Abstract
Calcineurin, a conserved Ca2+/calmodulin-regulated protein phosphatase, plays a crucial role in Ca2+ signaling in a wide variety of cell types. In Saccharomyces cerevisiae, calcineurin positively regulates transcription in response to stress by dephosphorylating the transcription factor Crz1p/Tcn1p. Dephosphorylation promotes Crz1p nuclear localization in part by increasing the efficiency of its nuclear import. In this work, we show that calcineurin-dependent dephosphorylation of Crz1p also down-regulates its nuclear export. Using a genetic approach, we identify Msn5p as the exportin for Crz1p. In addition, we define the Crz1p nuclear export signal (NES) and show that it interacts with Msn5p in a phosphorylation-dependent manner. This indicates that calcineurin regulates Crz1p nuclear export by dephosphorylating and inactivating its NES. Finally, we define a motif in Crz1p, PIISIQ, similar to the PxIxIT docking site for calcineurin on the mammalian transcription factor NFAT, that mediates the in vivo interaction between calcineurin and Crz1p and is required for calcineurin-dependent regulation of Crz1p nuclear export and activity. Therefore, in yeast as in mammals, a docking site is required to target calcineurin to its substrate such that it can dephosphorylate it efficiently.
Keywords: S. cerevisiae, calcium signaling, calcineurin, Crz1p, Msn5p, nuclear transport
Ca2+-dependent signal transduction pathways elicit diverse physiological responses. One way that Ca2+ exerts an effect is through activation of the Ca2+-binding protein calmodulin, which, in turn, activates the serine/threonine protein phosphatase calcineurin. In mammalian cells, calcineurin regulates the subcellular localization of the transcription factor NFAT (nuclear factor of activated T-cells) that is required for T-cell activation in response to antigen. Dephosphorylation of NFAT by calcineurin promotes its nuclear localization and, as a result, gene transcription and T-cell proliferation (Flanagan et al. 1991; Jain et al. 1993; Shaw et al. 1995). Activation of NFAT is blocked, however, when calcineurin phosphatase activity is inhibited by the immunosuppressive drugs FK506/FK520 and cyclosporin A (Liu et al. 1991a; Clipstone and Crabtree 1992; O'Keefe et al. 1992). Calcineurin interacts with NFAT through a defined consensus binding site, PxIxIT; when this site is mutated, dephosphorylation and nuclear translocation of NFAT do not occur (Aramburu et al. 1998). Calcineurin similarly regulates NFAT family members in a variety of cell types to play a role in other processes including cardiac and skeletal muscle development and angiogenesis (Chin et al. 1998; de la Pompa et al. 1998; Molkentin et al. 1998; Ranger et al. 1998; Graef et al. 2001).
In the budding yeast Saccharomyces cerevisiae, calcineurin is activated by extracellular stresses. Calcineurin mutants deficient for either the catalytic subunits (CNA1/CNA2; Cyert et al. 1991; Liu et al. 1991b) or the regulatory subunit (CNB1; Kuno et al. 1991; Cyert and Thorner 1992) are viable under standard growth conditions but are sensitive to high concentrations of ions such as Na+, Li+, Mn+, and OH− (Nakamura et al. 1993; Mendoza et al. 1994; Farcasanu et al. 1995; Pozos et al. 1996). They also lose viability during sustained treatment with mating pheromone (Moser et al. 1996; Withee et al. 1997). Under these conditions calcineurin is required to activate a number of genes, including PMC1, PMR1, and PMR2, which encode ion pumps (Rudolph et al. 1989; Haro et al. 1991; Cunningham and Fink 1994, 1996; Mendoza et al. 1994), and FKS2, which encodes a major cell wall biosynthetic enzyme (Mazur et al. 1995). Calcineurin regulates transcription by activating Crz1p/Tcn1p, which, in turn, binds a 24-bp promoter element termed the CDRE (calcineurin-dependent response element; Matheos et al. 1997; Stathopoulos and Cyert 1997) to activate transcription of its target genes.
Crz1p is regulated by calcineurin in a remarkably similar manner to NFAT; upon Ca2+ addition to the media, Crz1p rapidly translocates from the cytosol to the nucleus in a calcineurin-dependent fashion (Stathopoulos-Gerontides et al. 1999). Previous studies established that when Crz1p is dephosphorylated, its nuclear import rate increases because Crz1p binds to its importin Nmd5p more efficiently (Polizotto and Cyert 2001). Crz1p and NFAT are not homologous proteins but both do contain an SRR (serine-rich region) domain (Beals et al. 1997; Stathopoulos-Gerontides et al. 1999). In Crz1p mutants lacking the SRR, calcineurin-independent nuclear localization is observed, indicating that the SRR is important for regulation of Crz1p nuclear transport (Stathopoulos-Gerontides et al. 1999).
In this study we further characterize the calcineurin-dependent regulation of Crz1p nuclear transport. We establish that calcineurin negatively regulates Crz1p nuclear export and identify Msn5p as the required exportin. Furthermore, we define the Crz1p NES and show that its phosphorylation state is critical for function. Finally, we show that calcineurin interacts with Crz1p through a site, PIISIQ, that is similar to the calcineurin-binding site in NFAT; this motif is required for proper regulation of Crz1p nuclear transport by calcineurin and, thus, for wild-type Crz1p function.
Results
Crz1p nuclear export is regulated by calcineurin
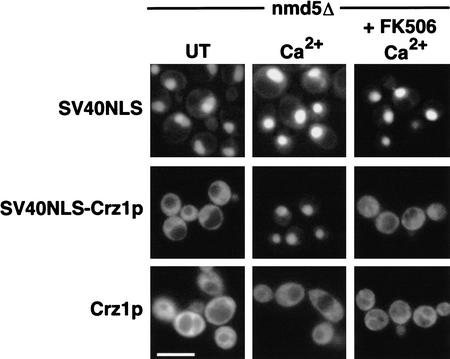
Ca2+/calcineurin-dependent dephosphorylation of Crz1p up-regulates nuclear import and causes rapid translocation of Crz1p from the cytosol to the nucleus of the cell (Polizotto and Cyert 2001). We wished to determine whether nuclear export of Crz1p was also regulated by calcineurin. To address this question, we used a strain deleted for the importin NMD5 such that we could observe Crz1p localization in the absence of calcineurin-regulated nuclear import. Because Crz1p does not localize to the nucleus with Ca2+ addition in this strain (Polizotto and Cyert 2001), to facilitate Crz1p nuclear import we used an exogenous nuclear localization signal (NLS) from the SV40 large T antigen (Kalderon et al. 1984a,b). The SV40 NLS targeted GFP to the nucleus; this targeting was unaffected by Ca2+ or FK506 addition to the media (Fig. 1), and thus is not regulated by calcineurin. The GFP–SV40NLS–Crz1p fusion localized to the cytosol of untreated cells, showing that Crz1p contains an NES and suggesting that under these conditions the rate of Crz1p nuclear export is greater than the rate of import directed by the SV40 NLS. However, when Ca2+ was added, GFP–SV40NLS–Crz1p localized to the nucleus of nmd5Δ cells (Fig. 1). This change did not occur when calcineurin was inhibited by FK506. Because the SV40 NLS provides constant nuclear import, we concluded that the Ca2+-induced nuclear localization of GFP–SV40NLS–Crz1p was owing to decreased export. Thus, dephosphorylation of Crz1p by calcineurin leads to decreased nuclear export.
Figure 1.
