Abstract
Brca1 C-terminal (BRCT) domains are a common protein–protein interaction motif in proteins involved in the DNA damage response and DNA repair. The DNA-damage response protein 53BP1 has two BRCT domains that bind to the DNA-binding domain of p53. The 53BP1 tandem-BRCT region is homologous to the tandem-BRCT region of Brca1, which is involved in double-strand break repair and homologous recombination and which binds BACH1, a member of the DEAH helicase family. Here we report the structures of a human 53BP1–p53 complex and of the rat Brca1 BRCT repeats. The 53BP1–p53 structure shows that the two BRCT repeats are arranged tandemly and pack extensively through an interface that also involves the inter-repeat linker. The first BRCT repeat and the linker together bind p53 on a region that overlaps with the DNA-binding surface of p53 and involves p53 residues that are mutated in cancer and are important for DNA binding. Comparison with the structure of the tandem-BRCT region of Brca1 shows a remarkable conservation of the repeat arrangement and of the inter-BRCT repeat interface. Analysis of human BRCA1 tumor-derived mutations and conservation identifies a potential protein-binding site that we show through mutagenesis is involved in BACH1 binding. The BACH1-binding region of Brca1 consists of a unique insertion in the first BRCT repeat and the inter-repeat linker and is analogous to the region of 53BP1 that binds p53.
Keywords: 53BP1, p53, DNA damage, BRCT domain, Brca1, BACH1
The Brca1 C-terminal (BRCT) sequence motif, first identified in the breast cancer-susceptibility gene product Brca1 as an ∼95 amino acid tandem repeat, is found in a large number of proteins involved in the response to DNA damage (Koonin et al. 1996; Bork et al. 1997; Callebaut and Mornon 1997), including in Saccharomyces cerevisiae Rad9, Schizosaccharomyces pombe Crb2, mammalian DNA ligases III and IV, XRCC1, p53-binding protein 1 (53BP1) and Brca1. Over fifty proteins from bacteria to mammals contain BRCT motifs, ranging from a single copy to as many as eight copies (Yamane et al. 1997).
Brca1 is involved in the repair of double strand breaks (DSBs) by homologous recombination (Moynahan et al. 1999, 2001). In response to DSBs, Brca1-containing nuclear foci disperse and redistribute to new foci that contain Rad51, which is the mammalian homolog of the Escherichia coli RecA protein that mediates DNA recombination, PCNA (Scully et al. 1997) and the Rad50/Mre11/NBS1 complex (Zhong et al. 1999; Wang et al. 2000; Wu et al. 2000; Chiba and Parvin 2001). The redistribution of Brca1 foci is accompanied by its hyperphosphorylation by kinases that are critical for the DNA damage response, including ATM (Cortez et al. 1999), hCds1/CHK2 (Lee et al. 2000), and ATR (Tibbetts et al. 2000).
53BP1, which was first identified as a p53-interacting protein in a yeast two-hybrid screen (Iwabuchi et al. 1994), responds to DNA damage in a manner similar to Brca1. Like Brca1, 53BP1 is hyperphosphorylated by ATM and colocalizes with the Rad50/Mre11/NBS1 complex in response to DSBs (Schultz et al. 2000; Anderson et al. 2001; Rappold et al. 2001; Xia et al. 2001). The entire tandem-BRCT repeat region of 53BP1 is more homologous to the corresponding BRCT regions of Brca1, S. pombe Crb2 and S. cerevisiae Rad9 DNA damage checkpoint proteins than to most other BRCT domain-containing proteins, suggesting that 53BP1 may share some functional similarities with these proteins in the DNA damage response. In support of this, the Xenopus laevis 53BP1 ortholog was identified independently in a screen for suppressors of mitotic catastrophe resulting from premature entry into mitosis, suggesting that it has the ability to arrest or slow cell cycle progression (Xia et al. 2001).
The function of the p53 tumor suppressor in the DNA damage response is well established. In response to a variety of DNA-damaging agents, p53 is phosphorylated by ATM and related kinases, leading to its stabilization (Caspari 2000; Khanna and Jackson 2001). Acting as a transcription factor, p53 then activates a transcription program that causes cell-cycle arrest or apoptosis. The 53BP1–p53 interaction is mediated by the DNA-binding domain of p53 (residues 94–292) and the tandem-BRCT repeats of 53BP1 (residues 1702–1972). Although in vitro studies have indicated that p53 cannot bind to sequence-specific DNA and 53BP1 simultaneously (Iwabuchi et al. 1994), 53BP1 has been shown to enhance transactivation by p53 in a transient transfection assay (Thukral et al. 1994; Datta et al. 1996). P53 binds 53BP1 in X. laevis oocytes, but an interaction in somatic cells has not yet been observed (Schultz et al. 2000; Xia et al. 2001), suggesting that the role of the 53BP1–p53 complex may be limited to certain stages of development (Xia et al. 2001).
The BRCT domain has been shown to mediate protein–protein interactions in several other proteins (Zhang et al. 1998; Aravind 1999). XRCC1 and DNA ligase III bind each other through their single BRCT domains, and DNA ligase IV uses its tandem-BRCT domains to bind XRCC4 (Grawunder et al. 1998). The Brca1 BRCT domain has been shown to interact with CtBP-interacting protein (CtIP; Li et al. 1999; Yu et al. 1998), and also with a DEAH-helicase family member Brca1-associated C-terminal helicase (BACH1; Cantor et al. 2001). Germline mutations in BACH1 have been identified in two cases of early onset familial breast cancer not involving human BRCA1 mutations, further supporting a role for the Brca1–BACH1 interaction in the DNA damage response (Cantor et al. 2001).
The structures of a single BRCT repeat from the XRCC1 protein (Zhang et al. 1998) and tandem-BRCT repeats from human BRCA1 (Williams et al. 2001) showed that the BRCT motif consists of a four-stranded parallel β-sheet surrounded by three α-helices. However, there has been no structural information on how single or multiple BRCT domains function in protein–protein interactions. Here we present the structure of the 53BP1 BRCT region bound to the DNA-binding domain of p53, the structure of the rat Brca1 tandem-BRCT region, and mutagenesis studies that identify a BACH1-binding site on Brca1. These structures, in conjunction with the mutagenesis data, reveal a remarkable similarity in the way the tandem-BRCT domains are arranged, and in the way they are used to mediate protein–protein interactions in 53BP1 and Brca1.
