Abstract
During body morphogenesis precisely coordinated cell movements and cell shape changes organize the newly differentiated cells of an embryo into functional tissues. Here we describe two genes, gex-2 and gex-3, whose activities are necessary for initial steps of body morphogenesis in Caenorhabditis elegans. In the absence of gex-2 and gex-3 activities, cells differentiate properly but fail to become organized. The external hypodermal cells fail to spread over and enclose the embryo and instead cluster on the dorsal side. Postembryonically gex-3 activity is required for egg laying and for proper morphogenesis of the gonad. GEX-2 and GEX-3 proteins colocalize to cell boundaries and appear to directly interact. GEX-2 and GEX-3 are highly conserved, with vertebrate homologs implicated in binding the small GTPase Rac and a GEX-3 Drosophila homolog, HEM2/NAP1/KETTE, that interacts genetically with Rac pathway mutants. Our findings suggest that GEX-2 and GEX-3 may function at cell boundaries to regulate cell migrations and cell shape changes required for proper morphogenesis and development.
Keywords: Epidermal morphogenesis, cell migration, tissue formation, C. elegans
Embryogenesis requires two at least partially distinct processes, the generation of cellular diversity, or differentiation, and the organization of cells into a functional body, or morphogenesis. A great deal is known about the mechanisms of cellular differentiation, but the mechanisms controlling cell shape changes and the cell movements that occur during morphogenesis are less well understood. For example, little is known about the factors needed for sending and receiving the signals that initiate cell migrations; for changing cell adhesiveness to allow both migrations past neighboring cells and anchoring of cells to new locations; and for initiating, executing, and strengthening cell shape changes in the embryo.
The Caenorhabditis elegans embryo is an ideal system for exploring the mechanisms of morphogenesis. C. elegans has an almost invariant cell division pattern (Sulston et al. 1983) in which both the origin and the fate of each embryonic cell is known. In addition, the physical process of body morphogenesis has been described (Priess and Hirsh 1986). Gastrulation in C. elegans begins when the daughters of the E blast cell, the precursors of the intestine, migrate from the exterior to the interior of the cell mass at the 28-cell stage. At the 400-cell stage, dramatic cell shape changes and cell movements reorganize the dorsal cap of epidermal cells, also known as hypodermal cells, arranging them first in six rows, and then five, as the two dorsal rows interdigitate to form one row in a process known as dorsal intercalation (Williams-Masson et al. 1998). While the dorsal-most epidermal cells are involved in intercalation, the ventral-most epidermal cells begin to migrate ventrally to enclose the embryo. Once the cells reach the ventral midline, they form adherens junctions enclosing the embryo (Priess and Hirsh 1986; Williams-Masson et al. 1997). After enclosure is completed, the nearly round embryo is squeezed into a tubular worm by a circumferential constriction of the embryo surface. This constriction, also known as elongation, appears to be mediated by actin bundles in the hypodermal cells on the surface of the embryo (Priess and Hirsh 1986).
Several genes have been described that specifically affect C. elegans embryogenic migrations and morphogenesis (Hedgecock et al. 1990; Costa et al. 1998; Shelton et al. 1999; for reviews, see Culotti and Merz 1998; Montell 1999; Chin-Sang and Chisholm 2000; Simske and Hardin 2001). Appropriate closure of the gastrulation cleft requires the genes vab-1, an Ephrin RTK, and vab-2, an Ephrin (George et al. 1998; Chin-Sang et al. 1999). The putative transcription factor die-1 has been reported to specifically affect dorsal intercalation (Williams-Masson et al. 1998; Heid et al. 2001), whereas mutations in the Adenomatous Polyposis Coli homolog apr-1 have been shown to affect both dorsal intercalation and ventral migrations (Frohli Hoier et al. 2000). The ventral enclosure process is disturbed in mutants lacking C. elegans homologs of α-catenin, β-catenin, and cadherin, as well as in vab-1 and vab-2 mutants (Costa et al. 1998; George et al. 1998; Chin-Sang et al. 1999). Embryos lacking the semaphorin gene mab-20 often fail to enclose owing to inappropriate contacts between hypodermal cells (Roy et al. 2000). Circumferential constriction and body elongation are dependent on the genes let-502 and mel-11, which are similar to Rho kinase and a regulatory subunit of myosin phosphatase (Costa et al. 1997; Wissmann et al. 1997, 1999). The PDZ and LRR (leucine rich repeat) containing protein let-413 affects the integrity of adherens junctions and therefore affects ventral enclosure as well as circumferential constriction (Legouis et al. 2000).
In previously described mutants, the epidermal cells successfully initiate ventral cell migrations, but fail to form adherens junctions at the ventral midline. An important unanswered question is what gene products are required for the earliest movements of the epidermal cells as they initiate their migrations to enclose the embryo. Here, we describe two gene products, GEX-2 and GEX-3, that are required for this process. Not only is dorsal intercalation disrupted when these products are missing, but the initial ventral migrations of the lateral epidermal cells appear to be completely absent. In addition, GEX-2 and GEX-3 are required for proper morphogenesis of other embryonic tissues. Antibodies to GEX-2 and GEX-3 reveal that they are both expressed at cell boundaries in all cells. We present evidence that GEX-2 and GEX-3 bind to each other, supporting the idea that they act together as a protein complex. gex-3 mutant animals also show postembryonic defects in egg laying and in gonadal morphogenesis (Kimble and White 1981). Similar gonadal morphogenesis defects are observed in animals mutant for ced-10, a C. elegans Rac homolog that is also required for the engulfment of apoptotic cell corpses and that has recently been shown to be required maternally for embryogenesis (Reddien and Horvitz 2000; Lundquist et al. 2001). We show that arresting embryos produced by ced-10 null homozygous mothers are severely defective in body morphogenesis including defects in some but not all of the morphogenetic events that require gex gene function. GEX-2 and GEX-3 proteins have close homologs in other organisms, including a Drosophila GEX-3 homolog that affects cell migrations and interacts with Rac pathway mutants (Hummel et al. 2000), raising the possibility that these molecules are important regulators of tissue morphogenesis and cell migration in many animal systems.
Results
Identification of gex genes
gex-2 was identified in screens using RNA-mediated interference (RNAi) to investigate the function of C. elegans homologs of mammalian Rac1 interactors. gex-2 has an RNAi embryonic lethal phenotype that appeared similar to a previously identified mutant called gex-3(zu196). gex-2(RNAi) embryos and gex-3(zu196) mutant embryos arrest development with apparently well differentiated cell types that fail to become organized properly. As a result, by the end of development several cell types that are normally internal are found spread out on the ventral surface of the embryo. We call this phenotype Gex and the mutants gex for gut on the exterior, their most obvious terminal phenotype. We describe this phenotype in more detail below. First, we report on the cloning of these two gex genes.