Crz1p nuclear export is regulated by calcineurin. nmd5Δ (HFY133) cells expressing 3xGFP–SV40NLS (LMB134), 3xGFP–SV40NLS–Crz1p (LMB148), or 3xGFP–Crz1p (LMB127) were grown to log phase at 21°C. Cells were treated with 200 mM CaCl2 or pretreated with FK506 (2 μg/mL) for 30 min before CaCl2 addition and visualized using fluorescence microscopy. Bar, 20 μm.
A genetic screen identifies MSN5 as the Crz1p exportin
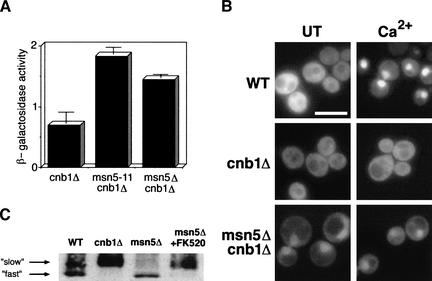
To establish the mechanism of Crz1p nuclear export, we devised a genetic screen to identify the Crz1p exportin. Previous studies revealed that expression of the CDRE::lacZ reporter element is dependent on both calcineurin and Crz1p (Stathopoulos and Cyert 1997). We reasoned that a mutation in the exportin for Crz1p might result in constitutive nuclear localization of Crz1p and consequent activation of the CDRE::lacZ reporter in a calcineurin mutant background (cnb1Δ). We identified 40 recessive mutants that expressed CDRE::lacZ in the absence of calcineurin activity, and a single complementation group containing 5 strains was chosen for characterization (see Materials and Methods). These strains were complemented by plasmids containing MSN5. Furthermore, an msn5Δ cnb1Δ strain failed to complement the mutants, confirming that each contained a mutation in MSN5. The msn5Δ cnb1Δ strain also had the same phenotype as the mutant isolated from our screen (msn5-11cnb1Δ). In a quantitative β-galactosidase assay, both strains showed a small but reproducible increase in basal CDRE::lacZ activity over the cnb1Δ strain (Fig. 2A). Therefore, we used the null allele of MSN5 for all further experiments.
Figure 2.
msn5 mutants activate Crz1p-dependent transcription in a cnb1Δ background and alter Crz1p subcellular localization. (A) Strains harboring an integrated CDRE::lacZ reporter were grown to log phase at 30°C and harvested. β-Galactosidase activity was measured for cnb1Δ (DD12), msn5-11cnb1Δ (ASY11), and msn5Δ cnb1Δ (LBY196) strains. For each strain two cell extracts were prepared and assayed in triplicate. The standard deviation is the error between the samples. (B) WT (ASY472), cnb1Δ (ASY475), and msn5Δ cnb1Δ (LBY172) cells expressing GFP–Crz1p (AMS463) were grown to log phase at 21°C. Cells were treated with 200 mM CaCl2, and GFP–Crz1p was visualized using fluorescence microscopy. Bar, 20 μm. (C) Msn5p does not affect Crz1p phosphorylation state. Whole cell extracts from wild-type (YPH499), cnb1Δ (DD12), and msn5Δ (ASY788) strains containing HA–Crz1p (AMS446) were treated with or without 2 μg/mL FK520 and analyzed by Western blot using anti-HA antibody.
Mutations in MSN5 affect Crz1p cellular localization
Because Msn5p had been previously characterized as a nuclear transport factor (Kaffman et al. 1998; Blondel et al. 1999; DeVit and Johnston 1999), we tested whether mutations in MSN5 affected Crz1p localization. GFP–Crz1p localizes to the cytosol of untreated cells, but translocates to the nucleus after Ca2+ addition to the media (Fig. 2B; Stathopoulos-Gerontides et al. 1999). This change in localization is completely dependent on calcineurin as GFP–Crz1p remains cytosolic in a cnb1Δ strain. In contrast, GFP–Crz1p was partially localized to the nucleus of msn5Δ cnb1Δ cells (Fig. 2B). Ca2+ addition had no further effect on GFP–Crz1p localization in this strain, as would be expected in the absence of calcineurin activity. The nuclear accumulation of Crz1p observed in untreated msn5Δ cnb1Δ cells is consistent with a defect in Crz1p nuclear export.
Crz1p subcellular localization is regulated by its phosphorylation state (Stathopoulos-Gerontides et al. 1999; Polizotto and Cyert 2001); therefore, we examined whether Msn5p affects Crz1p phosphorylation. Crz1p shows a faster electrophoretic mobility in wild-type strains than it does in cnb1Δ strains (Fig. 2C; Stathopoulos-Gerontides et al. 1999). This difference is due to phosphorylation because treatment of purified Crz1p with calcineurin in vitro eliminates the mobility shift (Stathopoulos-Gerontides et al. 1999). We determined that the electrophoretic mobility of HA–Crz1p also showed a calcineurin-dependent shift in msn5Δ strains. When msn5Δ cells were incubated with the calcineurin inhibitor FK520, Crz1p showed reduced mobility (Fig. 2C). Thus, in the absence of Msn5p and calcineurin activity, Crz1p is phosphorylated, yet it accumulates in the nucleus.
Msn5p is required for Crz1p nuclear export
We next sought to determine whether Msn5p is required for Crz1p export from the nucleus. To address this question, wild-type or msn5Δ cells expressing GFP–Crz1p were first treated with the protein synthesis inhibitor cycloheximide to ensure that we followed the same pool of GFP–Crz1p throughout the experiment. Ca2+ was added to promote nuclear localization of GFP–Crz1p, and cells were subsequently incubated with FK506 for 30 min to inhibit calcineurin activity. In wild-type cells, FK506 treatment caused GFP–Crz1p to return to the cytosol (Fig. 3), indicating that continual dephosphorylation of Crz1p by calcineurin is required to maintain its nuclear localization. However, in msn5Δ cells, GFP–Crz1p remained nuclear even after treatment with FK506 (Fig. 3), suggesting that Msn5p is required for Crz1p to exit the nucleus. Notably, the addition of Ca2+ to msn5Δ cells led to increased nuclear localization of GFP–Crz1p (Fig. 3). Because export is blocked in these cells, this change in localization is likely caused by increased nuclear import of Crz1p, and provides further evidence for the role of Ca2+/calcineurin in regulating this process (Polizotto and Cyert 2001).
Figure 3.
Msn5p is required for Crz1p nuclear export. WT (ASY472) and msn5Δ (ASY788) cells expressing GFP–Crz1p (AMS463) were grown to log phase at 21°C. Cells were treated with cycloheximide (10 μg/mL) for 20 min. CaCl2 (200 mM) was added for 10 min followed by FK506 (5 μg/mL). Images were captured 30 min after FK506 treatment. Bar, 20 μm.