Results
Overall structure of the 53BP1 BRCT–p53 complex
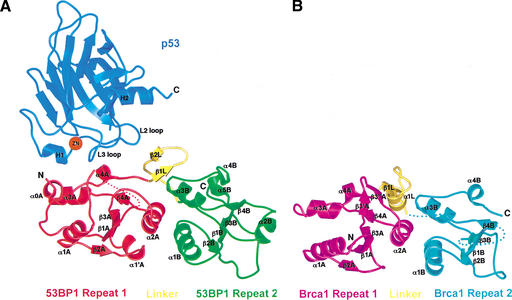
The C-terminal 271-residue fragment of 53BP1, which is necessary and sufficient for p53 binding, consists of an N-terminal extension (residues 1702–1727), two BRCT repeats, and an intervening linker (residues 1851–1867). The two BRCT repeats and the linker pack together extensively, and the entire fragment of 53BP1, adopts a globular structure (Fig. 1A). The BRCT repeats are arranged in tandem and are related by an ∼30 Å translation and a 30° rotation about an axis roughly colinear with the direction of translation, resulting in a screw-like motion (Fig. 1A).
Figure 1.
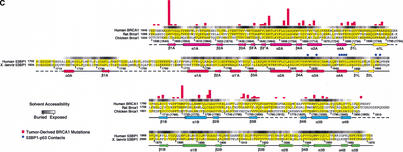
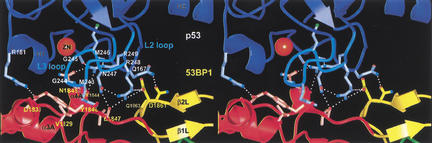
Structures of the human 53BP1–p53 complex and the rat Brca1 C-terminal tandem-BRCT region. (A) Schematic representation of the 53BP1–p53 complex showing the DNA-binding domain of p53 (blue) interacting with the C-terminal helix of the first BRCT repeat (red) and the inter-repeat linker (yellow), but not the second BRCT repeat (green) of 53BP1. The red dotted line indicates a 30-amino-acid insertion between β1A and α1A that does not have interpretable electron density. (B) Structure of rat Brca1 C-terminal tandem-BRCT region with the first BRCT repeat colored magenta, the linker yellow, and the second BRCT repeat cyan. A portion of the linker and the region between β3B and β4B are missing in the electron density and are represented by cyan dotted lines. (C) Sequence alignments of the first BRCT repeats and linkers (top) or the second BRCT repeats (bottom) from Brca1 and 53BP1. Secondary structure elements are colored as in A and B, with the disordered regions in dashes. Residue numbering of rat Brca1 or human BRCA1 (in parentheses) are indicated below the sequences. Red bars above the Brca1 sequences indicate a relative frequency of tumor-derived missense mutations as obtained from BIC database. Residue numbering of 53BP1 from human is shown below the 53BP1 sequences. Blue dots above the 53BP1 sequences represent the residues that contact p53.
As seen in previous structural studies (Cho et al. 1994; Gorina and Pavletich 1996), the p53 DNA-binding domain (referred to as the p53 core domain) consists of a β-sandwich scaffold that supports two large loops (L2 and L3) and an α-helix (H2 helix; Fig. 1A). The L3 loop, which is stabilized by a tetrahedrally coordinated zinc atom shared with the L2 loop, and the H2 helix form the DNA-binding surface of p53. In the 53BP1–p53 complex, the C-terminal portion of the first BRCT repeat and the linker region of 53BP1 form a continuous surface that binds part of the DNA-binding surface of the p53 core domain, mostly through contacts to the L3 loop and a smaller number of contacts to the L2 loop (Fig. 1A). The 53BP1–p53 complex formation buries a total of 1275 Å2 of mostly polar surface area.
The 53BP1 tandem-BRCT structure shows remarkable structural similarity to the tandem-BRCT region of Brca1 (residues 1589–1817 of rat Brca1 and residues 1646–1859 of human BRCA1; Williams et al. 2001; Fig. 1C) in the way the two repeats are arranged, in the details of the inter-BRCT repeat interface, and in the way the linker region participates in the interface (Fig. 1A,B).
Structure of the 53BP1 BRCT region and the interface between the tandem BRCTs
The individual BRCT repeats of 53BP1 have the same overall α/β fold observed in the structure of a single BRCT domain of XRCC1 (Zhang et al. 1998) but contain several sequence insertions as well as deviations in the positions of secondary structure elements (Fig. 1A). The BRCT repeats of 53BP1 consist of a four-stranded parallel β-sheet (strands β1A to β4A and β1B to β4B; the suffixes A and B indicate the first and second 53BP1 repeats, respectively) flanked by two α-helices on one side (α1A, α1B and α3A, α3B) and one on the other (α2A, α2B; Fig. 1A). The C-terminal portion of the first BRCT repeat of 53BP1 contains a 310 helix near its C terminus (α4A in residues 1843–1847; Fig. 1A,C).
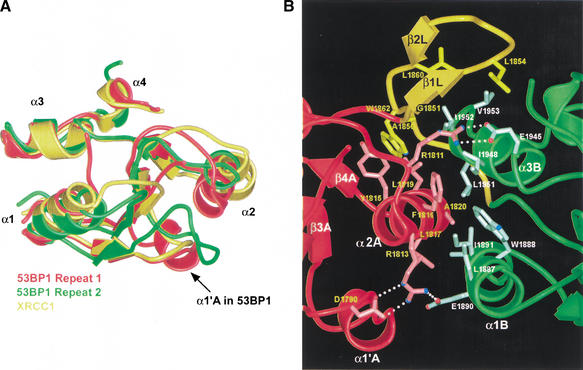
The positions and orientations of individual helices in the 53BP1 BRCT repeats show considerable deviations from a canonical structure, as defined by the superposition of the BRCT structures from 53BP1, Brca1, and XRCC1 (Fig. 2A,C). The most divergent structural elements are the α2A and α1B helices. Relative to the canonical structure, α2A is shifted by ∼6 Å along its helical axis and tilted by ∼30°, and α1B is tilted by ∼11°. These helices pack in the inter-BRCT repeat interface (Figs. 1A, 2B) and their deviation from the canonical structure is critical for the packing. The loops between the secondary structure elements also vary in size and precise structure (Figs. 1C, 2A). The loop between β2 and β3, termed the c2 loop (Zhang et al. 1998), contains a five-residue helical insertion (α1‘A) in repeat 1 of 53BP1 and an eight-residue insertion in repeat 2 (Figs. 1C, 2A).
