Abstract
Telomerase elongates chromosome ends by addition of telomeric DNA repeats. The telomerase ribonucleoprotein can copy only a short template sequence within the telomerase RNA subunit. Here, we identify a region of telomerase RNA that is necessary for both correct 5′ template boundary definition and high affinity telomerase reverse transcriptase (TERT) interaction. We also demonstrate that TERT mutants in the RNA binding domain compromise both 5′ boundary definition and RNA binding. Our results indicate that sequence-specific interaction of a telomerase RNA element with the TERT RNA binding domain, not the active site motifs, defines the template boundary.
Keywords: Telomerase, RNA, template, reverse transcriptase, ribonucleoprotein
Telomeres are structures at the ends of chromosomes that protect them from illegitimate recombination, end-to-end fusion, and degradation (for review, see McEachern et al. 2000). In most eukaryotes, the DNA portion of the telomeres is comprised of a tandem array of simple sequence repeats. A de novo addition of sequence repeats to chromosome ends is accomplished by the telomerase ribonucleoprotein (RNP) reverse transcriptase. This addition compensates for the sequence loss at the ends that takes place with each round of chromosome replication (for review, see Greider 1996). Although the telomeric repeat sequence varies from organism to organism and in a few organisms has some natural degeneracy, very few sequence changes can be made without deleterious affects on cell growth and viability. Synthesis of inappropriate telomeric repeat sequences rapidly induces chromosome instability and compromises cellular viability in yeast, ciliate, and human cells (McEachern and Blackburn 1995; Kirk et al. 1997; Guiducci et al. 2001).
Assembly of two telomerase components, telomerase RNA and telomerase reverse transcriptase (TERT), is sufficient to reconstitute telomerase activity in vitro. All telomerase RNAs identified contain a short template sequence complementary to a telomeric DNA repeat (for review, see Greider 1996). TERT proteins contain reverse transcriptase motifs that form the enzyme active site (Lingner et al. 1997; for review, see Nakamura and Cech 1998). In addition, TERTs contain an RNA binding domain within the N-terminal region of the protein, separable from the active site motifs, that is both necessary and sufficient for high affinity telomerase RNA binding in the ciliate and mammalian enzymes (Bryan et al. 2000; Lai et al. 2001).
One intriguing question about telomerase mechanism is how a specific region within the telomerase RNA is defined as the template. Accurate repeat synthesis requires a precise delineation of the template boundaries. Although the template 3′ boundary can be determined at least in part by hybridization of primer substrate with the 3′ end of the template, there is no obvious marker for the template 5′ boundary. For the endogenously assembled Kluyveromyces lactis telomerase RNP, an RNA stem located 5′ of the template is crucial for establishing the 5′ template boundary (Tzfati et al. 2000). In contrast, for recombinant Tetrahymena telomerase RNA assembled with MNase-treated endogenous proteins in vitro, a single-stranded region 5′ of the template is important in 5′ template boundary definition (Autexier and Greider 1995).
Expression of Tetrahymena TERT and telomerase RNA in rabbit reticulocyte lysate (RRL) reconstitutes an entirely recombinant enzyme that can elongate a single-stranded DNA primer by addition of radiolabeled nucleotides (Collins and Gandhi 1998). Using telomerase reconstituted in this system, we describe here the regions of telomerase RNA and TERT that are necessary for accurate 5′ template boundary definition. On the RNA side, both single-stranded and double-stranded regions of the RNA are required. We find that reduced affinity of TERT-telomerase RNA interaction is accompanied by bypass of the wild-type template 5′ boundary. Surprisingly, we show that it is the RNA binding domain of TERT, rather than the reverse transcriptase active site, which establishes the template 5′ end. Our results reveal that 5′ template boundary definition in Tetrahymena telomerase is governed by association of the TERT RNA binding domain with an RNA sequence 5′ of template.