Identification of the Crz1p NES
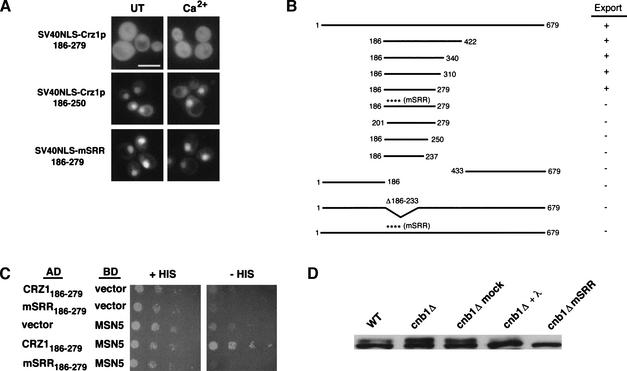
To further explore the mechanism of Crz1p nuclear export, we identified the Crz1p NES. We tested the ability of different portions of Crz1p to drive export of the constitutively nuclear GFP–SV40NLS fusion. Using this approach, we defined two distinct domains. One domain was required for calcineurin-dependent regulation of Crz1p nuclear export and is described in detail below. The other domain, amino acids 186–279, was sufficient to cause cytosolic localization of GFP–SV40NLS in the presence and absence of Ca2+ (Fig. 4A) as well as in a cnb1Δ strain (data not shown). In an msn5Δ background, however, GFP–SV40NLS–Crz1p186–279 is nuclear (data not shown). Thus, amino acids 186–279 of Crz1p contain an Msn5p-dependent NES. Residues 186–279 define the smallest region sufficient for nuclear export as GFP–SV40NLS–Crz1p186–250 and GFP–SV40NLS–Crz1p201–279 both failed to be exported (Fig. 4A,B). It is likely that this is the only NES in Crz1p because constructs encompassing the N terminus (amino acids 1–186) or the C terminus (amino acids 433–679) of the protein failed to be exported (Fig. 4B). Interestingly, the NES includes the SRR domain, which contains many putative Crz1p phosphorylation sites. The SRR is not sufficient for export, but it is necessary; when this region was deleted (Δ186–233), export activity was lost (Fig. 4B). In addition, when all serines and two threonines in the SRR domain were mutated to alanine (mSRR), export activity was again disrupted, suggesting that phosphorylation of the NES is required for its function (Fig. 4A, B). Mutating different subsets of serines in the SRR resulted in less severe export defects (data not shown); thus, elimination of multiple sites is required to disrupt the NES.
Figure 4.
Identification of the Crz1p NES. (A) WT (ASY472) cells expressing 3xGFP–SV40NLS–Crz1p186–279 (LMB162), 3xGFP–SV40NLS–Crz1p186–250 (LMB207), or 3xGFP–SV40NLS–mSRR186–279 (LMB 211) were grown to log phase at 21°C. Cells were treated with 200 mM CaCl2 and visualized using fluorescence microscopy. Bar, 20 μm. (B) Summary of export data for 3xGFP–SV40NLS–Crz1p constructs. (+) Cytosolic localization (export); (−) nuclear localization (no export). (C) The Crz1p NES interacts with Msn5p in vivo. A two-hybrid strain containing a HIS3 reporter (PJ69-4A) expressing AD–CRZ1186–279 (LMB193), AD–mSRR186–279 (LMB218), or GAL4BD–MSN5 (BM3694) alone or in combination was spotted onto media with or without histidine and grown at 21°C for 5 d. Cells were spotted using fivefold serial dilutions beginning at an OD600 of 0.05. (D) The Crz1p NES is similarly phosphorylated in WT and cnb1Δ cells. Whole cell extracts from WT (ASY472) or cnb1Δ (ASY475) strains containing LMB162 or LMB211 were analyzed by Western blot using anti-GFP antibody. Where indicated, extracts were treated with or without (mock) 200 units of λ phosphatase and incubated at 30° for 30 min.
Msn5p interacts with the Crz1p NES
We examined the interaction between the Crz1p NES and Msn5p using a directed yeast two-hybrid approach. Amino acids 186–279 of Crz1p fused to the Gal4p activation domain (AD–CRZ1186–279) interacted with Msn5p fused to the Gal4p DNA-binding domain (BD–MSN5; Fig. 4C; DeVit and Johnston 1999). Yeast containing a GAL4 UAS-driven HIS3 reporter showed robust growth on media lacking histidine when both fusions were expressed, whereas yeast expressing either construct alone showed little or no growth. Therefore, Msn5p physically interacts with the Crz1p NES in vivo. However, Msn5p failed to interact with AD–mSRR186–279 containing serine/threonine to alanine changes (Fig. 4C), suggesting that phosphorylation is required for binding between Crz1p and Msn5p and, thus, for nuclear export.
Our data indicate that phosphorylation within the SRR domain is required for NES activity; therefore, we investigated the phosphorylation state of the NES under different conditions. We examined GFP–SV40NLS–Crz1p186–279 protein isolated from wild-type and cnb1Δ backgrounds. Unlike full-length Crz1p (Fig. 2C), the NES showed similar electrophoretic mobility in wild-type and cnb1Δ cells (Fig. 4D). An upper band was evident in both samples that was missing in the GFP–SV40NLS–mSRR186–279 sample, where all putative phosphorylation sites had been mutated. When dephosphorylated in vitro, the protein collapsed to a single band, showing that the upper band was, indeed, a result of phosphorylation (Fig. 4D). Therefore, in this context, the NES directs export in wild-type and cnb1Δ cells and shows similar phosphorylation in both strains. These observations are consistent with our hypothesis that phosphorylation of the Crz1p NES is required for interaction with Msn5p.
Identification of the Crz1p export regulatory region
Full-length Crz1p shows calcineurin-dependent regulation of nuclear export, but the NES defined above, amino acids 186–279, is exported independently of calcineurin activity. To explore this difference, we used additional GFP–SV40NLS–Crz1p fusions to examine the mechanism by which calcineurin regulates Crz1p nuclear export. GFP–SV40NLS–Crz1p186–340 was the smallest fusion that behaved like full-length Crz1p. It localized to the nucleus of wild-type cells upon Ca2+ addition but remained cystosolic in cnb1Δ cells (Fig. 5A). A smaller fusion, GFP–SV40NLS–CRZ1186–310, was cytosolic in both wild-type and cnb1Δ backgrounds with or without Ca2+ addition (Fig. 5A). Therefore, amino acids 311–340 are required for calcineurin-dependent regulation of Crz1p export.
Figure 5.
Identification of the export regulatory region. (A) WT (ASY472) or cnb1Δ (ASY475) cells expressing 3xGFP–SV40NLS–Crz1p186–340 (LMB160), 3xGFP–SV40NLS–Crz1p186–310 (LMB180), or 3xGFP–SV40NLS–PIISIQΔ186–340 (LMB228) were grown to log phase at 21°C. Cells were treated with 200 mM CaCl2 and visualized using fluorescence microscopy. Bar, 20 μm. (B) Overexpression of activated calcineurin affects the localization of GFP–SV40NLS–Crz1p186–340. ASY472 cells expressing CNA2 trunc or vector (pVT100-L) and LMB160, LMB180, or LMB228 were grown to log phase at 21°C and visualized using fluorescence microscopy. Bar, 20 μm. (C) Constructs containing the Crz1p export regulatory region interact with calcineurin in vivo. A two-hybrid strain (PJ69-4A) containing a HIS3 reporter and expressing GAL4AD–Crz1p fusions LMB189, LMB226, or LMB224 and GAL4BD–CNA1 (BJP2014) alone or in combination were spotted on plates with or without histidine plus 100 mM CaCl2 and grown at 30°C for 2–3 d. Cells were spotted using fivefold serial dilutions beginning at an OD600 of 1.0. (D) The Crz1p/Msn5p interaction is dependent on Crz1p phosphorylation state. WT and cnb1Δ two-hybrid strains (PJ69-4A and ASY3) containing a HIS3 reporter and expressing LMB189 and GAL4BD–MSN5 (BM3694) alone or in combination were spotted on media with or without histidine plus 100 mM CaCl2 and grown at 21°C for 5 d. Cells were spotted using fivefold serial dilutions beginning at an OD600 of 1.0.