Figure 2.
The rat Brca1 BRCT domains have the same tandem arrangement as in 53BP1. (A) Superimposition of the two BRCT repeats (red and green) of 53BP1 and the single BRCT domain from XRCC1 (yellow). Divergent regions among the BRCT domains are located within loops (e.g., the insertion helix α1‘A in 53BP1) and also include the orientations and positions of the α1 and α2 helices. (B) Close-up view of the inter-BRCT repeat interface of 53BP1. Side chains from the first BRCT repeat are in light-red, those from the linker are in light-yellow, and those from the second are in light-green. The α2A helix of the first repeat (red) together with α1B and α3B helices of the second repeat (green) form a three-helix bundle that is stabilized by a β-hairpin linker (β1L and β2L, colored yellow) and an insertion helix α1‘A. White dotted lines indicate hydrogen bond interactions. (C) Superimposition of the tandem-BRCT repeats of 53BP1 (gray) and rat Brca1, with the first BRCT repeat colored magenta, the second BRCT repeat cyan, and the linker yellow. (D) Close-up view of the inter-BRCT repeat interface of rat Brca1. Side chains from the first repeat are in light-pink, those from the linker are in light-yellow, and those from the second repeat are in light-blue. White dotted lines indicate hydrogen bond interactions. The letter M indicates the tumor-derived missense mutations that occur at a high frequency (Met 1598, Arg 1645, Ala 1654, Met 1720, and Gly 1733 in rat).
The inter-BRCT repeat interface of 53BP1 contains both core BRCT motif elements and the inter-repeat linker. The interface buries a total of ∼2400 Å2, with one third of it being contributed by the linker (Fig. 2B). The central part of the interface forms through the packing of α2A of the first repeat with α1B and α3B of the second repeat, in a manner analogous to three-helix bundles (Fig. 2B). The three α-helices contribute eight hydrophobic residues to the interface (Fig. 2B). This three-helix bundle packing arrangement would not be possible if the positions and orientations of the α2A and α1B helices did not deviate from the canonical BRCT motif structure (Fig. 2A,B). The hydrophobic interface contacts are flanked by charge-stabilized hydrogen bond networks near the ends of the helices (Fig. 2B).
The inter-repeat linker adopts a β-hairpin structure (residues 1851–1860; β1L and β2L, where L indicates a linker element) followed by an extended segment (residues 1861–1867), both of which pack extensively with both BRCT repeats at the inter-BRCT repeat interface (Figs. 1A, 2B). One face of the hairpin contains hydrophobic residues that pack with residues from the α2A and α3B helices of the three-helix bundle (Fig. 2B).
A comparison of human and X. laevis 53BP1 sequences shows that many of the hydrophobic interface residues and all five amino acids that participate in salt bridges on the three-helix bundle and the linker β-hairpin are conserved (Figs. 1C, 2B), consistent with a model that the two BRCT-repeat region is a single globular domain. In contrast, the single BRCT repeat structure of XRCC1 differs from the portions of the 53BP1 repeats that form the interface in that (1) it does not show any deviations from a canonical structure in its α1 and α2 helices, and (2) it has only one exposed hydrophobic amino acid (Met 62) in its α1, α2, and α3 helices.
Comparison of 53BP1 and rat Brca1 tandem-BRCT structures
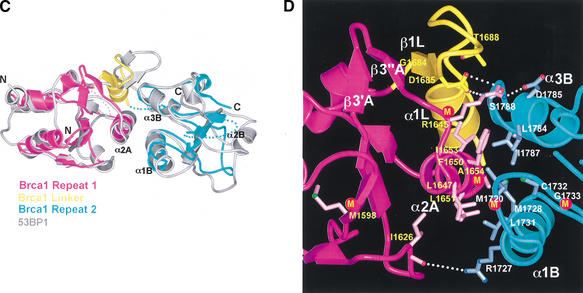
The entire tandem-BRCT structures of 53BP1 and rat Brca1 can be superimposed with a root mean square deviation (RMSD) of 1.71 Å in the positions of 138 out of 229 Brca1 Cα atoms (Fig. 2C), including all eight BRCT core β strands and seven of the eight core helices (the Brca1 α2B helix is disordered in our crystals; Figs. 1B, 2C). The most striking structural conservation is at the inter-BRCT repeat interface (Fig. 2C). The interface helices α2A of the first repeat and α1B and α3B of the second repeat have the same angular and positional deviations from the consensus structure as those of 53BP1, allowing them to form a three-helix bundle interface essentially identical to that of 53BP1 (Fig. 2C). The central interactions in the three-helix bundle interface of rat Brca1 involve hydrophobic residues that occur at positions on the helices equivalent to those in 53BP1 (Fig. 2B,D). Three of them are conserved between Brca1 and 53BP1 (Phe 1650, Leu 1651, and Ala 1654 of rat Brca1) and the rest are conservatively substituted (Figs. 1C, 2B,D). Just as in the 53BP1 interface, the Brca1 three-helix bundle is flanked by charge-stabilized hydrogen bond interactions near the ends of the helices (Fig. 2B,D). These hydrogen bond contacts (Arg 1645, Asp 1785, Arg 1727, and the backbone carbonyl of Ile 1626) occur at similar positions compared to those in 53BP1, but the detailed interactions differ somewhat due to insertions in the vicinity and differences in the linker region (Fig. 2B,D).
The 23-residue inter-repeat linker of rat Brca1 (residues 1682–1704) occupies the same position as that of 53BP1, packing on one side of the three-helix bundle and stabilizing the inter-BRCT repeat interface (Fig. 2B,D). Like 53BP1, the Brca1 linker contains a β-hairpin-like structure that packs with the α2A and α3B helices, although only the first segment has β-sheet backbone phi–psi angles (β1L; Fig. 2B,D). The linker hairpin forms a continuous β-sheet-like structure with β3′A–β3″ A (Fig. 2D), which corresponds to a seven-residue insertion between β3A and α2A of Brca1 compared to 53BP1 (Fig. 1C). The presence of the β3′A–β3"A insertion in Brca1 is associated with a rotation of the linker hairpin relative to that of 53BP1. Following the linker hairpin, Brca1 has a helix (α1L) compared to the extended segment of 53BP1 (Fig. 2B,D).