GEX-2 is the C. elegans homolog of Sra-1, a mammalian Rac1-interacting molecule
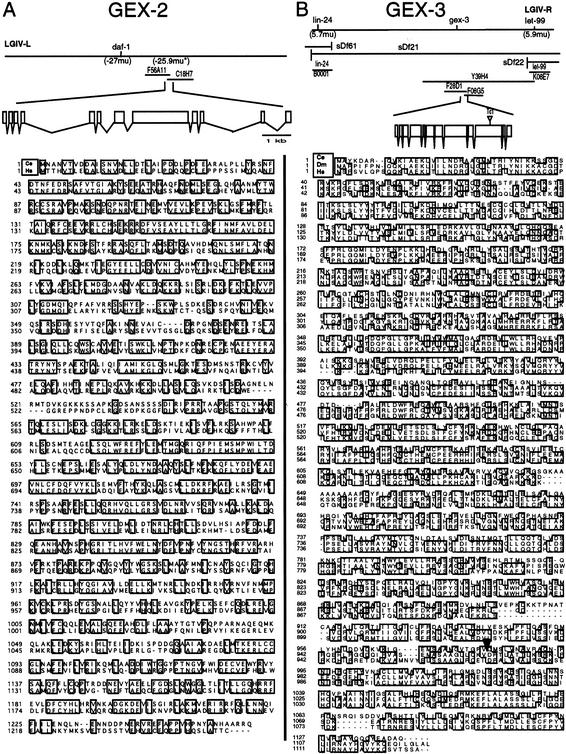
Kobayashi et al. (1998) used biochemical methods to identify the p140/Sra-1 protein as a direct interacting molecule of the small GTPase Rac1, but its physiological function is unknown. To explore the function of Sra-1 using C. elegans, we searched Yuji Kohara's EST library and found that the cDNA yk340a6 encodes the C. elegans Sra-1 homolog (Fig. 1A). The C. elegans Sra-1 homolog maps to cosmid F56A11 on the left arm of chromosome IV, and the F56A11.1 gene product predicted by Gene Finder is the C. elegans homolog of mammalian Sra-1. The complete cDNA sequence encodes a 1262-amino-acid protein with 51% identity to human Sra-1 (Fig. 1A). C. elegans Sra-1 has no homology to other proteins except for Sra-1 homologs in other proteins.
Figure 1.
(A) GEX-2 is the C. elegans Sra-1 homolog. The C. elegans Sra-1 homolog GEX-2 maps to cosmids F56A11 and C18H7 on the left arm of Chromosome IV, and the F56A11.1 gene product predicted by Gene Finder is the C. elegans homolog of mammalian Sra-1. The GEX-2 cDNA is composed of 10 exons. Amino acids of GEX-2 (Ce, Caenorhabditis elegans) and Sra-1 (Hs, Homo sapiens) are compared. Identical amino acids are shown boxed. (*) The approximate genetic position of gex-2 is indicated. (B) Cloning gex-3 indicates it is the HEM2/NCKAP1/KETTE homolog. gex-3(zu196) maps to the right arm of Chromosome IV at ∼+5.7 map units. gex-3 is rescued by YAC Y39H4 and by a PCR product consisting of the GEX-3 genomic region plus 1 kb of upstream and downstream DNA. C. elegans (Ce) GEX-3/HEM2 is aligned with Drosophila (Dm) HEM2/KETTE and human (Hs) NCKAP1 proteins. Identical amino acids are shown boxed. The zu196 allele contains a Tc1 insertion in exon 10 at amino acid 982, between the A and T, which causes a frameshift at the C terminus of the protein. An arrowhead marks the insertion spot.
To examine the function of C. elegans Sra-1, we disrupted its function by RNA-mediated interference (Fire et al. 1998). dsRNA was synthesized with an almost full-length cDNA clone (yk335f6) as template and injected into adult hermaphrodites. The resulting progeny had an embryonic lethal phenotype indistinguishable from that of gex-3 mutant embryos. We therefore named the C. elegans Sra-1 gene gex-2 based on the RNAi phenotype.
GEX-3 is homologous to HEM2/NAP1/NCKAP1, a mammalian Rac1 interacting molecule
The gex-3(zu196) mutation was discovered in a screen for maternal-effect lethal mutants (Mello et al. 1994). Homozygous gex-3 mothers appear wild-type except for a zygotic 100% Egl (egg laying defective) phenotype. Of the progeny made by gex-3 homozygous mothers 100% die during embryogenesis with the Gex phenotype described below, indicating a strong maternal contribution. The heterozygous progeny of homozygous gex-3 mothers mated to wild-type males are viable and fertile, indicating that the activity of this gene is sufficient either maternally or zygotically. The gex-3(zu196) mutation when placed over a deficiency results in the same zygotic Egl and maternal-effect Gex phenotypes observed in the homozygous mutant animals, suggesting that this allele is likely to represent a simple loss-of-function mutation.
The gex-3(zu196) mutation was mapped to ∼+5.7 on the genetic map of Chromosome IV (Fig. 1B; Materials and Methods). Candidate genes in the relatively small lin-24 to let-99 interval were tested for Gex phenotypes by RNAi. One gene, the C. elegans homolog of the vertebrate Rac1 interactor Hem-2 (Kobayashi et al. 1998), gave the Gex phenotype when targeted by RNAi. Because the zu196 mutation was induced in the mut-6 Tc1 transposon-mobilized strain, we asked if the C. elegans HEM-2 homolog, the predicted gene F28D1.10, might be disrupted by a Tc1 transposon in gex-3(zu196) (Fig. 1B). Consistent with this possibility, we found a single Tc1 transposon inserted into exon 10 of F28D1.10 in zu196. Furthermore, we found that gex-3 could be rescued by a yeast artificial chromosome (YAC), Y39H4, containing the corresponding physical interval, or by a PCR product predicted to contain only the CeHem-2 gene (see Materials and Methods). Therefore, we conclude that F28D1.10 is gex-3.
The gex-3 gene is predicted to encode a highly conserved but novel protein (Fig. 1B). There is close conservation along the entire length of the protein in C. elegans, Drosophila, mouse, and humans (Fig. 1B; Baumgartner et al. 1995). Sequence analyses of the GEX-3 protein revealed no recognizable motifs. It has been suggested that four hydrophobic regions could act as transmembrane regions (Baumgartner et al. 1995), but there is no evidence the GEX-3 is found in membranes, as opposed to near membranes, and mammalian GEX-3/HEM2 can be isolated from brain cytosol (Kobayashi et al. 1998). The closest homologs to GEX-3 are the human NAP1/NCKAP1 and Drosophila HEM2/NAP1/KETTE, which are, respectively, 39% and 40% identical over the entire length of the protein (Fig. 1B). NCKAP1 has been shown to interact with activated Rac, and to be down-regulated in the brains of Alzheimer's disease patients (Kitamura et al. 1997; Suzuki et al. 2000). Drosophila HEM2/NAP1/KETTE is required for axonal pathfinding and for appropriate actin cytoarchitecture. It genetically interacts with the SH2–SH3 domain adapter protein NCK/DOCK and can be phenocopied by overexpression of the GTPases Rac1 and CDC42 (Hummel et al. 2000).
