residues important for cleavage of HAC1 mRNA by Ire1p (−1G, −3C, and +3G) are conserved in the 5′ and 3′ loops. (D) RT–PCR analysis of XBP1 mRNA splicing using RNA templates from tunicamycin-treated wild-type and IRE1α-null MEFs. (E) Western blot analysis of XBP1. Cell extracts were prepared from wild-type and IRE1α-null MEFs cultured in the presence or absence of tunicamycin (10 μg/mL) with MG132 (10 μM) for increasing times as indicated. (F) Northern blot analysis of XBP1 mRNA in IRE1α-null MEFs. Wild-type and IRE1α-null MEFs were treated with or without 10 μg/mL tunicamycin for 6 h prior to harvesting total RNA for Northern blot analysis. The blots were probed with the [α-32P]-labeled 0.94-kb XhoI fragment of XBP1-u and β-actin cDNA. Quantification of the results showed 3.1-fold and 4.0-fold induction with tunicamycin treatment in wild-type and IRE1α-null MEFs, respectively. (G) Western blot analysis of XBP1. The 5× ATF6 reporter plasmid and β-galactosidase under control of the CMV promoter were cotransfected into COS-1 cells in the presence of the CMV-promoter-driven unspliced form of XBP1 (XBP1-u), the spliced form of XBP1 (XBP1-s), or the first ORF of XBP1 (XBP1–ORF1) as indicated. Cells were treated with or without tunicamycin (2 μg/mL) for 8 h before harvest. Lactacystin (10 μM) was added to the media for the final 2 h. An XBP1-reactive polypeptide likely derived from using a cryptic 3′ splice site is indicated with an asterisk. (H) The 5× ATF6 reporter is activated by IRE1α-dependent XBP1 mRNA splicing. Wild-type and IRE1α-null MEFs were transfected and assayed as described in Figure 2 in the presence of the CMV-promoter-driven unspliced form of XBP1 (XBP1-u), the spliced form of XBP1 (XBP1-s), or the first ORF of XBP1 (XBP1–ORF1).
If the defect in XBP1 mRNA splicing is responsible for the defect in 5× ATF6 reporter induction in the IRE1α-null MEFs, then expression of the spliced form of XBP1 mRNA, but not the unspliced form, should complement the 5× ATF6 reporter defect in the IRE1α-null MEFs. cDNAs encoding XBP1–ORF1 alone, XBP1-u (unspliced form of XBP1), and XBP1-s (spliced form of XBP1) were inserted behind the CMV promoter to direct their expression in transiently transfected COS-1 cells. Western blot analysis with antibody reactive to the N terminus of XBP1 detected a polypeptide of ∼35 kD in COS-1 cells transfected with the XBP1–ORF1 expression vector (Fig. 4G, lane 3). The 35-kD polypeptide decreased upon tunicamycin treatment (Fig. 4G, lane 4), likely a consequence of decreased mRNA encoding the 35-kD polypeptide owing to splicing of XBP1 mRNA. In addition, a 48-kD species (Fig. 4G, lane 4, asterisk) was induced upon tunicamycin treatment. The 48-kD species may represent products from aberrantly spliced mRNAs that use the 5′-splice-site junction in XBP1 mRNA and a downstream cryptic 3′ splice site. Similar analysis of XBP1-u-transfected cells detected the 35-kD polypeptide in addition to a heterogeneous 55-kD species representing XBP1-s. In contrast, cells transfected with XBP1-s produced only the latter 55-kD species, and its expression level did not change with tunicamycin treatment, likely because the CMV promoter is not induced by the UPR. These results showed that each of the expression plasmids directed the expression of the expected polypeptide.
We then measured the effect of these expression vectors when cotransfected with the 5× ATF6 luciferase reporter gene into wild-type and IRE1α-null MEFs. Expression of either XBP1–ORF1 or intact unspliced XBP1-u slightly increased both the basal and tunicamycin-induced expression from the 5× ATF6 luciferase reporter gene in wild-type MEFs. In contrast, expression of XBP1-s greatly increased 5× ATF6 reporter gene expression in the wild-type MEFs, even in the absence of ER stress (Fig. 4H). Qualitatively similar results were obtained from cotransfection experiments in COS-1 cells (data not shown). Strikingly, only XBP1-s complemented the 5× ATF6 reporter expression in the IRE1α-null MEFs. These results show that expression of the spliced form of XBP1 mRNA is necessary and sufficient to activate the 5× ATF6 reporter gene in the IRE1α-null MEFs.