The structural conservation in the inter-BRCT repeat interfaces of 53BP1 and Brca1 suggests that their tandem-BRCT regions diverged from a common ancestral protein. Although it is possible that the two tandem-BRCT domains emerged separately through convergent evolution starting with a single ancestral BRCT domain, the similarities in the BRCT–BRCT contacts, such as those made by Arg 1645 of Brca1 and Arg 1811 of 53BP1 (Fig. 2B,D), would make this less likely. The S. pombe Crb2 and S. cerevisiae Rad9 proteins have a pattern of hydrophobic and polar residues characteristic of the 53BP1 and Brca1 inter-BRCT repeat interfaces. Although the overall level of conservation is much lower compared to that between Brca1 and 53BP1, especially in the second BRCT repeats, the conservation in Crb2 and Rad9 of a few key interface residues, such as the arginines at positions corresponding to Arg 1645 of Brca1 and Arg 1811 of 53BP1, suggests that they may be evolutionarily related to Brca1 and 53BP1 and supports the hypothesis that they may share functional similarities.
53BP1–p53 interactions
The 53BP1–p53 interactions are concentrated on the L3 loop of p53. The L3 loop has a rigid conformation that is stabilized by zinc binding, by backbone hydrogen bonds, and by side chain interactions with the rest of the p53 structure (Cho et al. 1994). The seven-residue tip (residues 243–249) of the L3 loop binds a contiguous region of 53BP1 consisting of the α3A and α4A helices at the C terminus of the first BRCT repeat and the inter-repeat linker (Fig. 3). The interactions rely primarily on networks of hydrogen bonds, involving both side chain and backbone groups, and to a lesser extent on van der Waals contacts (Fig. 3). The L3 loop is one of the two key structural elements that make up the DNA-binding surface of p53, containing an arginine residue (Arg 248) that binds in the minor groove of the DNA (Cho et al. 1994). The other structural element on the DNA-binding surface of p53 is the loop-sheet-helix motif (H2 helix), which is not involved in binding to 53BP1 (Figs. 1A, 3).
Figure 3.
Stereo view of the 53BP1–p53 interface. The secondary structure of p53 is colored blue except for the L3 loop, which is cyan. The side chains of p53, first BRCT repeat, and the linker of 53BP1 are colored light-blue, light-red, and light-yellow, respectively. Hydrogen bonds are indicated by white dotted lines.
The 53BP1–p53 hydrogen bond networks are centered on Arg 248 and Arg 249 of p53. Arg 248 hydrogen bonds with the side chain of Asp 1861 from the β-hairpin of the inter-repeat linker and with the backbone carbonyl group of Leu 1847 at the start of the linker (Fig. 3). The Arg 248 side chain is stabilized by an intramolecular hydrogen bond with the OD1 group of Asn 247 (L3) whose ND2 group hydrogen bonds with the backbone carbonyl group of Leu 1847 from the α4A helix of 53BP1 (Fig. 3). The Arg 249-centered hydrogen bond network involves the OD1 group of Asn 1845 and the backbone carbonyl of Arg 1844, both from the α4A helix of 53BP1 (Fig. 3). In the structures of the p53–DNA complex and of the isolated p53, Arg 249 has an important role in stabilizing the structure of the L3 loop by hydrogen bonding with the backbone carbonyl groups of Gly 245 and Met 246 and with the side chain of Glu 171 from the L2 loop (Cho et al. 1994). In the 53BP1–p53 complex, the hydrogen bonds between Arg 249 and the backbone carbonyl groups are maintained, but the hydrogen bonds with Glu 171 are disrupted, with the NH1 and NH2 groups of Arg 249 now interacting with 53BP1 instead (Fig. 3). The Glu 171 side chain is displaced from its position in monomeric p53, and is disordered in the crystals.
Van der Waals contacts are made by the side chain of Met 243 and Cα of Gly 244, which wedge in between the α3A and α4A helices of 53BP1 and contact the side chains of Val 1829 and Tyr 1846 (Fig. 3). These p53 residues pack closely with 53BP1, allowing the backbone amide of Gly 244 of p53 to form a hydrogen bond with the backbone carbonyl group of Asn 1845 (Fig. 3). The L3 loop contacts are augmented by a few additional contacts from the adjacent L2 loop to α3A and the linker (Fig. 3).
The 53BP1 residues that contact p53 are either conserved (Val 1829, Tyr 1846, and Asn 1845) or are conservatively substituted (Asp 1861 to Glu) in X. laevis 53BP1 (Fig. 1C). The residues that make up the α3A, α4A, and the β-hairpin part of the linker are also highly conserved (Fig. 1C), indicating that these structural elements will adopt a very similar structure in X. laevis 53BP1. Relative to the overall ∼68% identity between the X. laevis and human 53BP1 BRCT regions, the high conservation of these residues supports an important functional role for the p53-binding site of the 53BP1 structure.
Implications for the p53 pathway
The L3 loop has a central role in p53’s tumor suppressor function because three of the six mutation hotspots (Gly 245, Arg 248, and Arg 249) and ∼30% of the tumor-derived missense mutations map to this region (Cho et al. 1994). In addition, the L3 loop region corresponds to one of the five evolutionarily conserved regions of p53. Mutation of the Arg 248 hotspot, which makes key contacts to both DNA and 53BP1, disrupts both DNA binding and 53BP1 binding (Thukral et al. 1994). Similarly, mutations of Gly 245 and Arg 249, which have central roles in the structural integrity of the L3 loop (Cho et al. 1994), also disrupt both DNA and 53BP1 binding (Thukral et al. 1994). The involvement of these L3 loop residues in both DNA and 53BP1 binding makes it difficult to estimate the contribution, if any, that loss of 53BP1 binding may make to tumorigenesis. We note however, that Met 243, which is a key 53BP1 contact but has no role in DNA binding or stabilizing the structure of the L3 loop, is also found mutated in cancer although at a much lower frequency (six of 3393 missense mutations in the database), compared to the residues that contact DNA. Although another role for Met 243 cannot be ruled out, this observation suggests that loss of 53BP1 binding but not of DNA binding may contribute to the deregulation of the p53 DNA-damage response pathway in certain contexts.