Embryonic maternal-effect phenotypes of gex mutants
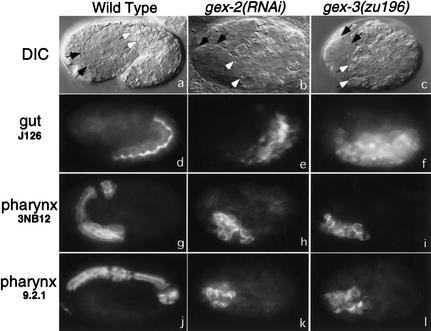
gex mutant embryos arrest development with their intestinal cells spread over the ventral surface of the embryo (Fig. 2b,c), yet the embryos produce the normal number of 20 gut cells. In wild-type embryos the entire gut is derived from the eight-cell-stage E blastomere. Ablation of the E blastomere in gex-3 mutant embryos resulted in partial embryos without intestine, suggesting that E is the only source of intestine. Lineage studies revealed that the E cells gastrulate properly and divide at the correct times (data not shown). Antibodies to intestine, pharynx, valve cells, and muscle indicated that these tissues are present and differentiate correctly in Gex embryos (Fig. 2; see Materials and Methods). However, during embryogenesis all tissues fail to organize properly, and by the end of embryogenesis are found in aberrant positions and with aberrant morphology. For example, the intestine appears flat rather than tubular, and the pharynx does not elongate (Fig. 2). In addition, both the cells of the pharynx and of the intestine are initially found at their normal internal positions (data not shown), yet at the end of embryogenesis they end up on the surface of the embryo (Fig. 2). Therefore, although cell specification appears normal in Gex embryos, tissue morphogenesis appears abnormal.
Figure 2.
gex mutant embryos fail to undergo morphogenesis but do differentiate specific tissues. (Top panels) Nomarski images. All other panels are fluorescent images using monoclonal antibodies to pharynx (3NB12 and 9.2.1) and intestine (J126). Anterior is left, and dorsal is up. Wild-type N2 embryos at this stage have the internal organs of the pharynx and intestine fully enclosed by the hypodermis. Black-tipped arrows point to the anterior of the pharynx, and white-tipped arrows point to the anterior of the intestine. Antibodies 9.2.1 and J126 illustrate that the pharynx and intestine form elongated tubular structures. In gex-2 and gex-3 embryos the hypodermis never encloses the embryo and instead constricts on the dorsal side. Embryos end embryogenesis with internal organs that lie on the outside and do not form the correct elongated tubular structures.
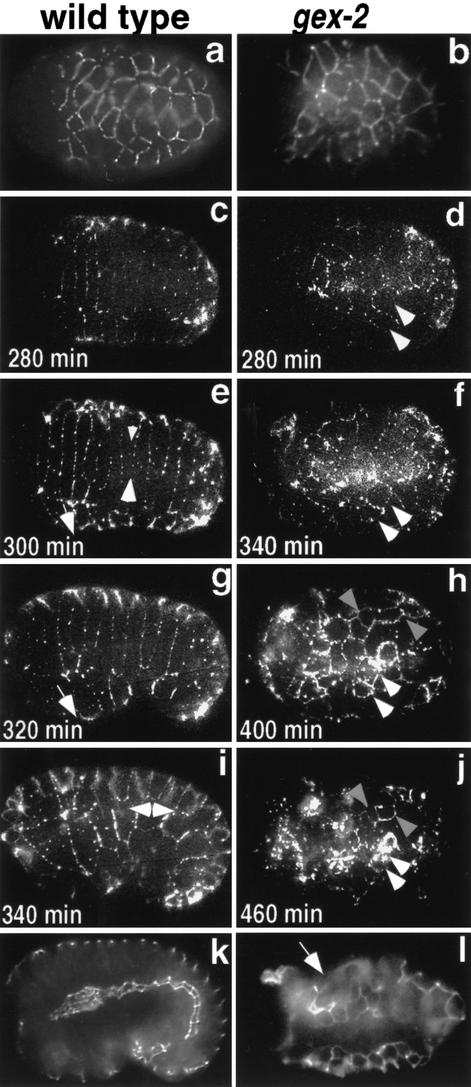
To further analyze the embryonic phenotype of gex mutants, we analyzed the expression of the AJM-1 protein, previously known as JAM-1, which is localized to the junctions between the epidermal, pharyngeal, and intestinal cells (Priess and Hirsh 1986; Podbilewicz and White 1994; Koppen et al. 2001). Although the hypodermal cells are initially formed properly in gex-2 and gex-3 mutants (Fig. 3b; data not shown) and express AJM-1 protein, they fail to migrate over and enclose the embryo. Instead, the hypodermal cells contract to form a tight cluster on the dorsal surface (Fig. 3l, white arrow). To analyze the dynamic localization of AJM-1 in living embryos, we used a transgenic line carrying AJM-1::GFP (Mohler et al. 1998). Wild-type embryos carrying AJM-1::GFP illustrate three hypodermal morphogenic steps in real time: dorsal intercalation (Fig. 3c,e), the beginning of ventral migrations, which we refer to here simply as ventral migration (Fig. 3e,g), and the end of ventral enclosure, which we refer to here as complete ventral enclosure (Fig. 3i). In gex mutant embryos, AJM-1::GFP and an antibody to AJM-1, MH27, are expressed and localized to the junctions of the hypodermal cells, as in wild-type embryos (Fig. 3, right panels; data not shown; Podbilewicz and White 1994). However, hypodermal cells in gex mutants fail to undergo dorsal intercalation (Fig. 3, right panels; note that panel i [WT] and panel f [gex-2] are at the same stage, 340 min). In addition, the ventral migration of epidermal cells fails to occur in gex mutant embryos (Fig. 3, right panels; note that lateral hypodermal cells do not elongate or move ventrally). Thus at a time when the wild-type hypodermis has fully enclosed the embryo and begins circumferential constrictions that elongate the animal into a worm (Fig. 3i; see also Priess and Hirsh 1986), the Gex hypodermis has remained essentially unchanged in shape and thereafter contracts apparently equally on all axes to form a tight ball of hypodermal cells on the dorsal side of the embryo (Fig. 3f–l). These data indicate that gex genes are needed in the earliest events of epidermal morphogenesis.
Figure 3.
Time course to illustrate the hypodermal Gex defect using antibody MH27 (a,b,k,l) and AJM-1::GFP (c–j). gex-2(RNAi) embryos are shown. Similar results were obtained with gex-3(zu196) and gex-3(RNAi) embryos. (a–f,h,j) Dorsal views; (g,i,k,l) lateral views, because wild-type embryos undergo a rotation as they finish morphogenesis. (a,b) The hypodermal cell junction marker AJM-1 (shown with antibody MH27) is present. For the AJM-1::GFP series, the wild-type embryos are shown at 20-min intervals, and the gex embryos are shown at 60-min intervals. (c,d) By 280 min after first cleavage, the gex embryo shows hypodermal abnormalities, having failed to initiate dorsal intercalation and ventral migrations. Comparing i with f, both at 340 min, shows that in gex embryos the hypodermis fails to become organized. In gex embryos the hypodermis fails to form rows (as is apparent in c,e,g,i), the cells do not change shape or intercalate (as the white arrowheads indicate in e), and the lateral cells do not migrate ventrally (as the white-tailed arrow shows in e,g). Instead, the disorganized cells begin to constrict on the dorsal side (f,h,j; e.g., follow the two gray arrows in h and j and the two white arrows in d–j). (Panel l) An embryo at a similar stage as the terminal embryos shown in Figure 2. l (MH27 antibody) shows the hypodermis constricted on the dorsal side (up; white arrow) and the internal organs exposed on the ventral side (down).