Results
A sequence element conserved among ciliate telomerase RNAs is important for 5′ template boundary definition
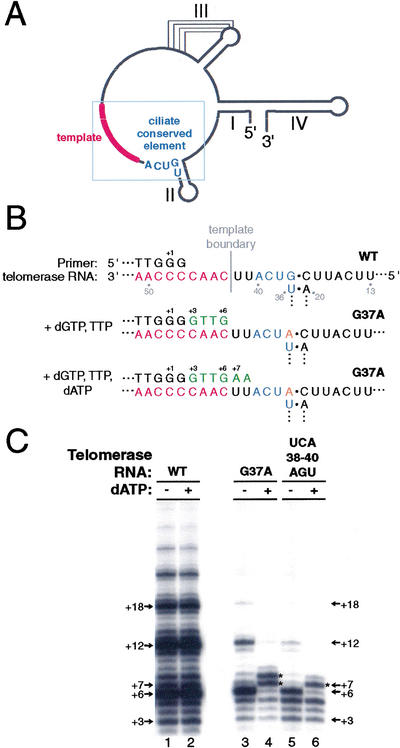
Ciliate telomerase RNAs share a conserved 5′-(U)GUCA-3′ sequence (McCormick-Graham and Romero 1995) positioned two nucleotides from the 5′ end of the template (Fig. 1A,B). The role of this conserved sequence was analyzed previously by assaying the activity of recombinant Tetrahymena telomerase RNA reconstituted with partially purified, micrococcal nuclease (MNase) treated, endogenous telomerase proteins (Autexier and Greider 1995). The nucleotide substitution UCA38-40AGU within this region resulted in primer extension past the normal 5′ template boundary. To determine if telomerase reconstituted in a fully recombinant system requires the ciliate conserved sequence for 5′ template boundary definition, we expressed recombinant Tetrahymena TERT in RRL and assembled it with telomerase RNAs bearing sequence substitutions in this region. If only dGTP and TTP nucleotides are present in an activity assay, copying will proceed to the correct 5′ boundary of the wild-type template (Fig. 1B, middle). If dATP is also added to the reaction and proper template boundary definition is disrupted, copying will continue through residues 5′ of the wild-type template boundary (Fig. 1B, bottom).
Figure 1.
A telomerase RNA sequence conserved among ciliates is necessary for correct 5′ template boundary definition. (A) Secondary structure of Tetrahymena telomerase RNA (Romero and Blackburn 1991). The ciliate conserved element is in blue, whereas the template region is shown as a red line. A blue box indicates the region of the telomerase RNA shown in B. Roman numerals indicate stems I, II, III, and IV. (B) Schematics of the primer-template hybrid (top), the product produced by synthesis to the wild-type template 5′ boundary (middle) and the product produced by “boundary pass” synthesis two nucleotides beyond the wild-type template 5′ end (bottom). T. thermophila telomerase RNA positions 13–20 and 36–51 are shown. Potential product lengths are labeled from the 3′ end of the template, with the first nucleotide added to primer (TG)8T2G3 at +3. Template sequence is in red, the ciliate conserved sequence is in blue, dNTPs added by telomerase are in green, and the residue changed in the G37A RNA variant is shown in orange. (C) Telomerase activity assays with ribonucleoproteins (RNPs) containing the indicated telomerase RNA variants. Reactions in odd numbered lanes contained only 32P–dGTP and TTP whereas reactions in even numbered lanes contained 32P–dGTP, TTP, and dATP. Arrows indicate the sizes of primer extension products, corresponding to the positions labeled in B. WT is wild-type telomerase RNA. Asterisks indicate products at positions of template boundary bypass.