XBP1 mRNA is a substrate of RNase activity of IRE1α in vitro
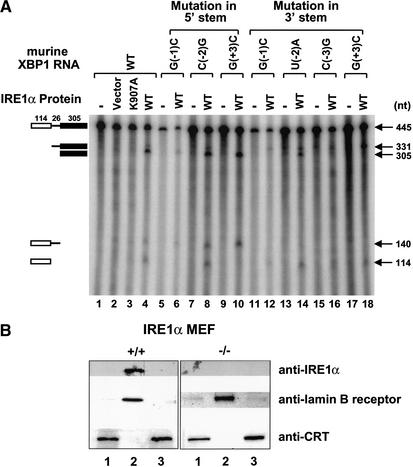
The predicted RNA structure of the XBP1 intron shows stem–loop hairpins with 7-membered rings at both the 5′- and 3′-splice-site junctions as observed in yeast HAC1 mRNA (Fig. 4C). Site-directed mutagenesis studies identified three residues (−1G, −3C, +3G) that are critical for cleavage of yeast HAC1 mRNA by Ire1p (Kawahara et al. 1998; Gonzalez et al. 1999). These bases are conserved in the 5′ and 3′ loops of XBP1 mRNA (boxed in Fig. 4C). We tested whether XBP1 mRNA is a direct substrate of the endoribonuclease activity of IRE1α in vitro and whether these conserved residues are required. Wild-type and mutant XBP1 RNA substrates were transcribed in vitro and incubated with human IRE1α protein expressed in transfected COS-1 cells. Wild-type substrate was cleaved at both 5′- and 3′-splice-site junctions (Fig. 5A, lane 4). Cleavage of the RNA at the 5′ or 3′ splice site was prevented by mutation of the conserved residues within the 5′ loop (−1G and +3G, Fig. 5A, lanes 6,10) or within the 3′ loop (−1G, −3C, and +3G, Fig. 5A, lanes 12,16,18), respectively. Mutation of the conserved residues in the 5′ loop did not prevent cleavage of the 3′ splice site and vice versa for mutations in the 3′ loop. In contrast, mutation of the nonconserved residue within the 5′ loop (−2C) or the 3′ loop (−2U) did not affect the cleavage of XBP1 RNA by IRE1α (Fig. 5A, lanes 8,14). Taken together, these results indicate that both 5′- and 3′-splice-site junctions in XBP1 RNA are cleaved by IRE1α upon ER stress to eventually generate a spliced product that encodes a larger translated protein with a greater transactivation potential.
Figure 5.
IRE1α cleaves both splice-site junctions in XBP1 RNA in vitro and is localized to the inner nuclear envelope. (A) 32P-labeled wild-type and mutant XBP1 RNAs were prepared and incubated with immunoprecipitated wild-type or RNase-defective (K907A) IRE1α protein in nuclease buffer and analyzed by electrophoresis on a denaturing polyacrylamide gel. The 5′ exon (114 nt), intron (26 nt), and 3′ exon (305 nt) cleavage products of the substrate are marked on the left. The numbers on the right are the expected nucleotide sizes. (B) Intracellular localization of IRE1α. Wild-type and IRE1α-null MEFs were fractionated as described in Materials and Methods. Western blot analysis was performed with mouse anti-IRE1α, human anti-lamin B receptor, or rabbit anti-calreticulin antibodies. (Lane 1) Cellular extract; (lane 2) nuclei with inner nuclear membrane; (lane 3) Triton X-100 soluble, microsomal, and outer nuclear membrane fraction.
IRE1α localizes to the inner nuclear envelope
Previous studies suggest that the IRE1-mediated HAC1 mRNA splicing reaction may occur within the cytoplasm or the nucleus (Chapman and Walter 1997; Ruegsegger et al. 2001). To provide insight into this question, we performed cell fractionation to localize IRE1α. Nuclei were isolated and their outer membranes were stripped as described in Materials and Methods. Western blot analysis of lamin B receptor showed enrichment in the Triton X-100-insoluble fractions containing nuclei with the inner nuclear membrane (Fig. 5B). Lamin B was absent from the microsomal fraction containing the outer nuclear envelope. In contrast, calreticulin, a lumenal ER protein, was associated with the microsomal fraction. These results indicated that the nuclear and microsomal fractions isolated did not have significant contamination. Interestingly, IRE1α was greatly enriched in the nuclear pellet that was stripped of outer nuclear membranes. Importantly, the immunoreactivity was not detected in fractions isolated from IRE1α-null MEFs. These results support the hypothesis that the majority of IRE1α is localized to the inner nuclear envelope.