To further investigate the effect of calcineurin activity on Crz1p nuclear export, we examined the localization of GFP–SV40NLS–Crz1p constructs in strains overexpressing a constitutively active, truncated form of calcineurin, CNA2 trunc (Withee et al. 1997).GFP–SV40NLS–Crz1p186–340 was nuclear in this background even in the absence of Ca2+ (Fig. 5B). This construct lacks a calcineurin-dependent NLS, hence, its nuclear localization must result from down-regulated export. In contrast, expression of activated calcineurin had no effect on the cytosolic localization of GFP–SV40NLS–Crz1p186–310 (Fig. 5B).
We hypothesized that residues 311–340 of Crz1p are required for its association with calcineurin. Using a directed yeast two-hybrid approach, we looked for an interaction between calcineurin (Cna1p) and Crz1p. AD–CRZ1186–340, the region that shows regulated nuclear localization, interacted with BD–CNA1 (Jiang and Cyert 1999), but AD–CRZ1186–310 did not (Fig. 5C). In addition, an examination of phosphorylation state revealed that, like amino acids 186–279, residues 186–310 were phosphorylated in both wild-type and cnb1Δ backgrounds (data not shown). These findings support the idea that residues 311–340 of Crz1p are required for its interaction with and efficient dephosphorylation by calcineurin.
A consensus binding site for calcineurin in NFAT family members has been defined, PxIxIT (Aramburu et al. 1998) We identified a similar site, PIISIQ (PxIxIQ), within amino acids 311–340 of Crz1p and examined its role in calcineurin-dependent regulation of Crz1p. First, GFP–SV40NLS–Crz1p186–340 lacking the PIISIQ motif (GFP–SV40NLS–PIISIQΔ186–340) no longer localized to the nucleus with Ca2+ addition (Fig. 5A). Next, we examined the effect of activated calcineurin on GFP–SV40NLS–PIISIQΔ186–340 localization and found that it remained cytosolic (Fig. 5B). Finally, we tested whether these residues were necessary for Crz1p/calcineurin interaction and found by two-hybrid analysis that AD–PIISIQΔ186–340 failed to interact with BD–CNA1 (Fig. 5C). These data suggest that the PIISIQ motif is a docking site for calcineurin on Crz1p and is required for calcineurin-dependent regulation of Crz1p nuclear export. We hypothesized that Crz1p fragments containing the PIISIQ motif are dephosphorylated by calcineurin, and thus, they show calcineurin-regulated nuclear export. In contrast, smaller pieces of Crz1p that lack the PIISIQ motif, such as 186–279 and 186–310, are not efficiently dephosphorylated by calcineurin, and therefore show unregulated nuclear export. If this model is correct, then Crz1p186–340 but not Crz1p186–279 should show a calcineurin-sensitive interaction with the exportin Msn5p. We confirmed this prediction using two-hybrid analysis. Amino acids 186–340 of Crz1p interacted more strongly with Msn5p in a cnb1Δ background than in a wild-type strain (Fig. 5D). However, we found that residues 186–279 (NES) interacted similarly with Msn5p in both wild-type and cnb1Δ strains (data not shown).
Deleting the PIISIQ motif affects Crz1p function
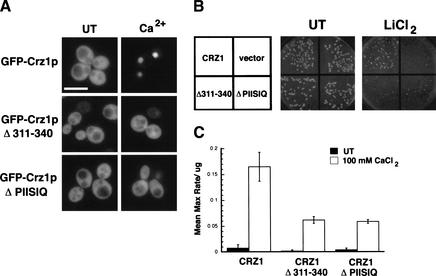
We established that amino acids 311–340 are required for calcineurin-dependent regulation of protein fusions containing small regions of Crz1p. To determine the function of this region in full-length Crz1p, we examined the localization of GFP–Crz1p constructs with in-frame deletions of either residues 311–340 or 331–336 (PIISIQ). These constructs lack the SV40NLS, relying instead on the endogenous Crz1p NLS for Nmd5p-mediated nuclear import. As previously shown, full-length GFP–Crz1p localized to the nucleus within 10 min of Ca2+ addition to the media (Fig. 6A). GFP–Crz1p311–340Δ and GFP–Crz1pPIISIQΔ, however, remained cytosolic under these conditions (Fig. 6A). At longer times, partial nuclear localization of GFP–Crz1p311–340Δ and GFP–Crz1pPIISIQΔ was observed (data not shown). Therefore, the PIISIQ motif is required for efficient nuclear localization of Crz1p in response to Ca2+. Next, we tested the ability of these constructs to complement the Li+ sensitivity of crz1Δ strains. When plated on 350 mM LiCl2, GFP–Crz1p fully complemented the crz1Δ, whereas GFP–Crz1p311–340Δ and GFP–Crz1pPIISIQΔ did not (Fig. 6B). Finally, we analyzed the transcriptional activity of the deletion constructs. Full-length GFP–Crz1p activated transcription of the CDRE::lacZ reporter in response to Ca2+ (Fig. 6C), whereas crz1Δ strains expressing vector alone showed no Ca2+ induction (data not shown). The ability of the deletion constructs to drive transcription of CDRE::lacZ was reduced (Fig. 6C), although all three fusion constructs were expressed at equivalent levels (data not shown). Together, these data indicate that the PIISIQ motif is required in vivo for maximal activation of Crz1p.
Figure 6.
Deletion of the PIISIQ motif affects Crz1p localization. (A) WT (ASY472) cells expressing 3xGFP–Crz1p (LMB127), 3xGFP–Crz1pΔ311–340 (LMB230), 3xGFP–Crz1pΔPIISIQ (LMB231), or GFP vector were grown to log phase at 21°C. Cells were treated with 200 mM CaCl2 for 10 min and visualized with fluorescence microscopy. Bar, 20 μm. (B) The PIISIQ motif is required for complementation of a crz1Δ strain. ASY472 cells expressing LMB127, LMB230, LMB231, or GFP vector were plated on selective media with or without 350 mM LiCl2 and grown at 30°C for 3 d. (C) Deletion of the PIISIQ motif affects Crz1p activity. β-Galactosidase activity was measured in crz1Δ cells harboring an integrated CDRE::lacZ reporter (ASY834). Strains expressed LMB127, LMB230, or LMB231 and were treated with (open bars) or without (black bars) 100 mM CaCl2. The numbers are the result of two replicates analyzed in triplicate, and the standard deviation is the error between samples.
Discussion
Calcineurin regulates nuclear export of Crz1p
In vivo, activation of yeast calcineurin causes rapid translocation of the Crz1p transcription factor from the cytosol to the nucleus, where it turns on transcription of specific target genes. Studies from this laboratory have characterized the nuclear transport of Crz1p to elucidate the mechanisms by which this change in localization occurs. In this report we examined export of Crz1p from the nucleus and showed that calcineurin down-regulates Crz1p export. To understand how calcineurin regulates this process, we identified the key elements of Crz1p nuclear export: its exportin and its NES.
Msn5p is the exportin for Crz1p
We have determined that Msn5p is the exportin for Crz1p. Crz1p is partially nuclear in msn5Δ cells even when calcineurin is inactive, and this allows calcineurin-independent expression of the CDRE::lacZ reporter. Furthermore, once Crz1p is in the nucleus, Msn5p is required for its return to the cytosol. Although we were unable to show direct binding between Crz1p and Msn5p in vitro, perhaps because it is a weak interaction or other stabilizing proteins are necessary, we do show that Msn5p interacts with the Crz1p NES by two-hybrid analysis. Msn5p has been identified as the exportin for several other proteins including Pho4p, Mig1p, Far1p, and Ste5p (Kaffman et al. 1998; Blondel et al. 1999; DeVit and Johnston 1999; Mahanty et al. 1999; Kunzler et al. 2001) and appears to also function in some cases as an importin (Yoshida and Blobel 2001), providing the first example of a karyopherin that functions bidirectionally.