Tetrahymena telomerase will add six nucleotides to a DNA primer ending with the sequence TTG before reaching the 5′ end of the template. The product DNA, also ending with TTG, can be released after first repeat synthesis and reposition at the template 3′ end for the synthesis of additional repeats. Multiple repeat addition to the primer (TG)8T2G3 resulted in the characteristic pattern of telomerase activity, with a ladder of product DNAs with six nucleotide periodicity (Fig. 1C, lane 1). Addition of dATP to the standard reaction did not alter the product profile (Fig. 1C, lane 2), indicating that, as expected, the correct template boundary is recognized even when the nucleotides that allow copying past this boundary are present. In the absence of dATP, telomerases with RNA substitutions within the ciliate conserved element, UCA38-40AGU or G37A, extended primer to the 5′ end of the wild-type template with some multiple repeat addition as well (Fig. 1C, lanes 3,5). In comparison, in the presence of dATP, telomerases with these RNA substitutions copied one to two nucleotides beyond the wild-type 5′ template boundary, producing predominant products that were +7 or +8 in length (Fig. 1C, lanes 4,6). These products would be incapable of reannealing at the template 3′ end and therefore would not be expected to be elongated by additional repeats. These results indicate that the ciliate conserved sequence GUCA, at RNA positions 37–40 in the T. thermophila telomerase RNA, is important for template 5′ boundary definition in the fully recombinant reconstituted RNP system.
Identification of telomerase RNA requirements for 5′ template boundary definition
Previous work has demonstrated that a stem-loop structure 5′ of the K. lactis telomerase RNA template is critical for establishing the template boundary (Tzfati et al. 2000). The G37A substitution in the Tetrahymena telomerase RNA assayed above alters the GUCA conserved sequence and would also unpair the proximal end of stem II (Fig. 2A). To determine if stem II of ciliate telomerase RNA is important in establishing the template boundary, we constructed several substitutions that affect stem II sequence and/or structure while maintaining a wild-type GUCA37-40 sequence.
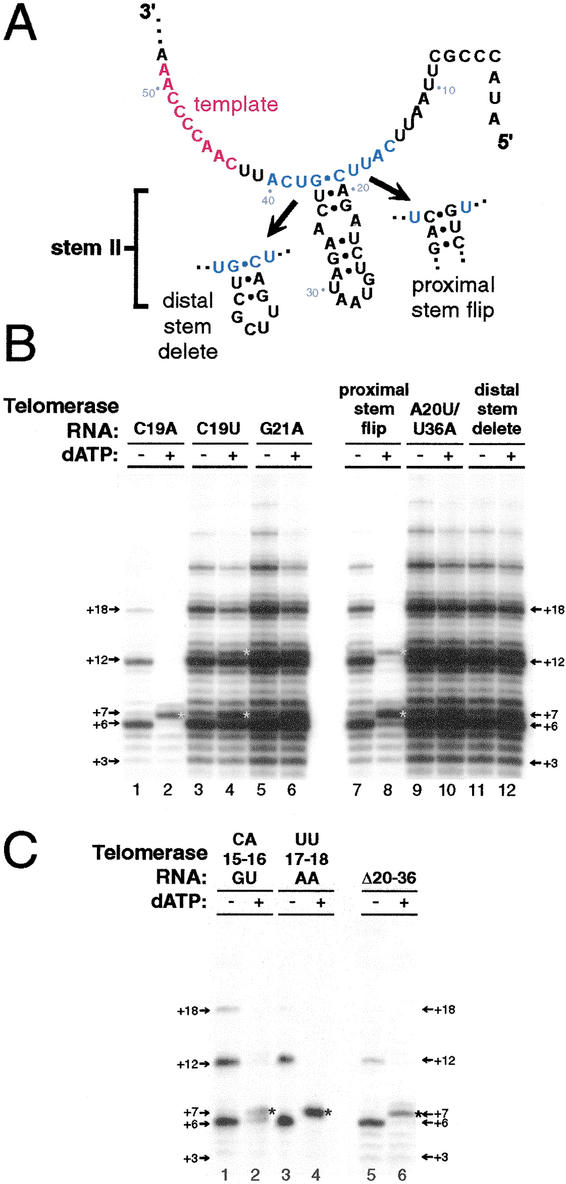
Figure 2.
The region of telomerase RNA flanking stem II is important for 5′ template boundary definition. (A) The phylogenetically predicted secondary structure of stem II and flanking single-stranded regions. Blue letters indicate nucleotides found to be important for correct 5′ template boundary definition. The proximal stem flip substitution includes both CAG19–21GUC and CUG35–37GAC substitutions. The distal stem delete substitution replaces nucleotides 22 to 34 with a UUCG tetraloop. (B, C) Telomerase activity assays with ribonucleoproteins (RNPs) containing the indicated telomerase RNA variants. Reactions in even numbered lanes have dATP added. Asterisks indicate products at positions of template boundary bypass.