IRE1α-mediated UPR transcriptional induction requires ATF6 cleavage
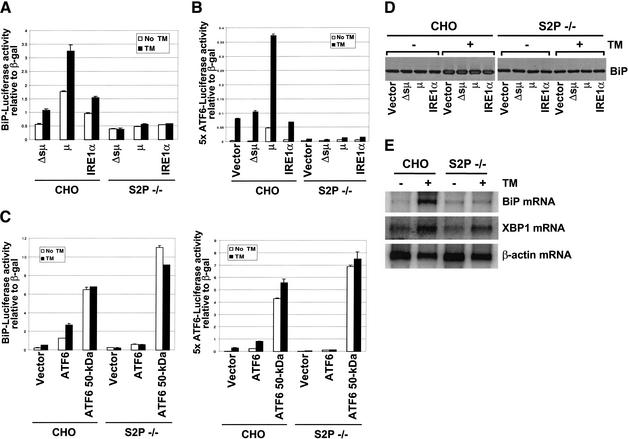
Site-1 protease (S1P) and site-2 protease (S2P) are implicated in the cleavage of ATF6 to generate the 50-kD cytosolic fragment upon ER stress. Indeed, ATF6 cleavage was not detected in S2P-deficient CHO cells upon activation of the UPR (Ye et al. 2000). To test the requirement for ATF6 cleavage in IRE1α-mediated UPR transcriptional induction, we studied IRE1α overexpression in S2P-deficient CHO cells. Overexpression constitutively activates IRE1α by promoting dimer/oligomer formation and trans-autophosphorylation. An IRE1α expression vector was introduced into S2P-deficient CHO cells with a BiP promoter reporter plasmid or the 5× ATF6 reporter plasmid. IRE1α transfection in wild-type CHO cells increased BiP-reporter expression by 70% compared to cells transfected with the immunoglobulin μ heavy chain deleted of the signal peptide (Δsμ; Fig. 6A; Wood et al. 1990). In contrast, IRE1α transfection increased BiP reporter expression by 38% in S2P-deficient CHO cells. IRE1α overexpression reproducibly increased BiP reporter expression to a lower level in S2P-deficient CHO cells, suggesting that maximal IRE1α-mediated transcriptional induction requires S2P-dependent cleavage of ATF6. BiP expression was further increased by tunicamycin treatment in wild-type cells, but not in S2P-deficient CHO cells (Fig. 6A). Similarly, overexpression of the immunoglobulin μ heavy chain, a known inducer of the UPR (Wood et al. 1990), increased BiP-reporter expression 208% in wild-type cells and only 23% in S2P-deficient CHO cells. In addition, overexpression of either IRE1α or the immunoglobulin μ heavy chain was not able to activate the 5× ATF6 reporter expression plasmid in S2P-deficient CHO cells, even in the presence of tunicamycin treatment (Fig. 6B). Northern and Western blot analysis of BiP in wild-type and S2P-deficient CHO cells revealed that S2P-dependent ATF6 processing is required for BiP induction upon tunicamycin-induced ER stress (Fig. 6D,E). Indeed, overexpression of the 50-kD processed form of ATF6, but not the full-length ATF6, rescued the UPR defect in S2P-deficient CHO cells (monitored by BiP-reporter or 5× ATF6-reporter expression) (Fig. 6C). BiP expression was not noticeably changed by overexpression of IRE1α or the immunoglobulin μ heavy chain even in wild-type CHO cells, probably because of the low transfection efficiency (Fig. 6D). These results indicate that ATF6 cleavage is required for induction of both IRE1α-dependent and ER-stress-activated target genes. Finally, XBP mRNA was induced in S2P-deficient CHO cells by tunicamycin treatment (Fig. 6E), suggesting that XBP1 mRNA expression is regulated by IRE1α-dependent XBP1 mRNA splicing, in addition to ATF6 cleavage (see Discussion).
Figure 6.
IRE1α-mediated induction of UPR genes requires ATF6 cleavage. (A–C) BiP reporter gene (A,C) and 5× ATF6 reporter gene (B,C) expression in S2P-deficient CHO cells. The reporter plasmids containing the luciferase gene under control of the rat BiP promoter or the 5× ATF6 binding sites were cotransfected with β-galactosidase under control of the CMV promoter and an IRE1α (A,B) or ATF6 (C) expression vector into S2P-deficient CHO cells. Immunoglobulin μ heavy chain (μ) and mutant immunoglobulin μ heavy chain deleted of the signal peptide (Δsμ) were used as positive and negative ER stress inducers, respectively. At 32 h posttransfection, cells were treated with 2 μg/mL tunicamycin for 16 h prior to harvest. The luciferase activities are presented relative to CMV β-galactosidase activities. Similar results were obtained from two independent experiments. (D) Western blot analysis of BiP. Wild-type and S2P-deficient CHO cells were transfected with plasmids as indicated. At 32 h posttransfection, the transfected cells were treated with 2 μg/mL tunicamycin for 16 h, harvested, and analyzed by Western blot analysis using anti-BiP antibody. (E) Northern blot analysis of BiP and XBP1 mRNA in S2P-deficient cells. Wild-type and S2P-deficient CHO cells were treated with or without 2 μg/mL tunicamycin for 16 h prior to harvesting total RNA for Northern blot analysis using hamster BiP, XBP1-u, and β-actin cDNAs as probes. Quantification of the results showed that tunicamycin induced BiP mRNA 34-fold and 2.6-fold and XBP1 mRNA 3.1-fold and 2.9-fold in wild-type and S2P-deficient CHO cells, respectively.