Interestingly, MSN5 was isolated both in a screen for mutants that activate a calcineurin/Crz1p-dependent reporter (this work) and in a screen for mutants that fail to do so (Matheos et al. 1997). In msn5 mutants, we find that Crz1p accumulates in the nucleus and allows calcineurin-independent expression of CDRE::lacZ. However, msn5 cells fail to induce expression of the Ca2+-activated calcineurin/Crz1p-dependent PMC1::lacZ reporter gene (Matheos et al. 1997). This apparent contradiction may result from differences between the two reporters: CDRE::lacZ consists of four tandem repeats of a 24-bp sequence that binds Crz1p (Stathopoulos and Cyert 1997), whereas PMC1::lacZ contains 585 bp of DNA upstream of the PMC1 start site (Cunningham and Fink 1996). Thus, although the CDRE reporter specifically reflects Crz1p activity, the PMC1 promoter could contain binding sites for numerous proteins. Msn5p is pleiotropic and affects localization of several proteins (Kaffman et al. 1998; Alepuz et al. 1999; Blondel et al. 1999; DeVit and Johnston 1999; Mahanty et al. 1999). Therefore, in msn5 cells, a protein that represses transcription of PMC1::lacZ may also accumulate in the nucleus and prevent its expression despite Crz1p nuclear localization.
The Crz1p NES
The Crz1p NES does not share the same characteristics as the classical NES. As defined in the protein kinase A inhibitor (PKI) and HIV-1 Rev, the classical NES is a short leucine-rich sequence in which the key residues are all hydrophobic (Fischer et al. 1995; Wen et al. 1995), and is recognized by the exportin Crm1p (Fornerod et al. 1997; Ossareh-Nazari et al. 1997; Stade et al. 1997). In Crz1p, we have instead defined a much larger 93-amino-acid region that is sufficient to drive nuclear export. This length is consistent with the sequences identified in other Msn5p cargoes (Blondel et al. 1999; DeVit and Johnston 1999). It is not yet understood why such a large NES is required for Msn5p-mediated export. Perhaps there are multiple NES sequences within the large region that work cooperatively. Alternatively, NES secondary structure rather than a specific amino acid sequence may be important for recognition by Msn5p. Interestingly, the Crz1p NES (amino acids 186–279) does contain one classical leucine-rich sequence, LDDLLSL (amino acids 257–263) that is necessary for its activity. The Crz1p NES also contains the SRR, a domain that is conserved between NFAT and Crz1p (see below). Although in NFAT the SRR was shown to be involved in regulating nuclear import (Beals et al. 1997; Okamura et al. 2000), our data show that it plays a key role in nuclear export, which is a novel function for the Crz1p SRR.
Mechanism of calcineurin-regulated Crz1p nuclear export
Calcineurin must interact with a conserved docking site in Crz1p to regulate its nuclear export. Crz1p fragments lacking the PIISIQ motif are constitutively exported and are unaffected by the overexpression of activated calcineurin. We propose that this motif is required for Crz1p to interact with and be efficiently dephosphorylated by calcineurin. In support of this model, we show using two-hybrid analysis that Crz1p186–340 interacts with calcineurin, but that it fails to do so when the PIISIQ motif is deleted. Also, fusion proteins containing residues 186–310 or 186–279, both missing the PIISIQ motif, are similarly phosphorylated in extracts of wild-type and calcineurin mutant cells, indicating that they are not dephosphorylated in vivo.
Several results suggest that the Crz1p NES must be phosphorylated to function. First, in the two-hybrid assay, Msn5p preferentially interacts with phosphorylated Crz1p186–340. Second, mutating potential phosphorylation sites within the Crz1p NES disrupts its interaction with Msn5p and nuclear export. Although specific phosphorylation sites in Crz1p have not been identified, mutating serines and threonines in the SRR reduces the calcineurin-dependent electrophoretic mobility shift (R. Polizotto, unpubl.), indicating that this region is phosphorylated. Combining these observations we propose that the PIISIQ motif, which is distinct from the NES, mediates a critical association between calcineurin and Crz1p that is required for calcineurin-dependent dephosphorylation of the NES. This dephosphorylation inhibits the interaction between the Crz1p NES and Msn5p, thereby decreasing Crz1p nuclear export. Similarly, Msn5p-mediated export of other cargoes, Pho4p and Mig1p, also requires their phosphorylation, and it has been suggested that Msn5p specifically exports phosphoproteins (Kaffman et al. 1998; DeVit and Johnston 1999). However, Far1p binding to Msn5p does not depend on phosphorylation state (Blondel et al. 1999).
Calcineurin-dependent regulation of Crz1p activity
In unstimulated cells, Crz1p resides in the cytosol because the rate of its nuclear export is greater than the rate of its nuclear import. When calcineurin-dependent signaling is activated, either in response to specific environmental stresses or by addition of Ca2+ to the media, Crz1p rapidly translocates to the nucleus and activates gene expression. Dephosphorylation of Crz1p increases its nuclear import, by causing a conformational change that exposes its nuclear localization signal (NLS) and promotes its interaction with the importin Nmd5p (Polizotto and Cyert 2001). At the same time, dephosphorylation of the Crz1p NES inhibits its interaction with Msn5p, thereby decreasing nuclear export. Thus, dephosphorylation of Crz1p by calcineurin performs two distinct functions: it both increases the rate of Crz1p nuclear import and decreases the rate of its nuclear export. These two effects combine to cause rapid and efficient nuclear accumulation of Crz1p in response to calcineurin activation. Continual dephosphorylation by calcineurin is required to maintain Crz1p nuclear localization. When signaling terminates or calcineurin is inhibited with FK506, Crz1p quickly returns to the cytosol because of its rephosphorylation. The kinases that phosphorylate Crz1p are as yet unidentified, although the rapidity with which Crz1p localization changes suggests that they may reside in the nucleus. Crz1p phosphorylation state oppositely affects its nuclear import and export; thus, the balance between calcineurin and kinase activity tightly regulates Crz1p localization and leads to efficient and reversible transcriptional activation in response to stress.
Although the accessibility of Crz1p to its target genes is clearly regulated through its subcellular localization, nuclear localization alone is apparently not sufficient to fully activate Crz1p. The increase in Crz1p-dependent transcription observed in msn5Δ cnb1Δ cells is low despite considerable nuclear accumulation of Crz1p in these cells. Therefore, calcineurin may regulate additional aspects of Crz1p function. Calcineurin activity modulates NFAT binding to DNA (Park et al. 1995; Shaw et al. 1995); however, the ability of Crz1p to bind DNA is not calcineurin-dependent; Crz1p purified from cnb1Δ cells associates well with the CDRE (Stathopoulos 1998). Alternatively, dephosphorylation of Crz1p by calcineurin may increase its ability to function as a transcriptional activator. Future investigations will address this possibility.