The telomerase RNA substitution C19A, like G37A, is predicted to unpair the proximal end of stem II (Fig. 2A). A wild-type profile of product DNA was generated by telomerase RNP with C19A telomerase RNA in the absence of dATP (Fig. 2B, lane 1). In the presence of dATP, however, telomerase with C19A RNA produced predominantly a +7 product, bypassing the wild-type template boundary by one nucleotide (Fig. 2B, lane 2). This result suggested that the secondary structure of stem II could be important for template boundary definition, independent of a requirement for the sequence of G37. We next tested a C19U telomerase RNA variant; this substitution would replace the predicted C19-G37 canonical base pair with a U·G wobble pair. In the presence of dATP, telomerase RNP containing C19U RNA generated products indicative of a slight boundary pass effect with some enhancement of +7 product, as confirmed by PhosphorImager quantitation comparing the ratio of +7 product to +6 product (Fig. 2B, lanes 3,4). To assess the role of proximal stem II pairing by substitution of a different base pair, we made G21A telomerase RNA. Although this substitution is predicted to unpair the proximal region of stem II (Fig. 2A), telomerase RNPs with G21A RNA did not demonstrate a substantial alteration of 5′ template boundary definition (Fig. 2B, lanes 5,6). To maintain proximal stem II base pairing potential while altering the sequence of the proximal stem, a proximal stem flip RNA that swapped the sides of the stem (Fig. 2A) was tested. The proximal stem flip substitution eliminated proper 5′ template boundary definition, evident by the formation of +7 product upon addition of dATP (Fig. 2B, lanes 7,8).
Almost all ciliate telomerase RNAs share a uridine residue immediately 5′ of the conserved GUCA motif. The notable exception is T. paravorax, which also lacks a predicted stem II element (McCormick-Graham and Romero 1995). To examine whether T. thermophila U36 contributes to template boundary definition, we tested the compensatory base pair substitution A20U/U36A. Telomerase RNP with A20U/U36A RNA and telomerase RNP with RNA lacking the distal end of stem II (Fig. 2A) both did not demonstrate a dATP-dependent boundary defect (Fig. 2B, lanes 9–12). From our analysis of these stem II telomerase RNA variants, we conclude that most of the sequence and the base pairing interactions of stem II are not essential for 5′ template boundary definition. Conversely, both the sequence and the base pairing at the proximal end of stem II appear critical at T. thermophila positions C19–G37. It is possible that a structure similar to a canonical base pair is stabilized by association with TERT rather than by stacking on a short RNA helix.
RNA sequence requirements are the same for template boundary definition and high affinity TERT binding
We have shown previously that the CAUU sequence at T. thermophila RNA positions 15–18 adjacent to stem II is critical for high affinity interaction between telomerase RNA and TERT. This sequence is also required for high affinity interaction between telomerase RNA and the N-terminal RNA binding domain of TERT alone (Lai et al. 2001). The CAUU15–18 sequence, unlike the GUCA37–40 sequence, is not conserved among ciliate telomerase RNAs. We therefore anticipated that adjacent but separable TERT–telomerase RNA interactions accomplished 5′ template boundary definition and high affinity interaction. To investigate whether the RNA sequence requirements for these two aspects of telomerase function were indeed distinct, we tested telomerase RNAs substituted in the region that mediate high affinity association to the TERT RNA binding domain for 5′ template boundary bypass.