Discussion
We have characterized the UPR in IRE1α-null MEFs and S2P-deficient CHO cells to elucidate how IRE1α-mediated signaling and ATF6 processing coordinate transcriptional activation of target genes. Our major findings support the following eight conclusions. (1) A UPR transcriptional defect exists in IRE1α-null MEFs. (2) The defect is caused by the absence of a novel ER-stress-induced translation product from the XBP1 locus. (3) The novel translation product requires IRE1α-mediated removal of a 26-nt intron within XBP1 mRNA. (4) IRE1 RNA cleavage specificity is conserved between mammalian and yeast substrates XBP1 and HAC1. (5) IRE1α is preferentially localized to the inner leaflet of the nuclear envelope. (6) IRE1α is not required for ATF6 cleavage or nuclear translocation and/or activation. (7) Both ER-stress and IRE1α signaling of the UPR require S2P-dependent cleavage of ATF6. (8) The two pathways signaling through the ER stress sensors IRE1 and ATF6 merge through qualitative and quantitative regulation of XBP1 to activate the UPR.
Previous data suggested that IRE1α is not required for signaling the mammalian UPR (Urano et al. 2000a,b; Fig. 1). However, our results described here, using a more specific UPR reporter having only one DNA sequence motif, is the first evidence to support a requirement for IRE1α in signaling the UPR. Although a promoter from a UPR-inducible cellular gene for which activation is defective in the IRE1α-null MEFs has yet to be identified, present ongoing microarray analysis should identify such genes. It is likely that cellular promoters have evolved multiple motifs and deletion of a single trans-acting factor may not significantly reduce UPR transcriptional induction. For example, it was recently shown that ire-1 deletion is synthetically lethal with pek-1 deletion and that these two pathways synergize for UPR transcriptional induction in C. elegans (Shen et al. 2001). In addition, the induction of approximately one-third of all UPR genes was defective in MEFs in the absence of the PERK signaling pathway (Scheuner et al. 2001). Our present understanding of the mammalian UPR leads us to propose that IRE1, PERK, and ATF6 are proximal sensors that regulate the production and/or quality of a class of bZIP-containing transcription factors that may form homo- and heterodimers. Combinatorial interactions of these factors, such as XBP1, ATF4, and ATF6, may generate diversity in responses for different subsets of UPR-responsive genes.
The 5× ATF6 reporter contains five copies of a consensus ATF6-binding site known to bind ATF6 in vitro (Wang et al. 2000). Transcriptional induction from these motifs was dependent on both IRE1α and S2P activity, suggesting that the endogenous XBP1 alone cannot activate the 5× ATF6 reporter in the absence of ATF6 cleavage. We propose that ATF6 is required to induce XBP1 transcription, thereby increasing the amount of substrate for IRE1α-mediated splicing. As a consequence, elevated levels of highly active XBP1 would activate this 5× ATF6 reporter gene. Under this hypothesis, IRE1α would primarily act on newly transcribed XBP1 mRNA, consistent with its preferential localization to the inner nuclear envelope. Alternatively, XBP1 and ATF6 heterodimers may be the most potent activators of UPR transcription; however, upon overexpression, either homodimer alone might activate the UPR. In support of both hypotheses, overexpression of XBP1-s activates the UPR in the absence of ATF6 cleavage (data not shown).
Our results support the idea that the spliced form of XBP1 (XBP1-s) is a better transcriptional activator than XBP1-u. Wild-type cells transfected with XBP1-u or XBP1–ORF1 showed similar levels of 5× ATF6 reporter gene expression in the absence of tunicamycin treatment, but higher induction was observed in XBP1-u-expressing cells upon tunicamycin treatment. These findings support recent results and conclusions (Yoshida et al. 2001b). Therefore, IRE1α-dependent RNA splicing produces a novel protein that is a more potent transcriptional activator. This is similar to observations in S. cerevisiae where Ire1p-mediated splicing of HAC1 mRNA generates a new C terminus on Hac1p that converts it into a better transcriptional activator (Mori et al. 2000).
Previously, XBP1-binding sites were also selected in vitro to generate a consensus 8-bp core motif that is very similar to CREB sites and identical to the ATF6-binding site (Fig. 4A; Clauss et al. 1996). Both ATF6- and XBP1-binding sites reside in the 3′ portion of the ERSE in reverse orientation (CCACG). Although ATF6 and XBP1 did not bind the ERSE directly in vitro, ATF6 and XBP1 binding was detected in the presence of NF-Y/CBF bound to the 5′ half site of the ERSE (CCAAT; Li et al. 2000; Yoshida et al. 2000, 2001b). Future studies are required to compare the binding affinities of XBP1 and ATF6 for the ERSE and to characterize the influence of NF-Y/CBF.