Egg-laying defective phenotypes of gex mutants
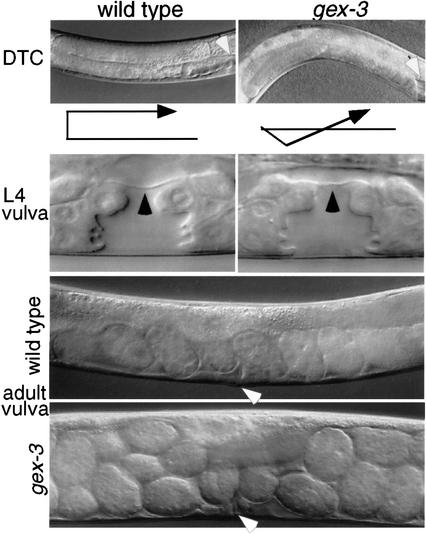
The gex-3 mutants are homozygous viable and maternal-effect lethal. Thus the maternally provided gex-3 products are sufficient for all of embryogenesis, but appear to be depleted by the time homozygous animals reach adulthood. We therefore asked if the development of postembryonic tissues is affected in gex-3 homozygous adults. We found that homozygous gex-3 hermaphrodites are severely egg-laying defective (Egl), laying at most a few embryos before becoming completely Egl (Fig. 4). We found that vulval cell fates are initially specified properly and that there are no defects in the vulval–uterine connection (Fig. 4; Materials and Methods). Using GFP lines we found that the key neurons and muscles required for egg laying are present and correctly positioned in gex-3(zu196) animals (Materials and Methods; data not shown). We conclude that the fully penetrant Egl phenotype in gex-3 mutants is not caused by major cell fate or vulval morphogenesis defects. Although the early vulva is indistinguishable from wild type, the adult vulva often assumes a slightly protruding appearance and fails to function, suggesting that some aspect of late vulval morphogenesis or maintenance may be defective.
Figure 4.
gex-3(zu196) zygotic phenotypes. Anterior is left, and dorsal is up. (Top panels) The DTC defect: In gex-3(zu196) embryos the DTCs (distal tip cells) often show migration defects including incorrect dorsal/ventral polarity. The black arrows below the photographs trace the migration of the anterior DTC. One or both DTCs can show defects in gex-3(zu196) animals. (Bottom panels) The vulval phenotype: gex-3(zu196) larvae form what appear to be wild-type mid-L4 vulvae, yet 100% of adult worms become Egl (Egg laying defective). (Middle panels) gex-3(zu196) mid-L4 vulvae have the Christmas tree structure and flattened-out thin laminar process (black arrowhead) separating the vulva and uterus, just as in wild-type, suggesting normal cell differentiation and cell fusions. Whereas the wild-type N2 adult worm contains one row of embryos, all at early stages, the adult gex-3 worm is beginning to fill up with unlaid embryos, many at late stages of development. The white arrowheads show the position of the vulva.
GEX-2 and GEX-3 are localized at cell boundaries
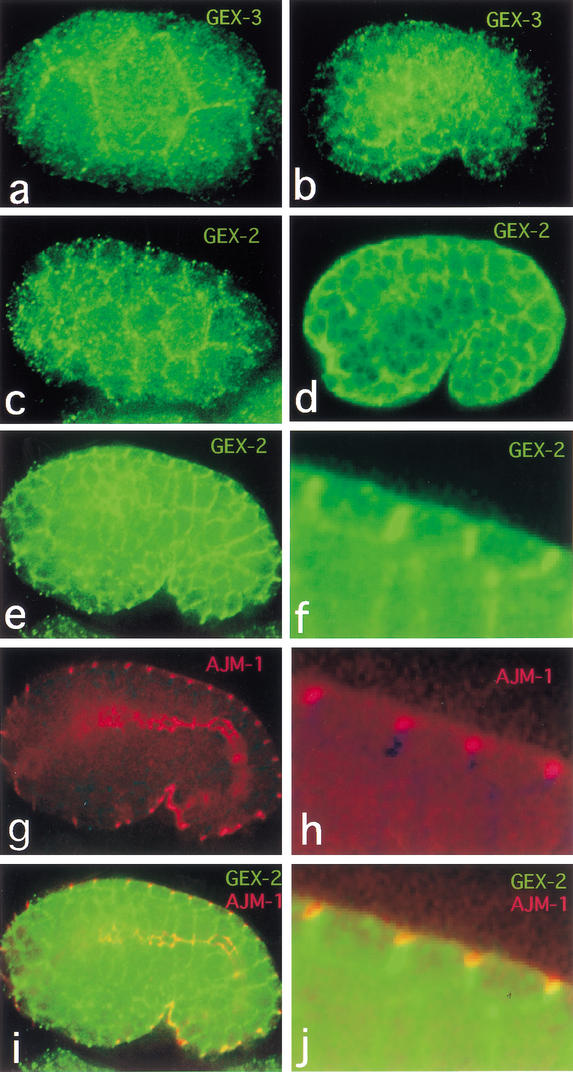
To gain further insight into the functions of GEX-2 and GEX-3, we sought to analyze the localization of these proteins in embryos. Anti-sera raised against GEX-2 and GEX-3 recognized major protein bands of 140 kD and 125 kD, respectively, in extracts of total C. elegans proteins (Materials and Methods; data not shown). These are the predicted sizes for GEX-2 and GEX-3. In wild-type embryos, both proteins appeared to be enriched at cell boundaries (Fig. 5). The GEX-2 immunostaining but not GEX-3 immunostaining disappears at all stages in gex-2(RNAi) embryos. Likewise, the GEX-3 immunostaining but not GEX-2 immunostaining disappears at all stages in gex-3(RNAi) embryos (data not shown). These findings suggest that these proteins do not depend on one another for their expression. GEX-2 and GEX-3 staining was observed in all cells beginning at the two-cell stage and extending throughout embryogenesis (Fig. 5a–f; data not shown). Therefore, GEX-2 and GEX-3 are present in the cell types, including the hypodermal cells, that are altered in gex-2 and gex-3 mutant embryos. A rescuing gex-3::gfp line shows expression similar to that of the antibody to GEX-3, with enrichment at the boundaries of all cells in the embryo starting at the 100-cell stage.
Figure 5.
Antibodies to GEX-2 and GEX-3 show localization to the boundaries of all cells starting at the two-cell stage. GEX-2 has a distinct localization pattern compared with AJM-1. Wild-type embryos are shown with dorsal up and anterior to the left. (a,b) Antibodies to GEX-3 on embryos at the 8-cell (a) and at the early comma stage (b) show GEX-3 expression in the hypodermis as well as at the boundaries of other cells. (c–f) Antibodies to GEX-2 on embryos at the 32-cell (c), 1.5-fold stage (d), and at the late comma stage (e,f) show GEX-2 is found at cell boundaries in the hypodermis, which has enclosed the embryo and is beginning to constrict to squeeze the embryo into a worm (d,e). (g,h) Antibodies to AJM-1 reveal that GEX-2 does not colocalize with the AJM-1 junctional domain. (i,j) Merged image. The focal plane in e–f is though the center of the embryo. f, h, and j are close-ups of a dorsal section of the images in e, g, and i.
To place the GEX-2 staining pattern in the context of proteins known to be expressed at cell junctions, we compared the localization of GEX-2 with that of AJM-1, using the MH27 antibody (Koppen et al. 2001). We found that whereas GEX-2 is enriched at all cell boundaries, it does not appear to overlap MH27 staining. Rather, the AJM-1 signal is located in a more apical region of the hypodermal cells, whereas GEX-2 appears to be located more basolaterally as well as all around the cell (Fig. 5e–j). GEX-3 showed the same localization relative to AJM-1 (data not shown).