To detect the activity of telomerase RNPs with RNAs that were substantially compromised in their ability to bind TERT, it was necessary to use high concentrations of RNA in the telomerase RNP assembly step. For consistency, all of the activity assays shown were performed with the same high concentration of telomerase RNA, an ∼150-fold molar excess of RNA over TERT (see Materials and Methods). To our surprise, we found that telomerases with the TERT binding site RNA substitutions CA15-16GU and UU17-18AA both demonstrated dATP-dependent 5′ template boundary bypass, producing +7 product in the presence of dATP (Fig. 2C, lanes 1–4). Telomerase RNPs with UU17-18AC or UU17-18CC RNA variants also exhibited the 5′ template boundary bypass defect as well (data not shown). We conclude that the RNA requirements for high affinity TERT binding are not distinct from the requirements for 5′ template boundary definition: The CAUU15-18 sequence is essential for both TERT binding and establishing the correct 5′ template boundary. This result is surprising considering that the CAUU15-18 sequence is not well conserved among ciliate telomerase RNAs but seems to play as crucial a role in template boundary definition as the conserved GUCA37-40 sequence.
In addition to CAUU15-18, previous studies have shown that high affinity TERT binding requires some portion of stem II. Several large deletions of stem II that retain C19 and G37 are nonetheless compromised for TERT association (Licht and Collins 1999). We tested for a 5′ boundary pass defect using a telomerase RNA variant with a deletion of nucleotides 20 to 36, removing most of stem II (Fig. 2A). Telomerase RNP with Δ20–36 RNA copied past the wild-type 5′ template boundary in a dATP-dependent manner (Fig. 2C, lanes 5,6). This deletion of stem II connects C19 and G37 with a phosphodiester linkage instead of a predicted base-pairing interaction, otherwise eliminating only residues that were shown to be unimportant for boundary definition. The fact that such a subtle disruption of the positioning of these two critical nucleotides can have major impact on template boundary definition demonstrates the significance of correct structure within this region of the telomerase RNA.
RNA substitutions that compromise TERT binding were shown above to alter the template 5′ boundary. We next tested whether all substitutions shown to impact template boundary definition also affect TERT binding. N-terminally HA epitope-tagged TERT was expressed in RRL and assembled with an ∼20-fold molar excess of telomerase RNA (see Materials and Methods). This RNA excess is less than that used in the activity assay to reduce the nonspecific background binding of RNA to the affinity resin. TERT protein was immunopurified with HA antibody, and the associated telomerase RNA was recovered and quantified by RNA blot hybridization. All telomerase RNA variants were tested for association with both full-length TERT (Fig. 3A) and the RNA binding domain of TERT alone (Fig. 3B).
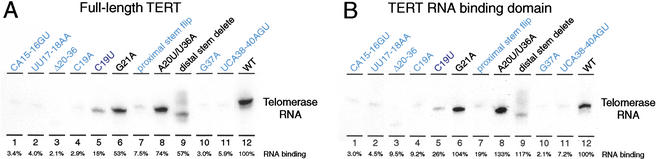
Figure 3.
RNA sequences important for template boundary definition also function in TERT binding. TERT expressed in rabbit reticulocyte lysate (RRL) was assembled with telomerase RNA then recovered by immunopurification. RNA coimmunoprecipitated with TERT was detected by RNA blot hybridization. Interaction of telomerase RNA with both full-length TERT (A) and the RNA binding domain of TERT (B) was examined in parallel. The amount of RNA coimmunoprecipitated is quantified relative to wild-type telomerase RNA (WT). Telomerase RNAs in ribonucleoproteins (RNPs) that resulted in strong boundary pass are shown in blue; telomerase RNAs in RNPs that resulted in weak boundary pass are shown in purple (C19U, lane 5) and telomerase RNAs in RNPs that resulted in no boundary pass are shown in black.
As reported previously (Lai et al. 2001), substitution of either nucleotides CA15-16 or UU17-18 substantially decreased telomerase RNA association to both full-length TERT and the TERT RNA binding domain (Fig. 3A,B, lanes 1,2). Similarly, the RNA deletion that removed most of stem II also strongly decreased TERT interaction (Fig. 3A,B, lane 3) as did RNA substitutions C19A, proximal stem flip, G37A and UCA38-40AGU (Fig. 3A,B, lanes 4,7,10,11). In contrast, the RNA substitutions G21A, A20U/U36A, and distal stem delete did not dramatically affect TERT association (Fig. 3A,B, lanes 6,8,9). The RNA substitution C19U had an intermediate impact, demonstrating reduced but still substantial interaction with full-length TERT and TERT RNA binding domain (Fig. 3A,B, lane 5). In sum, all RNA substitutions that strongly compromise accurate 5′ template boundary definition also strongly compromise high affinity TERT binding. In complementary fashion, none of the RNA substitutions that retain wild-type 5′ template boundary reduce TERT binding by >50%. The C19U RNA substitution results in both a moderate 5′ template boundary definition defect and a moderate TERT binding defect. This correlation between interaction affinity and template boundary definition is also evident in the activity assays themselves, because RNAs with reduced binding affinity for TERT reconstitute less telomerase RNP and therefore produce less product DNA. Assays of RNPs containing RNAs that promoted 5′ template boundary bypass generated less overall activity (Figs. 1, 2).