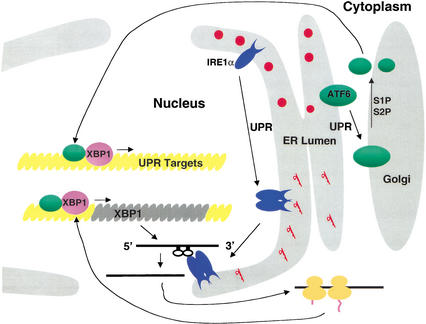
The UPR has evolved a positive feedback loop for trancriptional activation of XBP1 (Fig. 7). Expression of XBP1 mRNA was induced in S2P-deficient CHO cells upon activation of the UPR. Because XBP1 can bind to the ERSE (Yoshida et al. 2000, 2001b) and the XBP1 promoter contains an ERSE, we propose that the UPR induction of the XBP1 promoter in S2P-deficient cells is mediated through IRE1α splicing of XBP1 mRNA. Spliced XBP1 mRNA would produce a protein that activates its own promoter, as previously suggested (Reimold et al. 1996). Although this step is ATF6-independent, ATF6 cleavage and activation could further increase the rate and/or extent of XBP1 transcription or generate a more potent transcriptional activator by formation of heterodimers. These results support the conclusion that either IRE1α or ATF6 alone is not required for XBP1 induction.
Figure 7.
ATF6- and IRE1α-dependent UPR signaling pathways merge through regulation of the quantity and quality, respectively, of XBP1 protein. The model depicts the activation of two proximal sensors of the UPR, ATF6 and IRE1α, upon ER stress. Upon accumulation of unfolded proteins in the ER lumen, ATF6 leaves the ER to enter the Golgi apparatus, where it is cleaved by S1P and then S2P to release a 50-kD fragment that enters the nucleus through the nuclear pore. p50-ATF6 then interacts with ERSE motifs to activate transcription. Simultaneously and independently, the UPR induces dimerization, autophosphorylation, and activation of the RNase activity of IRE1α that is localized at the inner leaflet of the nuclear envelope. Activated IRE1α then initiates splicing of XBP1 mRNA to generate a potent transcriptional activator, XBP1-s, that also enters the nuclear pore to activate transcription from ERSE motifs. The status of XBP1-s and p50-ATF6 when bound to the ERSE is not known, but for simplicity they are depicted as heterodimers.
XBP1 is ubiquitously expressed in adult tissues, and disruption of the XBP1 gene revealed that it is essential for hepatocyte differentiation (Reimold et al. 2000), cardiomyocyte survival (Masaki et al. 1999), and plasma cell differentiation (Reimold et al. 2001). It is especially intriguing that XBP1 is the only factor identified to date that is required for B-cell differentiation into plasma cells. This differentiation involves a fivefold expansion of the ER compartment to accommodate the increased high level of immunoglobulin secretion (Wiest et al. 1990). Activated B-cells are driven to become plasma cells by signaling through the CD-40 receptor or through mitogens such as lipopolysaccharide (LPS), stimulation. Interestingly, XBP1 mRNA is increased by anti-CD40 antibody or LPS treatment, suggesting that the UPR and the signaling pathway for plasma cell differentiation cooperate in the induction and splicing of XBP1 mRNA (Reimold et al. 2001). Studies are in progress to determine whether IRE1α signaling and/or ATF6 activation are required for plasma cell differentiation and/or survival, and how signaling is transmitted. Comparison of gene expression patterns in cells lacking IRE1, XBP1, and ATF6 under various conditions will elucidate how these pathways cooperate in their signaling. The identification of XBP1 as a signaling molecule downstream of IRE1 and ATF6 in the UPR provides credence to the hypothesis that the UPR is required for expansion of the ER compartment upon differentiation of cells that secrete high levels of protein.
Materials and methods
Cell culture and transient DNA transfection
Culture methods and media for COS-1 monkey cells were previously described (Kaufman 1997). The same methods were applied to MEFs except that fetal bovine serum (FBS) was not heat-inactivated. Wild-type (K1) and S2P-deficient (clone M19) Chinese hamster ovary (CHO) cells were cultured as described (Ye et al. 2000). R1 murine embryonic stem (ES) cells (Joyner et al. 1989), generously provided by Linda Samuelson (University of Michigan), were plated onto mitomycin C-treated MEF feeder cells in ES cell medium (Dulbecco’s Modified Eagle Medium, GIBCO BRL) supplemented with 15% heat-inactivated FBS, 0.1 mM β-mercaptoethanol, and 1000 units/mL Leukocyte Inhibitory Factor (GIBCO BRL). COS-1 cells were transfected by either diethylaminoethyl(DEAE)-dextran (Kaufman 1997) or calcium-phosphate-BES methods (Ausubel et al. 1999). MEFs were transfected by either FuGENE6 (Roche) or Effectine (QIAGEN) according to the manufacturers' recommended procedures. CHO cells were transfected by FuGENE6 (Roche).