Interaction between GEX-2 and GEX-3
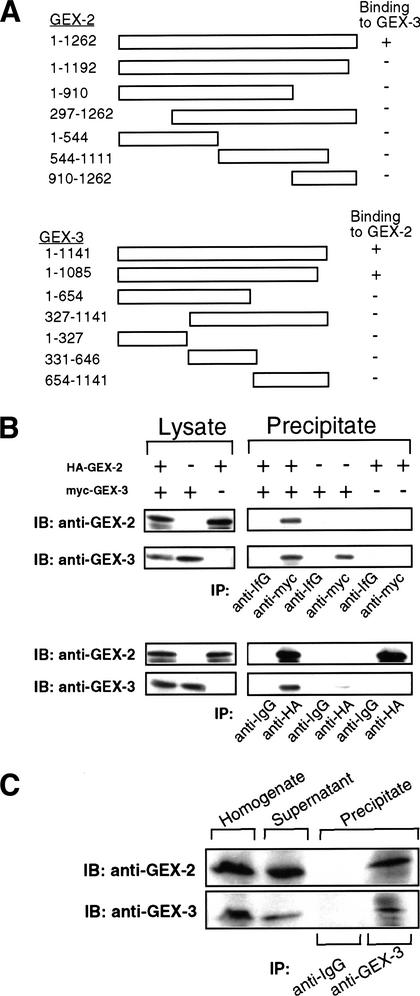
Given the similarity of the gex-2 and gex-3 phenotypes and the fact that both proteins appear to localize to cell boundaries, we asked if GEX-2 and GEX-3 interact. Using the yeast two-hybrid system, we found that the full-length GEX-2 could interact with the full-length GEX-3 (Fig. 6A). Surprisingly, we found that even small deletions of either protein abolished this interaction (Fig. 6A), suggesting that the physical interaction between GEX-2 and GEX-3 may involve extensive or multiple contacts. Alternatively, the folding of both proteins may be sensitive to truncations. An interaction between GEX-2 and GEX-3 was also observed in yeast cells expressing HA–GEX-2 and myc–GEX-3 by reciprocal coimmunoprecipitation experiments (Fig. 6B). When both myc–GEX-3 and HA–GEX-2 are expressed, antibodies to GEX-2 detect a protein that is immunoprecipitated by myc antibodies and that is the approximate size predicted for GEX-2. This protein is not detected in cells that do not express both myc–GEX3 and HA–GEX-2. Conversely, when myc–GEX-3 and HA–GEX-2 are expressed, antibodies to GEX-3 detect a protein that is immunoprecipitated by HA antibodies and that is the approximate size predicted for GEX-3. To detect an interaction in vivo, we performed an immunoprecipitation assay using C. elegans lysates (Fig. 6C). Immunoprecipitation with the GEX-3 antibody coprecipitates a protein that is recognized by the GEX-2 antibody. Therefore, the two-hybrid, coimmunoprecipitation and colocalization results most likely reflect an ability of the two proteins to interact.
Figure 6.
Direct interaction between GEX-2 and GEX-3. (A) Two-hybrid scheme including deletion constructs. The GEX-2 deletion series fused to the GAL4 activator domain and full-length GEX-3 fused to the GAL4 DNA-binding domain were tested in the two-hybrid assay. Only the combination of full-length GEX-2 and GEX-3 shows a direct interaction. Reciprocally, the GEX-3 deletion series fused to the GAL4 DNA-binding domain and full-length GEX-2 fused to the GAL4 activator domain were tested for interaction in the two-hybrid assay. Full-length and GEX-3 slightly deleted at the C terminus could bind to full-length GEX-2 in the two-hybrid assay. (B) Immunoprecipitation from yeast. Yeast lysates expressing HA–GEX-2 and myc–GEX-3 were prepared as described in Materials and Methods. Left lysate panels shows the expression of HA–GEX-2 and myc–GEX-3 recombinant proteins in yeast. These lysates were immunoprecipitated with anti-myc antibody (upper panels) and anti-HA antibody (lower panels). Coprecipitation was observed in the precipitate of the lysate prepared from yeast expressing both HA–GEX-2 and myc–GEX-3 as detected by anti-GEX-2 and anti-GEX-3 antibodies. (IB) Immunoblot; (IP) immunoprecipitate. (C) Immunoprecipitation from C. elegans. C. elegans lysates were immunoprecipitated with antibodies to GEX-3 and probed with anti-GEX-2 and anti-GEX-3 rabbit polyclonal antibodies. The GEX-3 immunoprecipitation includes a protein that can be detected by the GEX-2 antibody.
gex genes share phenotypes with C. elegans Rac-like genes
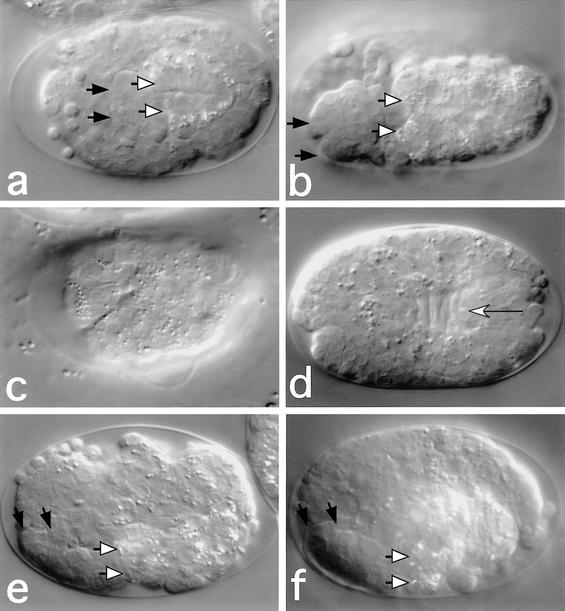
gex homologs show biochemical and genetic interactions with Rac signaling genes in other systems. We compared gex-2 and gex-3 with the known C. elegans Rac homologs ced-10/rac-1, mig-2, and rac-2 (Lundquist et. al 2001). mig-2 null mutants are homozygous viable with defects in certain postembryonic migrations. No alleles of rac-2 have as yet been described. However, a null allele of ced-10, n3417, was recently reported to cause maternal-effect embryonic lethality (Lundquist et al 2001). We therefore examined the embryos produced by ced-10(n3417) homozygous mothers for similarities to gex mutant embryos. We found that 71% (n = 132) of ced-10(n3417) embryos were defective in epidermal enclosure (Fig. 7). However, close examination of these embryos revealed that whereas gex mutant embryos never initiate morphogenesis, all of the ced-10(n3417) embryos examined undergo at least the initial stages of morphogenesis. For example, we found that 100% of ced-10(n3417) embryos examined undergo dorsal intercalation of the epidermis (n = 20) (Fig. 7d). Of the ced-10(n3417) embryos, 51% fail to undergo significant ventral migration of the epidermis (n = 132) (Fig. 7b,d,f), whereas another 18% (n = 132) were observed to undergo partial enclosure at the ventral midline. Thus ced-10 is required for at least some events in body morphogenesis in C. elegans and may function in these processes along with the Gex genes and perhaps other Rac gene family members (see Discussion).