For all RNA substitutions 5′ of the template examined here, if binding to TERT was reduced, binding to the isolated TERT RNA binding domain was reduced to a similar extent (cf. Fig. 3A and 3B). This suggests that interaction between TERT and the template boundary definition element of telomerase RNA takes place primarily or exclusively through the RNA binding domain of the protein. We conclude that telomerase RNA requirements for establishing the wild-type 5′ template boundary and for high affinity interaction with the TERT RNA binding domain overlap and are indeed not separable using the RNA substitutions and assays described here.
TERT mutations that affect high affinity RNA binding also affect template boundary definition
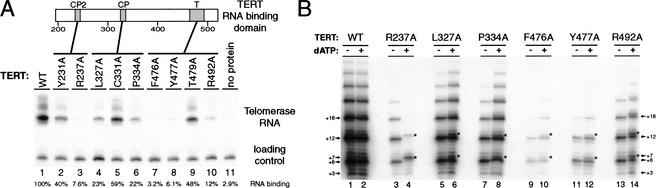
The RNA binding domain of Tetrahymena TERT spans from amino acids 195 to 516 (Lai et al. 2001) and contains two ciliate-conserved TERT motifs, CP and CP2, and one universally conserved TERT motif, motif T (Fig. 4A). Mutational analysis of TERT has previously identified two single amino acid substitutions in motif CP, L327A, and P334A, and three single amino acid substitutions within motif T, F476A, Y477A, and R492A, that affect telomerase RNA binding (Bryan et al. 2000). In our assay, the two motif CP substitutions each decreased telomerase RNA association to an intermediate level (Fig. 4A, lanes 4,6), and the three motif T substitutions each decreased telomerase RNA association more substantially (Fig. 4A, lanes 7,8,10). In addition, from an extensive single amino acid substitution screen of TERT, we previously identified one amino acid substitution within motif CP2 (R237A) that permits dATP-dependent synthesis past the 5′ template boundary when combined with wild-type telomerase RNA (Miller et al. 2000). The R237A TERT substitution strongly decreased telomerase RNA association (Fig. 4A, lane 3), whereas the other single amino substitutions in the RNA binding domain from the mutagenesis screen that did not affect template boundary definition also did not have a strong affect on TERT association (Fig. 4A, lanes 2,5,9).
Figure 4.
TERT substitutions that affect interaction with telomerase RNA also affect 5′ template boundary definition. (A) Effect of TERT substitutions on RNA binding. Wild-type TERT (lane 1) and single amino acid TERT substitutions (lanes 2–10) were expressed in rabbit reticulocyte lysate (RRL), assembled with wild-type telomerase RNA, and recovered by immunopurification. RNA was also coimmunoprecipitated with beads only as a test for nonspecific bead binding (lane 11). RNA coimmunoprecipitated with TERT was detected by RNA blot hybridization. One nanogram of a 123-nucleotide telomerase RNA variant was used as a precipitation and loading control. (B) Effect of TERT substitutions on 5′ template boundary definition. Reactions in odd numbered lanes contained only 32P–dGTP and TTP whereas reactions in even numbered lanes contained 32P–dGTP, TTP, and dATP. Asterisks indicate products at positions of template boundary bypass.