The complex structure shows that 53BP1 binding would block the L3 loop from interacting with DNA (Figs. 1A, 3). This would interfere with sequence-specific DNA binding, due to the unavailability of Arg 248 for the recognition of the A/T-rich region of the consensus sequence (Cho et al. 1994). However, the H2 helix of p53 remains accessible in the 53BP1–p53 complex (Figs. 1A, 3). In principle, the 53BP1–p53 complex could still use the H2 helix to bind in the major groove of the DNA in a nonspecific manner if the DNA is bent away from the L3 loop or is a DNA end that terminates before the L3 loop region. In this hypothetical model of a 53BP1–p53–DNA complex, 53BP1 could provide additional contacts to the DNA.
Mapping of the BACH1-binding site on Brca1
To start investigating which portions of the Brca1 BRCT structure are involved in protein–protein interactions, we first examined the distribution of the tumor-derived human BRCA1–BRCT missense mutations in the Breast Cancer Information Core (BIC) database (Fig. 1C). The majority of the mutations map to residues that have structure-stabilizing roles (Williams et al. 2001). Four of the five most frequently mutated residues are at the inter-BRCT repeat interface, and the fifth is in the hydrophobic core of the first BRCT repeat (Figs. 1C, 2D; Williams et al. 2001). These observations indicate that the integrity and precise structure of the inter-BRCT repeat interface are critical for the tumor suppressor function of the Brca1 BRCT region.
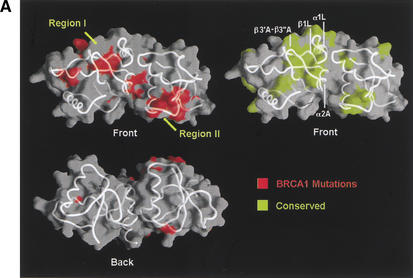
Although the majority of the mutated residues have structure-stabilizing roles, there are several mutated residues that are either fully or partially solvent-exposed and do not appear to have significant structural roles in the rat Brca1 structure. These residues tend to cluster in two regions of the molecular surface of Brca1. The first region (region I) consists of the insertion hairpin β3′A-β3"A of the first BRCT repeat, the adjacent linker hairpin, and a portion of the inter-BRCT repeat interface that is partially solvent-exposed. The second region (region II) consists of the β3B strand and the loop between β1B and α1B in the second BRCT repeat (Fig. 1C). However, in the structure of the human BRCA1 BRCT (Williams et al. 2001), most of region II is not solvent-exposed because it packs with the α2B helix that is disordered in our rat Brca1 BRCT structure. Thus, region I is the most likely candidate for a protein-binding site.
A protein-binding function for region I is also supported by its conservation in Brca1 orthologs (Fig. 1C). The highest degree of conservation closely corresponds to the structural elements that make up region I: the β3′A-β3"A insertion hairpin, the α2A interface helix, the linker hairpin and α1L helix (>85% identities compared to the ∼60% overall identity in the tandem BRCTs; Figs. 1C, 4A).
Figure 4.
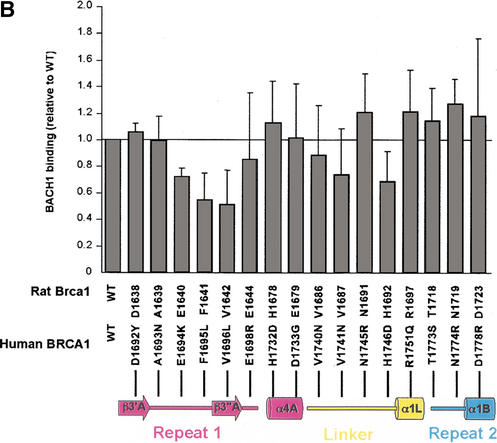
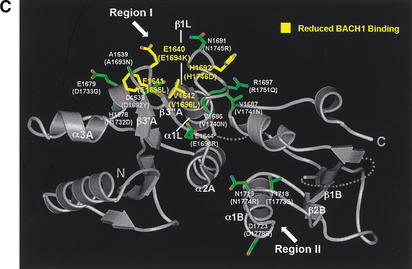
Tumor-derived mutations, sequence conservation and mutagenesis identify a BACH1-binding site on the rat Brca1 structure. (A) Composite figure showing the molecular surface representation (transparent gray) and backbone trace (white coil) of the Brca1 BRCT repeats. Red patches on the surface represent the tumor-derived mutations from the BIC database (left) and the yellow patches (right) represent residues conserved in the human, rat, and chicken Brca1 orthologs. Comparison of the front (top left) and the back (bottom left) views of the surface representations showing the clustering of mutations. (B) Region I binds to BACH1. 35S-labeled, in vitro-translated BACH1 fragment (residues 888–1063; Cantor et al. 2001) was tested for binding to purified human GST–BRCA1 (residues 1529–1863) fusion proteins carrying point mutations in surface residues within regions I and II. The binding activities of the mutants were calculated relative to the wild-type BACH1-binding activity set to 1. The data was averaged from five experiments. (C) Brca1 residues in Region I that interact with BACH1. The human residues with their amino acid changes in the mutants are indicated in parentheses. The mutated surface residues for which BACH1 binding was reduced are colored yellow; residues whose mutation did not produce a significant effect are colored green.
To address the possibility that region I may represent the binding site for BACH1, we introduced single amino-acid mutations in the human BRCA1 BRCT region (residues 1529–1863) and tested the ability of the purified mutant GST-fusion proteins to bind the in vitro translated Brca1-interacting domain of BACH1 (residues 888–1063; Cantor et al. 2001). We mutated not only the surface residues that are mutated in cancer, but also solvent-exposed residues that are conserved. We did not mutate residues that are likely to have significant structure-stabilizing roles (Arg 1699, Cys 1643, Gly 1684, and Asp 1685 of human BRCA1). We also mutated three region II-residues that are solvent-exposed in the structure of human BRCA1 (Williams et al. 2001), although none of these have been found mutated in cancer.