Parallels between calcineurin-regulated pathways in mammals and yeast
Calcineurin activity induces gene expression in both mammalian cells and yeast by regulating the subcellular localization of a transcription factor; both NFAT and Crz1p show calcineurin-regulated nuclear import and export (Beals et al. 1997; Zhu and McKeon 1999; Okamura et al. 2000; Polizotto and Cyert 2001). Despite their similar regulatory mechanisms, Crz1p and NFAT bind DNA through distinct motifs (Jain et al. 1995; Matheos et al. 1997; Stathopoulos and Cyert 1997) and share little sequence homology. However, two critical domains are conserved in these proteins, the SRR and a calcineurin docking site, both of which are required for calcineurin-dependent regulation. The SRR, or serine-rich region, contains multiple phosphorylated residues and is required in both proteins for calcineurin-dependent regulation of nuclear transport. Phosphorylation sites in the NFAT SRR contribute to regulation of its nuclear import (Beals et al. 1997; Okamura et al. 2000), whereas phosphorylation of the Crz1p SRR is required for NES activity. Also, both proteins contain a conserved calcineurin docking site. A consensus calcineurin-binding site, PxIxIT, has been defined for NFAT family members (Aramburu et al. 1998). Here we describe a similar motif in Crz1p, PIISIQ, which is required for its interaction with calcineurin in vivo. Crz1p lacking the calcineurin docking site, Crz1pPIISIQΔ, shows defects in Ca2+-induced nuclear accumulation and transcriptional activation. Therefore, the PIISIQ interaction motif is required for efficient dephosphorylation of Crz1p by calcineurin in vivo. Crz1pPIISIQΔ retains some activity, suggesting that other regions of Crz1p may also promote its interaction with calcineurin. Calcineurin forms stable associations with several other substrates, including the IP3 and ryanodine receptors as well as dynamin 1 (Cameron et al. 1995; Lai et al. 1999). In each case, when this interaction is perturbed, calcineurin-dependent regulation of the substrate is compromised. Our work shows that in yeast, as in mammalian cells, this interaction occurs at a site on the substrate that is distinct from the phosphorylated region, and is required to target calcineurin to its substrate.
Materials and methods
Yeast media and general methods
Culture conditions and yeast media were essentially as described (Sherman et al. 1986), except that in synthetic media the nutritional supplements were added to twice the indicated levels. When Ca2+ was added to synthetic media, 3.5 g of ammonium chloride was substituted for ammonium sulfate. Buffered YPD media was made with 40 mM succinate, and the pH was adjusted to 5.5 with KOH.
All recombinant DNA procedures were carried out according to standard protocols (Ausubel et al. 1987). Yeast cells were transformed using the lithium acetate method, and bacterial cells were transformed by electroporation (Ausubel et al. 1987). DNA templates for sequencing were prepared according to the manufacturer's instructions (Wizard Miniprep kit, Promega). Sequencing was also carried out according to the manufacturer's instructions (Sequenase, USB) using [α-35S]dATP (Amersham).
Yeast strains
The yeast strains used in this study are described in Table 1. ASY788 was created by homologous recombination using an msn5::loxP-kanMX-loxP disruption cassette generated by PCR (Guldener et al. 1996). LBY172 was made by crossing ASY788 to MCY3-1D. The resulting diploid was sporulated and the double mutant selected. pAMS367 containing the CDRE::lacZ reporter was integrated at the URA3 locus of LBY196, ASY456, and ASY569.
Table 1.
Yeast strains used in this study
| Strain
|
Relevant genotype
|
Source
|
|---|---|---|
| YPH499 | MATaura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 | Sikorski and Hieter 1989 |
| YPH500 | MATα ura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 | Sikorski and Hieter 1989 |
| DD12 | Same as YPH499 except cnb1::URA::hisG | Cyert and Thorner 1992 |
| MCY3-ID | Same as YPH500 except cnb1::LEU2 | Cyert and Thorner 1992 |
| PJ69-4A | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | James et al. 1996 |
| ASY3 | Same as PJ69-4A except cnb1::URA3::hisg | A. Stathopoulos-Gerontides |
| ASY456 | Same as YPH499 except ura3-52::URA3-4xCDRE | A. Stathopoulos-Gerontides |
| ASY472 | Same as YPH499 except crzl::loxP-kanMX-loxP | Stathopoulos and Cyert 1997 |
| ASY475 | Same as DD12 except crzl::loxP-kanMX-loxP | Stathopoulos and Cyert 1997 |
| ASY569 | Same as MCY3-1D except ura3-52::URA3-4xCDRE | A. Stathopoulos-Gerontides |
| ASY788 | Same as YPH499 except msn5::loxP-kanMX-loxP | A. Stathopoulos-Gerontides |
| ASY834 | Same as ASY472 except ura3-52::TRP1-4xCDRE | A. Stathopoulos-Gerontides |
| ASY835 | Same as ASY475 except ura3-52::TRP1-4xCDRE | A. Stathopoulos-Gerontides |
| LBY172 | Same as ASY788 except cnb1::LEU2 | This study |
| LBY196 | Same as LBY172 except ura3-52::URA3-4xCDRE | This study |
| HFY133 | ura3-1 ade2-1 trp1-1 his3-11,15 leu2-3,11 can1-100 nmd5::HIS3 | A. Jacobson |
Plasmids
The plasmids used in this study are described in Table 2. pLMB134 was created by first annealing complementary oligonucleotides encoding the SV40 large T antigen NLS (Kalderon et al. 1984a,b) flanked by SpeI and HindIII restriction sites. The annealed oligonucleotides were then ligated into pOM4, a 3xGFP vector (Polizotto and Cyert 2001), digested with SpeI and HindIII. A series of in-frame 3xGFP–SV40NLS–Crz1p fusions were constructed in the following manner: PCR was used to amplify the indicated regions of CRZ1 and to introduce HindIII and SalI restriction sites flanking the PCR product. The PCR product was first ligated into a bacterial vector, PCR2.1TOPO, using the TOPO-TA kit (Invitrogen), and then shuttled into pLMB134 predigested with HindIII and SalI. pLMB184 and pLMB186 were constructed as above except HindIII and ClaI restriction sites were used. pLMB189, pLMB193, pLMB218, pLMB224, and pLMB 226 were constructed using PCR to amplify the specified regions of CRZ1 with NcoI and BamHI sites flanking the PCR product. PCR products were then ligated into pACTII to create an in-frame fusion with the Gal4p activation domain. pLMB230 was constructed by a three-way ligation of base pairs 1–930 of CRZ1 flanked by HindIII and ClaI sites and base pairs 1021–2037 of CRZ1 flanked by ClaI and SalI sites into pOM4 digested with HindIII and SalI. pLMB231 was constructed similarly but using base pairs 1–990 and 1009–2037 of CRZ1.
Table 2.