Construction of IRE1α targeting vector and gene disruption
An XbaI–NotI fragment of a loxP neomycin resistance cassette under control of the phosphoglycerate kinase (PGK) promoter (Orban et al. 1992) was inserted into a murine IRE1α fragment to replace exons 7 to 14, yielding the BS-mIRE1α targeting vector. Trypsinized R1 ES cells were mixed with NotI-digested BS-mIRE1α targeting vector, and a high electric pulse (250 μF and 0.3 kV) was applied using a gene-pulser (Bio-Rad laboratories). The transfected cells were plated onto MEF feeder cells at a density of 106 cells/100-mm plate. Selection medium containing 300 μg/mL G418 (GIBCO BRL) was applied to the ES cells at 48 h posttransfection. G418-resistant colonies formed at 4–5 d after selection were isolated for screening.
RT–PCR and plasmid construction
XBP1 RNA splicing was detected by standard RT–PCR using total RNA templates isolated from MEFs treated with or without tunicamycin (10 μg/mL, 6 h) using oligo(dT)15 and specific primers; mXBP1-354 (5′-CCTTGTGGTTGAGAACCAGG-3′) and mXBP1-804-AS (5′-CTAGAGGCTTGGTGTATAC-3′). The spliced form of XBP1 cDNA was obtained by RT–PCR using RNA templates obtained from MEFs treated with tunicamycin and oligo(dT)15, mXBP1-354, and mXBP1–1150-R1 (5′CGAATTCTTAGACACTAATCAGC-3′) as primers. The spliced form of XBP1 cDNA, pcDNA3-XBP1-s, was constructed by subcloning the 0.7-kb BamHI–EcoRI RT–PCR fragment from XBP1 into the respective sites in pcDNA3–XBP1–ORF1 (from Laurie Glimcher, Harvard School Public Health, Boston). The unspliced form of full-length XBP1, pcDNA3-XBP1-u, was constructed using RNA templates obtained from IRE1α-null MEFs without tunicamycin treatment. DNA sequence analysis was performed to verify PCR-amplified DNA sequences.
Pulse-chase analysis of ATF6
Wild-type and IRE1α-null MEFs cultured on 100-mm plates were pulse-labeled with [35S]methionine and [35S]cysteine (0.5 mCi/100-mm dish, 1000 Ci/mmole, Amersham Pharmacia) for 40 min; then chase was performed with or without 10 μg/mL tunicamycin for the times indicated. Proteins were extracted and immunoprecipitated using anti-ATF6 antibody as previously described (Haze et al. 1999) and subjected to SDS-PAGE (10% gel). Radiolabeled proteins were analyzed using a PhosphorImager (Molecular Dynamics).
5× ATF6, BiP, and GAL4 reporter assays
The reporter plasmids containing the luciferase gene under control of five ATF6-binding sites or the GAL4 DNA-binding site (Wang et al. 2000) and the BiP promoter (Tirasophon et al. 1998) were previously described. Reporter assays were performed as previously described (Tirasophon et al. 2000) with the exception that a plasmid containing β-galactosidase under control of the CMV promoter was used to correct for transfection efficiency.
Southern and Northern blot analysis
Southern and Northern blot analysis followed standard procedures (Sambrook et al. 1989). 32P-labeled probes were prepared using a random prime labeling system (Amersham Pharmacia). A 0.5-kb BamHI–XhoI fragment from the BS-mIRE1α targeting vector or a 3.6-kb EcoRI–XbaI fragment from pED-hIRE1α cDNA (Tirasophon et al. 2000) was used for Southern and Northern analysis, respectively. The probes for Northern analysis of mXBP1, BiP, and GRP94 were a 0.94-kb XhoI fragment of pcDNA–mXBP1-u, the EcoRI–PstI fragment of hamster BiP (Ting et al. 1987), and a 146-bp PCR fragment of mouse GRP94 (142–287 of the coding region), respectively.
Immunoprecipitation and Western blot analysis
For analysis of ATF6, cells were directly harvested in SDS sample buffer lacking DTT and subjected to Western blot analysis or immunoprecipitation as previously described (Haze et al. 1999). ATF6 proteins were detected using purified anti-ATF6 antibody and anti-rabbit immunoglobulin conjugated with horseradish peroxidase (Amersham Pharmacia). For analysis of IRE1α, total cell extracts were prepared from MEFs, a pancretic β-cell line HIT-T15, or transfected COS-1 cells using Nonidet P-40 lysis buffer (1% NP-40, 50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.05% SDS) supplemented with protease inhibitors (Complete Mini, Roche), 0.1 mM sodium vanadate, and 1 mM sodium fluoride. Western blot analysis of IRE1α using anti-hIRE1α-lumenal-domain antibody and immunoprecipitation of T7-tagged IRE1α proteins using anti-T7 antibody were previously described (Tirasophon et al. 2000). XBP1 proteins were detected using anti-XBP1-s antibody (Fig. 4E; Yoshida et al. 2001b) or purified rabbit anti-XBP1 antibody (Fig. 4G; generously provided by Laurie Glimcher, Harvard School Public Health, Boston).