Figure 4B shows that three of the mutants on the β3′A-β3"A hairpin of region I have significant reductions in BACH1 binding. The Glu 1694 to Lys (Glu 1640 in rat), Phe 1695 to Leu (Phe 1641 in rat), and Val 1696 to Leu (Val 1642 in rat) mutations resulted in ∼30%, ∼45%, and 50% reductions in BACH1 binding, respectively (Fig. 4B). These three residues define a nearly continuous surface on rat Brca1 consisting of the β3′A-β3"A hairpin (Fig. 4C). The Phe 1695 to Leu and Val 1696 to Leu mutations correspond to tumor-derived mutations (Fig. 1C), suggesting that the role of these residues in BACH1 binding may be important for the tumor suppressor function of Brca1. The His 1746 to Asp (His 1692 in rat) mutation on the linker between β1L and α1L, also reduced BACH1 binding, but only to a modest extent (Fig. 4B,C). Mutations in region II did not result in any significant effects on BACH1 binding (Fig. 4A,B). Taken together, these results indicate that region I is a primary BACH1-binding site (Fig. 4A,C).
Discussion
The BRCT motif has been implicated in protein–protein interactions in several proteins involved in DNA repair and recombination. In addition to 53BP1–p53 and Brca1–BACH1 complexes, XRCC1 and DNA ligase III, which are involved in base excision repair, have been shown to interact with each other through their C-terminal single BRCT motifs (Ljungquist et al. 1994; Caldecott et al. 1995; Nash et al. 1997). In the structure of the 53BP1–p53 complex, both a core BRCT structural element (α4A helix) and a unique linker element mediate p53 binding (Fig. 3). The participation of the inter-repeat linker in p53 binding may help explain the conservation of the inter-BRCT repeat interface. As the linker structure folds on the interface, an important functional role for the linker would be reflected in the conservation and integrity of the interface.
Strikingly, the structure of the rat Brca1 tandem-BRCT repeats shows not only a near identical arrangement of the two repeats but also very similar inter-BRCT contacts and an analogous linker structure at the interface (Fig. 2C). Sequence conservation and tumor-derived mutations suggest that region I, consisting of the inter-BRCT repeat interface, the linker, and a β-hairpin insertion adjacent to the linker is important for Brca1 function, and mutations in solvent-exposed residues of this region demonstrate that it is involved in binding BACH1 (Fig. 4A–C). Interestingly, region I of Brca1 overlaps with the region of 53BP1 that binds p53 (Figs. 1A,B, 4C), revealing similarities not only in the structures of the Brca1 and 53BP1 BRCT repeats but also in the way they function in protein–protein interactions. Although both proteins use a similar region to bind their respective partners, the structural details in their protein-binding sites differ. The presence of the insertion hairpin in Brca1 would sterically prevent p53 from binding to this site.
The comparison of the 53BP1 and Brca1 BRCT structures suggests that the BRCT-containing proteins evolved specificity for different proteins not only by evolving different amino acids on structurally conserved portions of the core BRCT motif, but also by evolving divergent structural elements as insertions within or in between BRCT repeats. This is reminiscent of other regularly repeating protein-recognition motifs, where divergence from a canonical structure is often associated with a functional uniqueness and biological specificity. For example, in the four-ankyrin repeat protein p16INK4A, a divergent region of the second ankyrin repeat that has a helix replaced by a loop has a central role in binding Cdk6 (Russo et al. 1998).
The similar structures of the 53BP1 and rat Brca1 tandem-BRCT regions suggest that the two proteins may be evolutionarily related, possibly having diverged from a common tandem-BRCT ancestral protein. This observation, coupled to the conservation of residues in the inter-BRCT repeat interface between 53BP1, Brca1, and the S. pombe Crb2 and S. cerevisiae Rad9 DNA damage checkpoint genes, supports the proposed model that these proteins may share similarities in their cellular functions as well (Saka et al. 1997).
Materials and methods
Protein expression and purification
The DNA-binding domain of human p53 (residues 94–292) was expressed and purified as described previously (Cho et al. 1994). The human 53BP1 (residues 1702–1972) was overexpressed as a GST fusion protein in the BL21(DE3) strain of E. coli at room temperature and isolated from the soluble cell lysate by glutathione-Sepharose (Pharmacia) affinity chromatography. After cleavage from the GST by thrombin, the 53BP1 protein fragment was purified by anion-exchange and gel filtration chromatography. The purified p53 and 53BP1 polypeptides were combined and isolated as a complex from an additional round of gel-filtration chromatography. The purified complex was concentrated to ∼13 mg/mL in 50 mM BTP–HCl (pH 6.8), 200 mM NaCl, and 5 mM dithiothreitol (DTT) by ultrafiltration. The rat Brca1 (residues 1589–1817) containing the tandem-BRCT repeats was overexpressed as a GST fusion protein and purified as described for 53BP1. Purified rat Brca1 was concentrated to ∼75 mg/mL in 10 mM Na-HEPES (pH 7.5), 150 mM NaCl, and 5 mM DTT by ultrafiltration.
Crystallization and data collection
The 53BP1–p53 complex crystals were grown at 4°C using the hanging-drop vapor-diffusion method by mixing an equal volume of the purified protein complex and crystallization buffer containing 50 mM sodium citrate (pH 6.0), 125 mM ammonium acetate, 16% (w/v) polyethylene glycol (PEG) 4000, and 5 mM DTT. The best crystals were obtained in drops for which the reservoir solution lacked ammonium acetate. Diffraction data were collected at −170°C with crystals flash-frozen in the crystallization buffer containing 36% (w/v) PEG 4000 and lacking DTT. The crystals formed in space group P212121 with a = 73.1, b = 95.0, c = 133.7 Å and contained two complexes per asymmetric unit. Multi-wavelength anomalous diffraction (MAD) data were collected at the zinc edge at energies of 9662.5 electron Volts (eV), 9673.5 eV, and 9823.0 eV (corresponding to λ1, λ2, and λ3, respectively, in Table 1) at the X4A beamline of the National Synchrotron Light Source at Brookhaven National Laboratory (NSLS). The rat Brca1 crystals were grown at 4°C using the hanging-drop vapor-diffusion method from a crystallization buffer containing 50 mM Tris-HCl (pH 8.5), 150 mM NaCl, 18% (w/v) PEG 8000, and 5 mM DTT. The crystals formed in space group C2 with a = 84.5, b = 58.7, c = 65.4 Å, β = 111.2° and contained one Brca1 molecule per asymmetric unit. For multiple isomorphous replacement (MIR) experiments, crystals were soaked in either 1 mM thimerosal or 2 mM K2PtCl4 for 2 h. Diffraction data were collected at −170°C with the crystals flash-frozen in the crystallization buffer containing 20% (w/v) PEG 8000 and 20% (w/v) PEG 400 and lacking DTT. The native data were collected at the A1 beamline of Cornell High Energy Synchrotron Source (CHESS) and the derivative data were collected at the X9A beamline of NSLS. All diffraction data were processed using the HKL suite (Otwinowski and Minor 1997).