Figure 7.
ced-10(n3417) mutant embryos show morphogenesis defects similar to Gex defects. Black-tipped arrows point to the anterior of the pharynx, and white-tipped arrows point to the anterior of the intestine in a, b, e, and f. The embryos in a–d are at an earlier stage and are shown from a different angle from that of the embryos in e and f. (a,c) gex-2(RNAi) embryo (a) shown from the ventral side shows unenclosed pharynx and intestine and (c) shown from the dorsal side shows disorganized epidermis. (b,d) A ced-10(n3417) embryo similarly shows (b) unenclosed pharynx and intestine on the ventral side, but the dorsal view (d) reveals some organization of the dorsal epidermis, including dorsal intercalation (white-tipped arrow). (e) A gex-3(zu196) embryo shown laterally reveals unenclosed pharynx and intestine on the ventral (bottom of the image) side. (f) A ced-10(n3417) embryo, also shown laterally, at a later stage than the embryo in b and d has ended embryogenesis with a terminal phenotype resembling gex-2 and gex-3 (also compare with the embryos in Fig. 2b,c). The embryo in a shows several spherical refractile cells (to the left of the black arrows) that resemble persistent corpses found in ced-10 mutants, whereas similar cells in the other panels are out of the plane of focus.
We also examined gex mutants for phenotypes found in ced-10 mutants. The ced-10 gene is required for the engulfment of apoptotic cell corpses that arise during the normal course of embryogenesis and is also required for the migration of the distal tip cells (DTCs) that are necessary for formation of the symmetric U-shaped gonad (Reddien and Horvitz 2000). We first examined gex embryos for cell death abnormality (Ced) phenotypes. We found that persistent cell corpses were present in the tissues of gex-3(zu196) and gex-2(RNAi) embryos (Fig. 7) in numbers similar to those found in ced-10(n3417) embryos (Fig. 7; see Supplemental Fig. 1 at http://www.genesdev.org). In gex mutants and in unenclosed ced-10(n3417) mutants we observed that the corpses were progressively found outside of the tissues (Fig. 7a; Supplemental Fig. 1). We find that all these corpses are dependent on ced-3, a gene required for all programmed cell deaths in C. elegans, suggesting these are true apoptotic corpses (data not shown; Yuan et al. 1993). However, the significance of this finding is not clear because we also observed persistent cell corpses in apx-1 mutant control embryos. The apx-1 mutant is defective in Notch signaling in the early embryo and fails to undergo morphogenesis owing to a cell fate defect that leads to missing ventral epidermal cells (Mello et al. 1994). Therefore, we do not know if the cell corpse engulfment phenotype in gex mutants reflects a defect in the same phagocytosis mechanism that requires ced-10 or might instead reflect a secondary consequence of the morphogenesis defects in these mutants.
Finally, we examined gonadal morphology in gex mutants for evidence of defects in DTC migrations. In normal development each arm of the gonad grows, under the influence of an organizing cell called the distal tip cell (DTC), to form a symmetrical U-shaped structure. Initially each gonad arm extends along the ventral surface of the animal. During midlarval stages the arms execute a turn and reverse directions to grow along the dorsal side of the animal. Previous work has shown that ced-10 and mig-2 mutants perturb this process, causing the gonad to show abnormalities that are thought to reflect defects in DTC migration (Zipkin et al. 1997; Blelloch et al. 1999; Reddien and Horvitz 2000). We found that gex-3(zu196) homozygotes frequently showed a spectrum of gonadal abnormalities similar to those observed in ced-10(n1993) mutants. For example, ∼12% (n = 151) of gex-3(zu196) animals scored at the L3/early L4 stage had clear defects in gonad morphology (Fig. 4) versus 13% (n = 71) in ced-10(n1993) and 0% (n = 76) in wild-type animals. These defects included failure of the gonad to move to the dorsal side of the animal, premature bends in the gonad arms, and meandering back and forth between dorsal and ventral sides. These findings taken together with similarities in embryonic phenotypes (Fig. 7) suggest that gex genes function with ced-10/rac-1 and perhaps with other members of the C. elegans Rac-like gene family in both embryonic and postembryonic developmental events.
Discussion
The process of body morphogenesis must involve signals both within and between the tissues of the embryo. How these signals guide the assembly of functional organs remains largely mysterious at present. Here, we have described two genes, gex-2 and gex-3, that appear to be essential for proper morphogenesis in C. elegans. These genes affect the earliest cell movements of the hypodermis needed to enclose the embryo. Both genes encode large proteins that appear to interact and colocalize in vivo at cell boundaries. Closely related genes exist in Drosophila and vertebrates and have been implicated in signaling mediated by the small GTPase Rac (Baumgartner et al. 1995; Kitamura et al. 1997; Kobayashi et al. 1998; Hummel et al. 2000; for reviews, see Kaibuchi et al. 1999; Settleman 1999). Taken together, these findings suggest that GEX-2 and GEX-3 may be part of a conserved protein complex important for transducing the signals or forces that mediate cell migration and cell shape change during morphogenesis.
GEX-2 and GEX-3 are essential for body morphogenesis
Inactivation of GEX-2 and GEX-3 results in embryonic lethality with defects in morphogenesis of the hypodermis (Fig. 2). In gex mutants, neither dorsal intercalation nor ventral migration (including leading cell migration) occurs, resulting in a complete loss of ventral enclosure (Fig. 3). Because none of the cells meet at the ventral midline, we cannot address whether ventral adherens junctions could form. Previously identified mutants involved in ventral enclosure show at least partial ventral migration, but fail to form stable junctions at the ventral midline (Costa et al. 1998; George et al. 1998; Frohli Hoier et al. 2000). Given that Gex animals do not show even the initial cell shape changes required for ventral migration, we infer that gex mutations disrupt either the polarizing signal, which induces cell shape changes, or the ability of the cells to respond to a polarizing signal.
gex mutant embryos also show disorganization of embryonic tissues other than the hypodermis. The body muscles, which depend for their position on the hypodermis, are highly disorganized (data not shown). Both the pharynx and intestine fail to elongate into the appropriate tubular structures (Fig. 2). Some of these defects could reflect indirect consequences of the failure of hypodermal enclosure and body morphogenesis. However, other mutants that fail to undergo body morphogenesis, as well as wild-type embryos in which we ablated the ABp blastomere to prevent enclosure, end embryogenesis with a pharynx and intestine that are better organized than in gex mutant embryos (M. Soto and C. Mello, unpubl.).