To investigate whether all TERT substitutions that decrease telomerase RNA binding affinity also alter the 5′ template boundary, we performed activity assays on mutant TERTs with wild-type telomerase RNA, with and without dATP (Fig. 4B). Telomerase RNPs with the three mutant TERTs that did not strongly decrease telomerase RNA binding (Y231A, C331A, and T479A; >25% of wild-type level of telomerase RNA bound) did not produce noticeable template boundary bypass in the presence of dATP (data not shown). Telomerase RNPs with the two TERT mutants that had intermediate affect on telomerase RNA association (L327A and P334A; between 15% and 25% of wild-type level of telomerase RNA bound) showed a mild template boundary pass defect, producing only a slight increase in the ratio of +7 product to +6 product on dATP addition, as quantitated by PhosphorImager analysis (Fig. 4B, lanes 5–8). Finally, the TERT mutants that had the most substantial affect on telomerase RNA association (R237A, F476A, Y477A, R492A; <15% of wild-type level of telomerase RNA bound) all produced telomerase RNPs with a more substantial 5′ template boundary defect, generating a significant amount of +7/+13 products on dATP addition (Fig. 4B, lanes 3–4, 9–14). It is notable that the R237A substitution causes a more severe template boundary bypass defect than the other TERT mutants tested (Fig. 4B, lanes 3,4). This result suggests that motif CP2 in particular could play a unique role in 5′ template boundary definition or that it may have the greatest impact on telomerase RNA binding affinity. This study of TERT substitutions combined with the telomerase RNA substitution data above reveal that 5′ template boundary definition and TERT–telomerase RNA interaction affinity are established by linked mechanisms. We arrive at the unanticipated conclusion that both high affinity protein–RNA interaction and template 5′ boundary definition are provided by association of the TERT RNA binding domain with a region of the telomerase RNA 5′ of the template.
Discussion
We have identified specific residues of Tetrahymena telomerase RNA that are critical for both 5′ template boundary definition and for stable association with TERT. Substitutions of these residues affect the accuracy of the 5′ template boundary, TERT association, and association with the isolated RNA binding domain of TERT in a parallel fashion. The overlap in RNA sequence requirements detected in these three assays suggests that the interaction of the TERT RNA binding domain and RNA motifs 5′ of the template mediates high affinity binding and template boundary definition simultaneously. The simplest model to explain this result is that stable binding of the TERT RNA binding domain to the template boundary/high affinity binding element of the RNA sterically blocks copying of the template past the appropriate 5′ end. Although template boundary definition is the telomerase feature most faithfully recapitulated in the minimal recombinant RNP system, it is possible that there are additional unidentified components of the Tetrahymena telomerase holoenzyme that contribute to 5′ template boundary definition as well.
For telomerase of the yeast K. lactis, formation of a stem-loop structure immediately 5′ of the template is necessary for establishing the template boundary (Tzfati et al. 2000). Our results indicate that for the recombinant Tetrahymena telomerase RNP, formation of an RNA secondary structure alone is not sufficient. Instead, it is the ability of an RNA element to interact with the TERT RNA binding domain that is critical for establishing the wild-type template 5′ boundary. It will be interesting to determine if protein binding to the stem-loop structure 5′ of the template in K. lactis telomerase RNA accomplishes template boundary definition in an analogous manner.
Telomerase is an atypical reverse transcriptase. Unlike viral reverse transcriptases, which can copy any sequence of RNA polynucleotide into DNA, TERT can copy only the telomerase RNA. In addition, only a short region within the entire telomerase RNA is recognized as template. Finally, although other reverse transcriptases only transiently associate with the their templates, TERT is bound to its template as a highly stable complex. The combination of these unique telomerase features results in an enzyme specialized for the processive addition of telomeric repeats. In the evolution of TERT, it would seem likely that at some point the ancestral reverse transcriptase domain acquired an accessory RNA binding domain by gene fusion. This could have been the first step in the specialization of the TERT active site for copying telomerase RNA, because the increased effective concentration of telomerase RNA would have promoted its use as a template. An intriguing possibility is that this hypothesized gene fusion might have simultaneously accomplished a second specialization: defining a template 5′ boundary. Establishing a template boundary would have brought telomerase much closer to its current physiological function in telomeric repeat synthesis.