Table 1.
Summary of crystallographic analysis
| MAD data collection (53BP1–p53 complex) | MAD phasing statistics (15–2.7Å) | |||||||
|---|---|---|---|---|---|---|---|---|
| Wavelength (Å)
|
Resolution (Å)
|
Measured reflections
|
Unique reflections
|
Coverage (%)
|
Rsym (%)
|
Phasing power
|
Rcullis
|
Rcullis (ano)
|
| λ1 (1.2832) | 2.5 (2.6–2.5) | 169611 | 29872 | 89.4 (56.5) | 5.1 (17.8) | — | — | 0.84 |
| λ2 (1.2817) | 2.5 (2.6–2.5) | 170142 | 30000 | 89.4 (56.4) | 5.3 (18.6) | 0.71 | 0.88 | 0.79 |
| λ3 (1.2622) | 2.5 (2.6–2.5) | 172211 | 29847 | 89.5 (55.9) | 5.3 (21.7) | 0.88 | 0.84 | 0.83 |
| Refinement statistics | RMSD
|
|||||||
| Resolution range (Å) | Reflections (|F| > 0σ) | Atoms (total, water) | R-factor (%) | R-free (%) | bonds (Å) | angles (°) | B factor (Å2) | |
| 15–2.5 (2.6–2.5) | 28192 | 6988, 216 | 21.6 (32.1) | 25.6 (39.5) | 0.014 | 1.73 | 2.2 | |
| MIR data collection (rat Brca1) | MIR phasing statistics (15–3.4Å) | |||||||
| Crystal | Resolution (Å) | Measured reflections | Unique reflections | Coverage (%) | Rsym (%) | Phasing power | Rcullis | |
| Native | 2.1 (2.18–2.10) | 55198 | 17223 | 97.0 (97.9) | 8.2 (45.7) | — | — | |
| Thimerosal | 3.7 (3.87–3.70) | 16590 | 3662 | 98.9 (98.5) | 14.8 (22.8) | 1.48 | 0.75 | |
| K2PtCl4 | 2.8 (2.90–2.80) | 24596 | 7387 | 99.1 (99.0) | 8.7 (32.3) | 0.93 | 0.85 | |
| Refinement statistics | RMSD | |||||||
| Resolution range (Å) | Reflections (|F| > 0σ) | Atoms (total, water) | R-factor (%) | R-free (%) | bonds (Å) | angles (°) | B factor (Å2) | |
| 15–2.3 (2.4–2.3) | 12529 | 1702, 177 | 20.3 (26.2) | 25.8 (34.1) | 0.011 | 1.61 | 4.5 | |
Coverage (%) indicates the percentage of total theoretically observable reflections measured. Rsym = ΣhΣi |Ih,i − Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of reflection h. Phasing power = [FH(calc)2/(FPH(obs) − FPH(calc))2]1/2, where FPH(obs) and FPH(calc) are the observed and calculated derivative structure factors, respectively. Rcullis is the mean residual lack-of-closure error divided by the dispersive or anomalous difference. Figure of Merit (FOM) = <Σ P(α) exp(iα)/ΣP(α)>, where P(α) is the probability distribution for the phase α. R = Σ |Fobs − Fcalc|/ΣFobs, where Fobs = FP, and Fcalc is the calculated protein structure factor from the atomic model. R-free is the R factor calculated using 5% of the reflection data chosen randomly and omitted from the refinement process, and R-factor is calculated with the remaining data in the refinement. The root means square deviation (RMSD) values in bond lengths and angles are deviations from ideal values, and the RMSD values in B factors are calculated between covalently bonded atoms.
Structure determination and refinement
For the 53BP1–p53 complex structure, initial phases were obtained from a molecular replacement solution of the p53 core domain structure using AMORE (CCP4 1994), and were used to locate the two zinc atoms in the asymmetric unit (one zinc per p53 molecule) using the MAD diffraction data. MAD phases, calculated with the program MLPHARE (CCP4), had a mean figure of merit (FOM) of 0.48 to 2.7 Å resolution, and were improved by density modification with DM (CCP4). Despite the presence of only one zinc atom per 474 amino acids, the experimental map was clearly interpretable for both p53 and 53BP1, including electron density for the majority of the side chains. The initial model of 53BP1 was built with the program O (Jones et al. 1991), and was refined by CNS (Brünger et al. 1998). The refinement statistics are shown in Table 1. Nearly all residues (99.6%) are in the allowed regions of the Ramachandran plot. An N-terminal fragment (residues 1702–1713) and the region between β1A and α1A (residues 1741–1769) do not have interpretable electron density, and we presume are disordered. Residues 1741–1769 correspond to a 29 amino acid insertion that is not found in other BRCT repeats and is rich in polar and charged residues. For the BRCT region of rat Brca1, initial MIR phases were calculated with MLPHARE. They had a mean FOM of 0.42 to 3.4 Å, and were improved by solvent flattening and density modification using DM. The model was built with the program O and was refined by CNS. Five N-terminal residues (residues 1589–1593), a three-residue portion of the inter-repeat linker region (residues 1702–1704), and the C terminus (residues 1800–1817) do not have interpretable electron density, and we presume are disordered. In addition, residues 1757–1775, which form the α2B helix in the structure of the human BRCA1 BRCT repeats (Williams et al. 2001), are disordered in our crystals. The α2B helix is the least conserved structural element of the Brca1 BRCT repeats. The refinement statistics are shown in Table 1. All residues are in the allowed regions of the Ramachandran plot.