The nature of the GEX-2/GEX-3 interaction
GEX-2 and GEX-3 mammalian homologs were initially identified as activated Rac interactors. In this paper we show that, in fact, the two worm homologs have the same phenotype and appear to colocalize at the cell cortex. In addressing possible models for gex function, it would be useful to know if the two proteins are acting at a similar step in interpreting signals required for cell shape changes and cell migration. The two-hybrid results together with the in vivo coimmunoprecipitation data suggest that GEX-2 and GEX-3 are found in a common complex in C. elegans. This interaction could be direct, as suggested by the two-hybrid results, or it may require additional proteins. It is not clear why the two-hybrid interaction requires nearly full-length proteins.
gex-2, gex-3, and Rac-like genes affect several related developmental processes
The observation that gex-3 and ced-10/rac-1 show similar postembryonic defects in the migration of the DTC and both show embryonic defects in epidermal enclosure provides evidence in support of a role for Rac signaling in GEX-2/GEX-3 functions. Rac and other Rho family GTPases are known to be involved in cytoskeletal rearrangements and cell migration in other organisms (for reviews, see Kaibuchi et al. 1999; Montell 1999; Settleman 1999). Furthermore, homologs of GEX-2 and GEX-3 have been implicated in Rac signaling. Kobayashi et al. (1998) identified mammalian homologs of GEX-2 and GEX-3, Sra-1 and HEM-2, respectively, as interactors of Rac1 GTPase. Interestingly, the closest homolog to GEX-3, the human NAP1/ NCKAP1 protein, is thought to interact with activated Rac (Kitamura et al. 1997). There is clear evidence in Drosophila for a connection between the GEX-3 homolog HEM2/NAP1/KETTE and Rac signaling. The Drosophila kette mutation shows genetic interactions with Rac pathway genes for the control of neuronal migrations. The kette mutation is partially rescued by expression of activated Rac and can be phenocopied by overexpression of mutant DRAC1 or DCDC42 in the CNS midline (Hummel et al. 2000). GEX-3 and HEM2/NAP1/KETTE may be playing similar roles in C. elegans and Drosophila. Both proteins are involved in the migrations of cells in several tissues during late embryogenesis. kette is known to have genetic interactions with Rac mutations, and gex-3 likewise shows genetic interactions with C. elegans Rac homologs (see below).
In C. elegans, three Rac homologs have been identified; one of these, CED-10/RAC-1, was reported to localize to cell boundaries of the hypodermal cells (Chen et al. 1993, 1996). However, to date we have not detected fully penetrant Gex-like embryonic phenotypes in ced-10 or mig-2 mutants, and, instead, see partially penetrant and less severe enclosure defects (Fig. 7). RNAi experiments targeting ced-10 do not appear to inactivate ced-10 fully because they result in animals with milder embryonic and adult phenotypes than are observed in the ced-10 null embryos (Lundquist et al. 2001; M. Soto and C. Mello, unpubl.). Furthermore, RNAi targeting individual or combinations of the three C. elegans Rac-like GTPases also gave only partially penetrant phenotypes, suggesting that these genes may be partially refractory to RNAi (data not shown; for review of caveats regarding RNAi, see Tabara et al. 1998). Lundquist et al. (2001) reported that true null alleles of ced-10 are maternal-effect lethal. ced-10, therefore, has a significant maternal contribution, like the gex genes. Our examination of the dead embryos made by the ced-10 null animals revealed a morphogenesis defect similar to but less severe and less penetrant than the Gex phenotype (Fig. 7). Lundquist et al. have shown that there is redundancy between ced-10 and mig-2. However, to study double-mutant combinations, they must provide either maternal ced-10 or maternal mig-2 (Lundquist et al. 2001). The larval lethality in double homozygotes precludes examination of embryos completely lacking maternal ced-10 and mig-2 function. Thus we do not know if the less severe morphogenesis defects observed in ced-10 mutant embryos might reflect a partial redundancy with mig-2 and possibly rac-2, or might instead indicate that ced-10 functions only at relatively later steps in morphogenesis with respect to the gex genes.
The ced-10 gene is required for the engulfment of cell corpses that arise as a consequence of programmed cell death during embryogenesis. Hypomorphs of ced-10 undergo normal body morphogenesis but are defective in phagocytosis of the dying cells (Reddien and Horvitz 2000). As a result, the cell corpses persist within the tissues of the otherwise wild-type, viable ced-10 embryos. In gex mutants we observed persistent cell corpses produced via the programmed cell-death pathway; however, we also observed persistent cell corpses in apx-1 mutants that are thought to block morphogenesis by altering the proper specification of the epidermal cells required to enclose the embryo (Mello et al. 1994). It may be that body morphogenesis is required to ensure that cell contacts necessary for engulfment of corpses are established correctly. It is therefore possible that gex mutants only indirectly affect the engulfment process by causing dying cells to lose contact with cells capable of engulfing them. Further study will be needed to explore the possibility that gex genes function directly with ced-10 in the apoptotic cell-death engulfment process.
Finally, we observed postembryonic phenotypes in gex mutants that suggest a role for these genes in migrations and morphogenetic events that occur after hatching. These include a penetrant defect in egg laying as well as defects in morphogenesis of the gonad. In addition to these phenotypes, we have consistently observed an uncoordinated (Unc) phenotype associated with RNAi targeting gex-2 and gex-3 (data not shown). Furthermore, although single mutants in gex-3(zu196), mig-2(mu28), and ced-10(n1993) are not obviously uncoordinated, we have observed that double mutants between gex-3(zu196) and mig-2 and between gex-3(zu196) and ced-10(n1993) both show a similar Unc phenotype (data not shown). Postembryonic defects in egg laying, gonadal morphology, and motility are also observed in ced-10; mig-2 double mutants (Lundquist et al. 2001). It is difficult to interpret these genetic interactions between gex and Rac-like genes because the activities of each of these genes are maternally supplied, and, therefore, embryonic and larval functions in homozygous mutants may be rescued in part by the maternal products. Nevertheless, the overall similarity in the embryonic and postembryonic phenotypes suggests that gex and Rac genes do function together in several developmental processes.
In summary, the gex genes are highly conserved C. elegans genes essential for the earliest events in body morphogenesis. Future genetic study will be required to determine if the GEX-2 and GEX-3 proteins mediate their effects on cell shape and migration in C. elegans by regulating or responding to Rac signaling. Analysis of additional genes by RNAi and the molecular cloning of additional mutants with Gex phenotypes should shed light on how the process of morphogenesis uses components like GEX-2 and GEX-3 to initiate the migrations and cell shape changes essential for tissue and body morphogenesis.
Materials and methods
Strains, alleles, and genetic analysis
The Bristol strain N2 was used as the standard wild-type strain. C. elegans culture and genetics were as described in Brenner (1974). The strains used were as follows: unc-24(e138) dpy-20(e1282), let-99(it141), lin-24(n1057), sDf2, sDf22, sDf61, dpy-20(e1282) unc-31(e928), dpy-20(e1282); ayls6, ced-10(n1993), jcIs1 (SU93, the integrated AJM-1::GFP line), and gmIs1.
gex-3(zu196) was discovered by C. Mello in screens conducted in Jim Priess's Laboratory for maternal-effect embryonic lethals (Mello et al. 1994). gex-3(zu196) was mapped to LGIV, +5.7 map units in the dpy-20(e1282) unc-31(e928) interval. Because it complements sDf61 and sDf 22 but not sDf 21, it is to the right of lin-24 and to the left of let-99, a region containing only a few known complementation groups, all of which complement gex-3. The strain gex-3(zu196)/DnT1 was used for phenotypic analysis. DnT1 contains a dominant Unc mutation, so that non-Unc animals are gex-3 homozygotes.