Plasmids used in this study
| Plasmid
|
Description
|
Reference
|
|---|---|---|
| AMS367 | URA3-4xCDRE integrating plasmid | Stathopoulos and Cyert 1997 |
| AMS446 | HA-CRZ1 | Stathopoulos and Cyert 1997 |
| AMS463 | GFP-CRZ1 | Stathopoulos-Gerontides et al. 1999 |
| pVT CNA2 | CNA2 trunc | Withee et al. 1997 |
| pOM4 | 3xGFP vector | Polizotto and Cyert 2001 |
| LMB127 | 3xGFP-CRZ1 | This study |
| LMB134 | 3xGFP-SV40NLS vector | This study |
| LMB148 | CRZ1 in LMB134 | This study |
| LMB158 | CRZ1 (aa 186–422) in LMB134 | This study |
| LMB160 | CRZ1 (aa 186–340) in LMB134 | This study |
| LMB162 | CRZ1 (aa 186–279) in LMB134 | This study |
| LMB163 | CRZ1 (aa 186–237) in LMB134 | This study |
| LMB165 | CRZ1 (aa 433–679) in LMB134 | This study |
| LMB178 | CRZ1 (aa 1–186) in LMB134 | This study |
| LMB180 | CRZ1 (aa 186–310) in LMB134 | This study |
| LMB184 | CRZ1 (aa Δ186–233) in LMB 134 | This study |
| LMB186 | CRZ1 (mSRR) in LMB 134 | This study |
| LMB199 | CRZ1 (aa 201–279) in LMB134 | This study |
| LMB207 | CRZ1 (aa 186–250) in LMB134 | This study |
| LMB211 | mSRR (aa 186–279) in LMB134 | This study |
| LMB228 | PIISIQΔ (aa 186–340) in LMB134 | This study |
| LMB189 | GAL4AD-CRZ1 (aa 186–340) | This study |
| LMB193 | GAL4AD-CRZ1 (aa 186–279) | This study |
| LMB218 | GAL4AD-mSRR (aa 186–279) | This study |
| LMB224 | GAL4AD-PIISIQΔ (aa 186–340) | This study |
| LMB226 | GAL4AD-CRZ1 (aa 186–310) | This study |
| LMB230 | CRZ1 (aa Δ311–340) in pOM4 | This study |
| LMB231 | CRZ1 (aa Δ331–336) in pOM4 | This study |
| BM3694 | GAL4BD-MSN5 | DeVit and Johnston 1999 |
| BJP2014 | GAL4BD-CNA1 | Jiang and Cyert 1999 |
β-Galactosidase assays
Quantitative assay
Exponentially growing yeast cells in selective synthetic media were diluted to an OD600 of 0.2 in buffered YPD and grown at 30°C for 7 h. Cells were harvested and washed once, and the cell pellets were frozen. Cells were broken using glass bead lysis (Withee et al. 1997) in Breaking Buffer (100 mM Tris at pH 8, 20% glycerol, 1 mM DTT) plus protease inhibitors (1 mM PMSF, 1 mM benzamidine, 2 μg/mL leupeptin, 2 μg/mL aprotinin). Protein concentrations of the resulting cell extracts were determined using the Bio-Rad protein assay. β-Galactosidase activity was measured at 30°C in a microtiter plate using 75 μg of total protein, 100 μL of Z-buffer (100 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.027% β-mercaptoethanol), and 40 μL of 4 mg/mL ONPG (O-nitrophenyl-β-D-galactopyranoside, Sigma). Values result from the average of two independent extracts each measured in triplicate. Alternatively, cells were grown for 5 h in buffered YPD with or without 100 mM CaCl2 and measured at room temperature in a microtiter plate using 90 μL of Z buffer and 20 μL of ONPG.
Qualitative assay
Yeast colonies were scored for β-galactosidase activity as described (Stathopoulos and Cyert 1997). Blue colonies were selected after overnight incubation with 0.2 mg/mL X-gal (Sigma).
Genetic screening
Calcineurin mutant strains containing the CDRE::lacZ reporter gene (ASY456 or ASY569) were grown to log phase (OD600 = 0.8–1.0) and plated onto selective agar plates. Cells were subjected to UV mutagenesis to ∼50% killing in a UV Stratalinker (Stratagene). β-Galactosidase filter lifts were performed and blue colonies were selected as described (Stathopoulos and Cyert 1997). There were 40 mutants identified from 128,000 colonies screened. The mutants were tested for dominance or recessiveness by mating to a cnb1Δ strain and checking CDRE::lacZ reporter activation in the diploid. MATa and MATα mutants were crossed to each other to place them into complementation groups. The majority of mutants did not fall into groups, but one complementation group of five members was defined. Mutants were then tested for specificity by selecting those that failed to activate a mutant version of the CDRE reporter. In addition, a requirement for CRZ1 was established by deleting CRZ1 in the mutant strains and assaying CDRE::lacZ reporter activation. Mutants were cloned by complementation using the YPH1 genomic library (ATCC #77162); complementing plasmids were rescued, and the ends of the insert were sequenced. The resulting sequence was compared to the genome using BLAST to determine the ORFs contained on the plasmids. The sequence shared by both complementing plasmids contained 4 ORFs on Chromosome IV: YDR334w, MSN5, YDR336w, and MRPS28. Allelic analysis with an msn5Δ strain confirmed the mutation was in MSN5.
Immunoblot analysis
Yeast cultures were grown to log phase at 21°C, harvested, and the cell pellets were frozen. FK520 (Merck), in 90% ethanol, 10% Tween-20, was added to 2 μg/mL where indicated. Protein extracts were made in Buffer 88 (20 mM HEPES at pH 6.8, 150 mM KOAc, 250 mM Sorbitol, 2 mM MgOAc, 2 mM DTT, protease inhibitors) using glass bead lysis as described above. Samples (10 μg of total protein in wild-type and cnb1Δ strains, or 50 μg of total protein in msn5Δ strains) were resolved on a 7% reducing gel. Where indicated, extracts were treated with 200 units of λ phosphatase (NEB) at 30° for 30 min. HA–Crz1p was detected using monoclonal anti-HA 12CA5 antiserum (Roche Molecular Biochemicals) and anti-mouse IgG-coupled HRP secondary antibody (Amersham). GFP–Crz1p was detected using a monoclonal GFP antibody (Covance) and anti-mouse IgG coupled HRP secondary antibody (Amersham). Immunoblots were developed using ECL (Amersham).
Fluorescence microscopy
Living cells expressing green fluorescent protein (GFP) were visualized as described (Stathopoulos-Gerontides et al. 1999) but using an Eclipse E600 microscope (Nikon) with fluorescence optics and an HB100 mercury lamp. Fluorescein filter sets (Chroma) were used to visualize GFP, and digital images were captured with a CCD 4742-95 camera (Hammamatsu) and QED software (QED Imaging). Cells were treated with 10 μg/mL cycloheximide for 20 min to inhibit protein synthesis where noted. Also, CaCl2 was added to 200 mM, and FK506 (Fugisawa) in 90% ethanol, 10% Tween-20 was added to 5 μg/mL as indicated.
Two-hybrid analysis
For the Crz1p/Msn5p interaction, strains containing the two-hybrid constructs to be tested were spotted onto selective agar media using fivefold serial dilutions beginning at an OD600 of 0.05. Plates were incubated at 21°C for 5 d. The interaction was scored by assessing growth on media lacking histidine. For the Crz1p/calcineurin interaction, yeast were spotted onto selective agar media containing 100 mM CaCl2. Fivefold serial dilutions beginning at an OD600 of 1.0 were used, and plates were incubated at 30°C for 2–3 d.
Acknowledgments
We thank Renee Polizotto for providing reagents and for helpful discussions about Crz1p nuclear transport, and Rachel Smith and Victoria Heath for critical reading of the manuscript. We are grateful to the following people for providing strains and plasmids: Mark Johnston (GAL4BD-MSN5), Allan Jacobson (nmd5Δ), Bo Jiang (GBT9-CNA1), Phil James (PJ69-4A), and Angela Stathopolous-Gerontides for numerous strains and constructs. M.S.C. is supported by National Institutes of Health (NIH) research grant GM-48729. L.M.B. is supported by NIH training grant 5T32GM07276.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mcyert@stanford.edu; FAX (650) 725-8309.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.967602.