In vitro BACH1-binding assays
The human BRCA1 (residues 1529–1863) point mutants were constructed by overlap PCR mutagenesis as described previously (Sambrook et al. 1989) and sequenced. The wild-type and mutant proteins were overexpressed and isolated as GST fusion proteins as described for rat Brca1. The GST fusion proteins were purified by anion-exchange chromatography (Pharmacia Q-Sepharose Fast-Flow) and stored in 50 mM Tris-HCl (pH 8.5), 300 mM NaCl, and 5 mM DTT at −80°C.
An expression plasmid encoding the human BRCA1-interacting portion of BACH1 (residues 888–1063) was generated as described previously (Cantor et al. 2001). The 35S-labeled BACH1 products were synthesized by in vitro translation (TNT kit, Promega) and were tested for binding to GST fusion proteins containing either the wild-type human BRCA1 (residues 1529–1863) or 16 derivatives containing various point mutations. The GST fusion proteins bound to glutathione-Sepharose beads were incubated with the 35S-labeled BACH1 (888–1063) for 2 h at 4°C with agitation in NET-N buffer (20 mM Tris-HCl at pH 8.0, 0.5 M NaCl, 0.5% NP-40, and 1 mM EDTA). Following an extensive wash in NET-N buffer, the samples were run on SDS–polyacrylamide gel. The gel was then dried and exposed to the Molecular Dynamics Storm Scanner. The binding activities were quantitated using the ImageQuant software (Molecular Dynamics).
Acknowledgments
We thank H. Erdjument-Bromage of the Sloan-Kettering Microchemistry Facility for N-terminal sequence and mass spectroscopic analyses, the members of the Pavletich lab and K. Lee for helpful discussions, C. Murray for administrative assistance, the staff of the National Synchrotron Light Source X4A and X9A beamlines and of the Cornell High Energy Synchrotron Source (CHESS) for help with data collection. We also thank M. Bennett and R. Wiseman for providing us with the cDNA for rat Brca1. This work was supported by the NIH, the Howard Hughes Medical Institute, the Dewitt Wallace Foundation, and the Samuel and May Rudin Foundation. Coordinates have been deposited in the RCSB Protein Data Bank. PDB accession numbers are 1KZY for 53BP1–p53 and 1L0B for rat Brac1.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nikola@xray2.mskcc.org; FAX (212) 717-3135.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.959202.
References
- Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstieve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Caldecott KW, Tucker JD, Stanker LH, Thompson LH. Characterization of the XRCC1–DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP. From BRCA1 to RAP1: A widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- Caspari T. How to activate p53. Curr Biol. 2000;10:R315–R317. doi: 10.1016/s0960-9822(00)00439-5. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chiba N, Parvin JD. Redistribution of BRCA1 among four different protein complexes following replication blockage. J Biol Chem. 2001;276:38549–38554. doi: 10.1074/jbc.M105227200. [DOI] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor–DNA complex: Understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of Brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- Datta B, Li B, Choubey D, Nallur G, Lengyel P. p202, an interferon-inducible modulator of transcription, inhibits transcriptional activation by the p53 tumor suppressor protein, and a segment from the p53-binding protein 1 that binds to p202 overcomes this inhibition. J Biol Chem. 1996;271:27544–27555. doi: 10.1074/jbc.271.44.27544. [DOI] [PubMed] [Google Scholar]
- Gorina S, Pavletich NP. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- Grawunder U, Zimmer D, Leiber MR. DNA ligase IV binds to XRCC4 via a motif located between rather than within its BRCT domains. Curr Biol. 1998;8:873–876. doi: 10.1016/s0960-9822(07)00349-1. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Altschul SF, Bork P. BRCA1 protein products… Functional motifs. Nat Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- Li S, Chen PL, Subramanian T, Chinnadurai G, Tomlinson G, Osborne CK, Sharp ZD, Lee WH. Binding of CtIP to the BRCT repeats of BRCA1 involved in the transcription regulation of p21 is disrupted upon DNA damage. J Biol Chem. 1999;274:11334–11338. doi: 10.1074/jbc.274.16.11334. [DOI] [PubMed] [Google Scholar]
- Ljungquist S, Kenne K, Olsson L, Sandstrom M. Altered DNA ligase III activity in the CHO EM9 mutant. Mutat Res. 1994;314:177–186. doi: 10.1016/0921-8777(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Cui TY, Jasin M. Homology-directed DNA repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- Nash RA, Caldecott KW, Barnes DE, Lindahl T. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry. 1997;36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes & Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Thukral SK, Blain GC, Chang KK, Fields S. Distinct residues of human p53 implicated in binding to DNA, simian virus 40 large T antigen, 53BP1, and 53BP2. Mol Cell Biol. 1994;14:8315–8321. doi: 10.1128/mcb.14.12.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes & Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes & Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Green R, Glover JN. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat Struct Biol. 2001;8:838–842. doi: 10.1038/nsb1001-838. [DOI] [PubMed] [Google Scholar]
- Wu X, Petrini JH, Heine WF, Weaver DT, Livingston DM, Chen J. Independence of R/M/N focus formation and the presence of intact BRCA1. Science. 2000;289:11. doi: 10.1126/science.289.5476.11a. [DOI] [PubMed] [Google Scholar]
- Xia Z, Morales JC, Dunphy WG, Carpenter PB. Negative cell cycle regulation and DNA damage-inducible phosphorylation of the BRCT protein 53BP1. J Biol Chem. 2001;276:2708–2718. doi: 10.1074/jbc.M007665200. [DOI] [PubMed] [Google Scholar]
- Yamane K, Kawabata M, Tsuruo T. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur J Biochem. 1997;250:794–799. doi: 10.1111/j.1432-1033.1997.00794.x. [DOI] [PubMed] [Google Scholar]
- Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- Zhang X, Morera S, Bates PA, Whitehead PC, Coffer AI, Hainbucher K, Nash RA, Sternberg MJ, Lindahl T, Freemont PS. Structure of an XRCC1 BRCT domain: A new protein–protein interaction module. EMBO J. 1998;17:6404–6411. doi: 10.1093/emboj/17.21.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH. Association of BRCA1 with the hRad50–hMre11–p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]