In vitro cleavage of XBP1 mRNA
In vitro cleavage of murine XBP1 mRNA was performed as previously described by Sidrauski and Walter (1997). Briefly, a 404-bp BamHI and EcoRI fragment of XBP1 DNA that contains the intron was amplified by PCR and subcloned into the same sites of pSPT19 (Roche) that contain the T7 RNA polymerase promoter. Because of polylinker sites between the T7 promoter and 5′ end of the XBP1 fragment, a 445-base-long transcribed RNA fragment is expected. Mutant XBP1 DNA fragments were created by overlapping PCR using mutant oligonucleotides: mXBP1–5′G(−1)C, 5′-TCTGCTGAGTCCCCAGCAC-3′; mXBP1–5′G(−1)C-AS, 5′-GTGCTGGGGACTCAGCAGA 3′; mXBP1–5′C(−2)G, 5′-TCTGCTGAGTCGGCAGCAC-3′; mXBP1–5′C(−2)G-AS, 5′-GTGCTGCCGACTCAGCAGA-3′; mXBP1–5′G(+3)C, 5′-GTCCGCACCACTCAGACTAT-3′; mXBP1–5′G(+3)C-AS, 5′-ATAGTCTGAGTGGTGCGGAC-3′; mXBP1–3′G(−1)C, 5′-ATGTGCACCTCTCCAGCAG-3′; mXBP1–3′G(−1)C-AS, 5′-CTGCTGGAGAGGTGCACAT-3′; mXBP1–3′T(−2)A, 5′-ATGTGCACCTCAGCAGCAG-3′; mXBP1–3′T(−2)A-AS, 5′-CT GCTGCTGAGGTGCACAT-3′; mXBP1–3′C(−3)G, 5′-ATGTG CACCTGTGCAGCAG-3′; mXBP1–3′C(−3)G-AS, 5′-CTGCTGC ACAGGTGCACAT-3′; mXBP1–3′G(+3)C, 5′-CTCTGCACCAG GTGCAGGC-3′; mXBP1–3′G(+3)C-AS, 5′-GCCTGCACCTGGT GCAGAG-3′. XBP1 RNA was transcribed in vitro using T7 RNA polymerase (Roche) in the presence of [32P]UTP (3000 Ci/mmole, Amersham Pharmacia). The 32P-labeled XBP1 RNA was purified by electrophoresis in a 5% denaturing polyacrylamide gel, eluted, precipitated, and dissolved in endoribonuclease buffer (20 mM HEPES at pH 7.3, 1 mM dithiothreitol, 10 mM magnesium acetate, 50 mM potassium acetate, 2 mM ATP). Purified RNA (3 × 104 cpm) was added to the immunoprecipitated wild-type and endoribonuclease mutant K907A hIRE1α, which contain T7-epitope tags at their C termini (Tirasophon et al. 2000), and incubated at 30°C for 1 h. The reactions were terminated by extraction with phenol/chloroform, precipitated with ethanol, and analyzed by electrophoresis on 5% denaturing polyacrylamide gels. Gels were dried prior to autoradiography.
Isolation and extraction of nuclei
Nuclei were isolated from MEFs as described (Blobel and Potter 1966). Cells were homogenized in two volumes of a solution containing 250 mM sucrose, 25 mM KCl, 5 mM MgCl2, and 50 mM Tris (pH 7.5). The homogenate was overlaid on a step sucrose gradient consisting of 1.62 M and 2.3 M sucrose and centrifuged at 124,000g for 30 min using a Beckman SW50.1 rotor. The white pellet containing pure nuclei was collected, suspended in a solution containing 25 mM KCl, 5 mM MgCl2, and 50 mM Tris (pH 7.5), and centrifuged at 13,000g for 12 min. The pellet contained pure, intact nuclei. To remove the outer nuclear membranes, the purified nuclei were solubilized with 5% Triton X-100 in 25 mM KCl, 5 mM MgCl2, and 50 mM Tris (pH 7.5) before being centrifuged at 800g for 5 min. The supernatant (Triton X-100 soluble fraction) contained solubilized outer nuclear membrane. The final pellet, containing the outer membrane-stripped nuclei, was suspended in a solution containing 25 mM KCl, 5 mM MgCl2, and 50 mM Tris (pH 7.5) and centrifuged at 13,000g for 10 min. Quality of the isolated nuclei was monitored by electron microscopy (Blobel and Potter 1966).
Acknowledgments
We gratefully thank Joseph Goldstein for providing the S2P-deficient CHO cells and Laurie Glimcher for providing XBP1 reagents and for comments on this work. We thank the members of the Kaufman laboratory for critical input. Portions of this work were supported by NIH grant AI 42394 (R.J.K.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kaufmanr@umich.edu; FAX (734) 763-9323.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.964702.