References
- Alepuz PM, Matheos D, Cunningham KW, Estruch F. The Saccharomyces cerevisiae RanGTP-binding protein msn5p is involved in different signal transduction pathways. Genetics. 1999;153:1219–1231. doi: 10.1093/genetics/153.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramburu J, Garcia-Cozar F, Raghavan A, Okamura H, Rao A, Hogan PG. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley; 1987. [Google Scholar]
- Beals CR, Clipstone NA, Ho SN, Crabtree GR. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes & Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- Blondel M, Alepuz PM, Huang LS, Shaham S, Ammerer G, Peter M. Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes & Dev. 1999;13:2284–2300. doi: 10.1101/gad.13.17.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron AM, Steiner JP, Roskams AJ, Ali SM, Ronnett GV, Snyder SH. Calcineurin associated with the inositol 1,4,5-trisphosphate receptor–FKBP12 complex modulates Ca2+ flux. Cell. 1995;83:463–472. doi: 10.1016/0092-8674(95)90124-8. [DOI] [PubMed] [Google Scholar]
- Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes & Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- Cunningham KW, Fink GR. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J Cell Biol. 1994;124:351–363. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2226–2237. doi: 10.1128/mcb.16.5.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS, Thorner J. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol Cell Biol. 1992;12:3460–3469. doi: 10.1128/mcb.12.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS, Kunisawa R, Kaim D, Thorner J. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc Natl Acad Sci. 1991;88:7376–7380. doi: 10.1073/pnas.88.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E, Potter J, Wakeham A, Marengere L, Langille BL, et al. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature. 1998;392:182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- DeVit MJ, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- Farcasanu IC, Hirata D, Tsuchiya E, Nishiyama F, Miyakawa T. Protein phosphatase 2B of Saccharomyces cerevisiae is required for tolerance to manganese, in blocking the entry of ions into the cells. Eur J Biochem. 1995;232:712–717. [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Flanagan WM, Corthesy B, Bram RJ, Crabtree GR. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Graef IA, Chen F, Chen L, Kuo A, Crabtree GR. Signals transduced by Ca2+/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell. 2001;105:863–875. doi: 10.1016/s0092-8674(01)00396-8. [DOI] [PubMed] [Google Scholar]
- Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro R, Garciadeblas B, Rodriguez-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- Jain J, McCaffrey PG, Miner Z, Kerppola TK, Lambert JN, Verdine GL, Curran T, Rao A. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- Jain J, Burgeon E, Badalian TM, Hogan PG, Rao A. A similar DNA-binding motif in NFAT family proteins and the Rel homology region. J Biol Chem. 1995;270:4138–4145. [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Cyert MS. Identification of a novel region critical for calcineurin function in vivo and in vitro. J Biol Chem. 1999;274:18543–18551. doi: 10.1074/jbc.274.26.18543. [DOI] [PubMed] [Google Scholar]
- Kaffman A, Rank NM, O'Neill EM, Huang LS, O'Shea EK. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984a;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984b;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kuno T, Tanaka H, Mukai H, Chang CD, Hiraga K, Miyakawa T, Tanaka C. cDNA cloning of a calcineurin B homolog in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1991;180:1159–1163. doi: 10.1016/s0006-291x(05)81188-x. [DOI] [PubMed] [Google Scholar]
- Kunzler M, Trueheart J, Sette C, Hurt E, Thorner J. Mutations in the YRB1 gene encoding yeast ran-binding-protein-1 that impair nucleocytoplasmic transport and suppress yeast mating defects. Genetics. 2001;157:1089–1105. doi: 10.1093/genetics/157.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MM, Hong JJ, Ruggiero AM, Burnett PE, Slepnev VI, De Camilli P, Snyder SH. The calcineurin–dynamin 1 complex as a calcium sensor for synaptic vesicle endocytosis. J Biol Chem. 1999;274:25963–25966. doi: 10.1074/jbc.274.37.25963. [DOI] [PubMed] [Google Scholar]
- Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin–cyclosporin A and FKBP–FK506 complexes. Cell. 1991a;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ishii S, Tokai M, Tsutsumi H, Ohki O, Akada R, Tanaka K, Tsuchiya E, Fukui S, Miyakawa T. The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol Gen Genet. 1991b;227:52–59. doi: 10.1007/BF00260706. [DOI] [PubMed] [Google Scholar]
- Mahanty SK, Wang Y, Farley FW, Elion EA. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- Matheos DP, Kingsbury TJ, Ahsan US, Cunningham KW. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes & Dev. 1997;11:3445–3458. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur P, Morin N, Baginsky W, el-Sherbeini M, Clemas JA, Nielsen JB, Foor F. Differential expression and function of two homologous subunits of yeast 1,3-β-D-glucan synthase. Mol Cell Biol. 1995;15:5671–5681. doi: 10.1128/mcb.15.10.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo JM. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MJ, Geiser JR, Davis TN. Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol Cell Biol. 1996;16:4824–4831. doi: 10.1128/mcb.16.9.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Liu Y, Hirata D, Namba H, Harada S, Hirokawa T, Miyakawa T. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Aramburu J, Garcia-Rodriguez C, Viola JP, Raghavan A, Tahiliani M, Zhang X, Qin J, Hogan PG, Rao A. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- O'Keefe SJ, Tamura J, Kincaid RL, Tocci MJ, O'Neill EA. FK-506- and CsA-sensitive activation of the interleukin-2 promoter by calcineurin. Nature. 1992;357:692–694. doi: 10.1038/357692a0. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Park J, Yaseen NR, Hogan PG, Rao A, Sharma S. Phosphorylation of the transcription factor NFATp inhibits its DNA binding activity in cyclosporin A-treated human B and T cells. J Biol Chem. 1995;270:20653–20659. doi: 10.1074/jbc.270.35.20653. [DOI] [PubMed] [Google Scholar]
- Polizotto RS, Cyert MS. Calcineurin-dependent nuclear import of the transcription factor Crz1p requires Nmd5p. J Cell Biol. 2001;154:951–960. doi: 10.1083/jcb.200104078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozos TC, Sekler I, Cyert MS. The product of HUM1, a novel yeast gene, is required for vacuolar Ca2+/H+ exchange and is related to mammalian Na+/Ca2+ exchangers. Mol Cell Biol. 1996;16:3730–3741. doi: 10.1128/mcb.16.7.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T, Mickanin C, Baldwin HS, Glimcher LH. The transcription factor NF-ATc is essential for cardiac valve formation. Nature. 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- Rudolph HK, Antebi A, Fink GR, Buckley CM, Dorman TE, LeVitre J, Davidow LS, Mao JI, Moir DT. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell. 1989;58:133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- Shaw KT, Ho AM, Raghavan A, Kim J, Jain J, Park J, Sharma S, Rao A, Hogan PG. Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells. Proc Natl Acad Sci. 1995;92:11205–11209. doi: 10.1073/pnas.92.24.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stathopoulos AM. “Calcineurin regulates the CRZ1 transcription factor to control gene expression in Saccharomyces cerevisiae.” Ph.D. thesis. Stanford, CA: Stanford University; 1998. [Google Scholar]
- Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes & Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos-Gerontides A, Guo JJ, Cyert MS. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes & Dev. 1999;13:798–803. doi: 10.1101/gad.13.7.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Withee JL, Mulholland J, Jeng R, Cyert MS. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol Biol Cell. 1997;8:263–277. doi: 10.1091/mbc.8.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Blobel G. The karyopherin kap142p/msn5p mediates nuclear import and nuclear export of different cargo proteins. J Cell Biol. 2001;152:729–740. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, McKeon F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature. 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]