Materials and methods
TERT and telomerase RNA synthesis
Expression constructs for telomerase RNA and TERT variants were created by site-specific mutagenesis. Transcription reactions were treated with DNase I before RNA purification by denaturing gel electrophoresis. RNA concentration was determined by UV spectrophotometry and/or fluorometry. RNA purity was verified by gel electrophoresis. TERT proteins were expressed in RRL as described by the manufacturer (Promega TNT).
Telomerase activity assay
Four microliters of RRL expression reaction was immunopurified, combined with 1.5 pmole of telomerase RNA, then brought up to 10 μL in T2MG (20 mM Tris-HCl at pH 8.0, 1 mM MgCl2, 10% glycerol). HA antibody was bound to GammaBind Protein G-Sepharose (Pharmacia) and preblocked with 20 μg/mL tRNA and 20 μg/mL BSA in T2MG + 0.1 M NaCl. Activity assay reactions contained final concentrations of 50 mM Tris-acetate at pH 8.0, 10 mM spermidine, 5 mM ß-mercaptoethanol, 1 mM DTT and 2 mM MgCl2. To initiate the reaction, 200 μM TTP, 3.5 μM unlabeled dGTP, 1.2 μM 32P-dGTP (800 Ci/mmol) and 2 μM of the primer (TG)8T2G3 were added; 200 μM of dATP was also added to some activity assays as indicated. Reactions were incubated at 30°C for 1 h followed by extraction with phenol:chloroform:isoamyl alcohol and precipitation with ethanol. The radiolabeled product DNA was resolved by denaturing gel electrophoresis.
Immunopurification of TERT–telomerase RNA complexes
Full-length TERT or the TERT RNA binding domain with an N-terminal HA tag was expressed in RRL at 30°C for 90 min. The amount of protein expression was measured by 35S-methionine incorporation and equalized for each sample. Ten microliters of expression reaction was mixed with 0.56 pmole telomerase RNA, 5 μg BSA, and 5 μg tRNA then incubated at 30°C for an additional 20 min. Samples were incubated with 15 μL HA antibody resin prepared as described above in 400 μL final volume of T2MG + 0.1 M NaCl for 4 h at 4°C. Bound TERT complexes were washed with T2MG + 0.1 M NaCl, extracted with phenol:chloroform:isoamyl alcohol and then precipitated with ethanol. Purified nucleic acid was resolved by denaturing gel electrophoresis and transferred to Hybond N+ (Amersham) for hybridization. Blots were probed with a 32P end-labeled DNA oligonucleotide complimentary to wild-type RNA sequence 3′ of the template. Quantification was performed by PhosphorImager (Fuji).
Acknowledgments
We thank Doreen Cunningham, Jill Licht, and Aubree Gordon for TERT and telomerase RNA expression constructs. We also thank James Berger, Don Rio, Steph Yonker, and members of the Collins lab for discussion on this manuscript. This work was funded by a grant from the National Institutes of Health (GM54198) and a Burroughs Wellcome Fund New Investigator Award to K.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kcollins@socrates.berkeley.edu; FAX (510) 642-6062.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.962602.
References
- Autexier C, Greider CW. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes & Dev. 1995;9:2227–2239. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Goodrich KJ, Cech TR. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol Cell. 2000;6:493–499. doi: 10.1016/s1097-2765(00)00048-4. [DOI] [PubMed] [Google Scholar]
- Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;66:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Guiducci C, Cerone MA, Bacchetti S. Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities and rapid loss of viability. Oncogene. 2001;20:714–725. doi: 10.1038/sj.onc.1204145. [DOI] [PubMed] [Google Scholar]
- Kirk KE, Harmon BP, Reichardt IK, Sedat JW, Blackburn EH. Block in anaphase chromosome separation caused by a telomerase template mutation. Science. 1997;275:1478–1481. doi: 10.1126/science.275.5305.1478. [DOI] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes & Dev. 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Gen. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- Miller MC, Liu JK, Collins K. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J. 2000;19:4412–4422. doi: 10.1093/emboj/19.16.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Cech TR. Reversing time: Origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]