References
- Aridor M, Balch WE. Integration of endoplasmic reticulum signaling in health and disease. Nat Med. 1999;5:745–751. doi: 10.1038/10466. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: Wiley; 1999. [Google Scholar]
- Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, West AB, Ron D. Increased sensitivity to dextran sodium sulfate colitis in IRE1β-deficient mice. J Clin Invest. 2001;107:585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Potter VR. Nuclei from rat liver: Isolation method that combines purity with high yield. Science. 1966;154:1662–1665. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- Chandler JM, Cohen GM, MacFarlane M. Different subcellular distribution of caspase-3 and caspase-7 following Fas-induced apoptosis in mouse liver. J Biol Chem. 1998;273:10815–10818. doi: 10.1074/jbc.273.18.10815. [DOI] [PubMed] [Google Scholar]
- Chapman RE, Walter P. Translational attenuation mediated by an mRNA intron. Curr Biol. 1997;7:850–859. doi: 10.1016/s0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- Chapman R, Sidrauski C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus. Ann Rev Cell Dev Biol. 1998;14:459–485. doi: 10.1146/annurev.cellbio.14.1.459. [DOI] [PubMed] [Google Scholar]
- Clauss IM, Chu M, Zhao JL, Glimcher LH. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 1996;24:1855–1864. doi: 10.1093/nar/24.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Gonzalez TN, Sidrauski C, Dorfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y, Tsuru A, Kohno K. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat Cell Biol. 2001;3:158–164. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Skarnes WC, Rossant J. Production of a mutation in mouse En-2 gene by homologous recombination in embryonic stem cells. Nature. 1989;338:153–156. doi: 10.1038/338153a0. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Overview of vector design for mammalian gene expression. Methods Mol Biol. 1997;62:287–300. doi: 10.1385/0-89603-480-1:287. [DOI] [PubMed] [Google Scholar]
- ————— Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translation controls. Genes & Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Yanagi H, Yura T, Mori K. Unconventional splicing of HAC1/ERN4 mRNA required for the unfolded protein response. Sequence-specific and non-sequential cleavage of the splice sites. J Biol Chem. 1998;273:1802–1807. doi: 10.1074/jbc.273.3.1802. [DOI] [PubMed] [Google Scholar]
- Li M, Baumeister P, Roy B, Phan T, Foti D, Luo S, Lee AS. ATF6 as a transcription activator of the endoplasmic reticulum stress element: Thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol. 2000;20:5096–5106. doi: 10.1128/mcb.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- Masaki T, Yoshida M, Noguchi S. Targeted disruption of CRE-binding factor TREB5 gene leads to cellular necrosis in cardiac myocytes at the embryonic stage. Biochem Bioph Res Comm. 1999;261:350–356. doi: 10.1006/bbrc.1999.0972. [DOI] [PubMed] [Google Scholar]
- Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signalling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes to Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc Natl Acad Sci. 2000;97:4660–4665. doi: 10.1073/pnas.050010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- Nikawa JI, Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol Micro. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Ponath PD, Li YS, Hardy RR, David CS, Strominger JL, Glimcher LH. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J Exp Med. 1996;183:393–401. doi: 10.1084/jem.183.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, et al. An essential role in liver development for transcription factor XBP-1. Genes & Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Ruegsegger U, Leber J, Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Schroder M, Chang JS, Kaufman RJ. The unfolded protein response represses nitrogen-starvation induced developmental differentiation in yeast. Genes & Dev. 2000;14:2962–2975. doi: 10.1101/gad.852300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ellis R, Lee K, Liu C, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit D, Mori K, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Ting J, Wooden SK, Kriz R, Kelleher K, Kaufman RJ, Lee AS. The nucleotide sequence encoding the hamster 78-kDa glucose-regulated protein (GRP78) and its conservation between hamster and rat. Gene. 1987;55:147–152. doi: 10.1016/0378-1119(87)90258-7. [DOI] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes & Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes & Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000a;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Urano F, Bertolotti A, Ron D. IRE1 and efferent signaling from the endoplasmic reticulum. J Cell Sci. 2000b;113:3697–3702. doi: 10.1242/jcs.113.21.3697. [DOI] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- Welihinda AA, Kaufman RJ. The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J Biol Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- Wiest DL, Burkhardt JK, Hester S, Hortsch M, Meyer DI, Argon Y. Membrane biogenesis during B cell differentiation: Most endoplasmic reticulum proteins are expressed coordinately. J Cell Biol. 1990;110:1501–1511. doi: 10.1083/jcb.110.5.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CR, Dorner AJ, Morris GE, Alderman EM, Wilson D, O'Hara RM, Jr, Kaufman RJ. High level synthesis of immunoglobulins in Chinese hamster ovary cells. J Immunol. 1990;145:3011–3016. [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6α and 6β that activates the mammalian unfolded protein response. Mol Cell Biol. 2001a;21:1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001b;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]