Molecular analysis and plasmids
cDNAs of gex-2 and gex-3 were predicted by the genome-wide EST project (Y. Kohara, National Institute of Genetics, Mishima, Japan). yk340a6 contained full-length gex-2 cDNA. The gex-2 cDNA sequences differ slightly from the genome center's GeneFinder predictions for the corresponding open reading frame F56A11.1 and are detailed in GenBank accession number AB073209. yk408a8 contained almost full-length gex-3 cDNA. Missing sequence was determined from a PCR product amplified from the cDNA library (R. Barstead, Oklahoma Medical Research Foundation, Oklahoma City). The gex-3 cDNA showed no variations from the genome center's GeneFinder predictions for the corresponding open reading frame F28D1.10. gex-3 allele zu196 was generated in a mut-6 mutator strain. Sequencing of the zu196 allele revealed a Tc1 insertion at amino acid 982, between the A and T, which causes a frameshift at the C terminus of the protein. The GenBank accession number for gex-3 is AB073210.
Rescue experiments
The strain unc-24 gex-3(zu196)/nT1 was injected with either YAC Y39H4, which contains the gex-3 gene, or with a genomic PCR product including 1 kb of 5′ UTR and 1 kb of 3′ UTR, amplified from YAC Y39H4, with GFP inserted at the 3′ end, as well as the injection marker pRF4. Unc Roller progeny with viable progeny were tested for the presence of the gex-3 mutation and for the loss of rescue by gex-3 RNAi.
For expression in yeast Saccharomyces cerevisiae, full-length GEX-2 and GEX-3 were tagged at the N terminus with HA and myc epitopes, respectively, and cloned into yeast expression vectors containing the YCp backbone. Resultant plasmid pKK442 contains HA–GEX-2 and TRP1 as markers, whereas pKK836 contains myc–GEX-3 and URA3 as markers. The vectors pGAD-C1 and pGBDU-C1 were used for the two-hybrid assays (James et al. 1996). Corresponding regions of gex-2 or gex-3 cDNA were digested with restriction enzymes and cloned into pGAD-C1 or pGBDU-C1.
Microinjection
RNAi was performed as described in Fire et al. (1998) and Rocheleau et al. (1997). The gex-2 cDNA clone yk335f6 and the gex-3 cDNA clone yk29h5 were used to prepare dsRNA. Injections for rescue were performed as described (Mello et al. 1991).
Immunostaining and microscopy
Immunostaining was performed as described in Shi and Mello (1998). Merged fluorescent images were obtained using a confocal scanning microscope (Zeiss LSM510). Analysis of zygotic phenotypes and of the embryonic lethal phenotype was performed using light and Nomarski microscopy and UV fluorescence.
Analysis of embryonic tissue differentiation
Tissue differentiation of gex embryos was analyzed using morphological criteria at the light microscope and with antibodies as described in Bowerman et al. (1992) and Mello et al. (1994). Antibodies used include monoclonal antibodies (MAb) 3NB12 and 9.2.1 to visualize pharyngeal muscle and intestinal valve cells; MAb MH27 and a polyclonal antibody to LIN-26 (Labouesse et al. 1996) to visualize the hypodermis; and MAb 5.6.1 to visualize body wall muscles.
Phenotypic assays
Scoring gonadal morphogenesis
Animals were scored at the L3/early L4 stage. Abnormalities scored included early bends in the gonad arms, and dorsal/ventral defects. Gonads were scored as abnormal if the gonad arms bend too soon, stay on the ventral side, or move back and forth from ventral to dorsal.
Phenotypic assays to test the Egl defect
The vulva forms during the third and fourth larval stages. We and others observed that the vulva appears normal in L3 and L4 gex-3 animals, adopting the distinctive mid-L4 stage morphology (Fig. 4). Moreover, the anchor cell fuses with the uterine seam cell, and their nuclei migrate away from the vulval orifice as in wild type (Fig. 4; Newman et al. 1996; Hanna-Rose and Han 1999; Wendy Hanna-Rose, pers. comm.).
Scoring egg-laying neurons
gex-3 L1 animals were scored for correctly positioned hermaphrodite-specific neurons (HSNs) between the cells P5/6 and V4. The position of HSNs in adult hermaphrodites was monitored using the arrestin::GFP line gmIs1. The HSNs, which are required for egg laying (Sulston and Horvitz 1981; Desai et al. 1988; Garriga et al. 1993), are correctly positioned, and their projections appear to connect to the ventral cord as in wild type (data not shown), even after Gex animals become Egl.
Scoring egg-laying muscles
Animals were scored for final position of the sex myoblasts (SMs), muscles necessary for egg laying (Chen and Stern 1998; Harfe et al. 1998), at the L3 stage by monitoring the expression of ayls6 (TWI::GFP; Harfe et al. 1998).
Egg-laying assays
Single worms were placed for 90 min in 50 μL of M9 solution with the following drugs added: 10 mg/mL serotonin, 100 μg/mL levamisole, and 0.75 mg/mL imipramine (Trent et al. 1983; Weinshenker et al. 1995).
Immunoprecipitation
In vitro IPs
Wild-type yeast strain YPH500 (Sikorski and Hieter 1989) was transformed with pKK442 and pKK836, expressing HA–GEX-2 and myc–GEX-3, respectively, resulting in three yeast strains harboring pKK442, pKK996, and both. Yeast cells were lysed with Lysis buffer (1 mM EDTA, 20 mM Tris-Cl at pH 7.5 and 4°C, 0.5% Triton X-100, 50 mM NaCl, 1 mM CaCl2, 10 mg/mL leupeptin, 10 mM APMSF) and glass beads and centrifuged. The resulting supernatant was subjected to immunoprecipitation with antibodies to myc, 9E10, or HA, 12CA5. Following the immunoprecipitation, GEX-2 and GEX-3 were detected with polyclonal anti-GEX-2 and anti-GEX-3 rabbit antibodies.
In vivo IPs
Immunoprecipitation using C. elegans lysates was performed as in Rocheleau et al. (1999). The resulting supernatant was subjected to immunoprecipitation with antibodies to GEX-3. Following the immunoprecipitation, GEX-2 and GEX-3 were detected with polyclonal anti-GEX-2 and anti-GEX-3 rabbit antibodies.
Antibody production
Polyclonal antibodies were generated against parts of GEX-2 or GEX-3 fused with MBP. Plasmids pKK204, pKK251, and pKK216 express amino acid regions 1057–1262 of GEX-2, 763–1061 of GEX-3, and 62–363 of GEX-2, respectively. Using these plasmids, MBP fusion proteins were expressed in Escherichia coli and purified with amylose resin (New England Biolabs). Rabbits were immunized with purified proteins derived from pKK204 and pKK251. Rats were immunized with purified proteins derived from pKK216. For immunostaining, these antibodies were affinity-purified with purified antigen proteins.
Two-hybrid assay
A yeast strain, PJ69-4A (James et al. 1996), was transformed with one of the pGAD GEX-2 plasmids or with pGAD-C1 together with either one of the pGBDU GEX-3 or pGBDU-C1 plasmids. Two-hybrid interactions were determined by colony formation on Ade(−) medium using the ADE2 reporter.
Acknowledgments
We thank Alan Coulson for cosmids and YAC clones; Yuji Kohara for cDNA clones; Philip James for providing the two-hybrid host yeast strain; Fumie Nishimura for technical assistance; Jeff Simske and Jeff Hardin for providing the SU93 strain; Gian Garriga, Jim Rader, Gage Crump, and Cori Bargmann for sharing the unpublished gmIs1 strain; Joe Culotti, Gian Garriga, Christina Borland, Mike Stern, Jim Priess, Andy Fire, Bob Horvitz, and the C. elegans Genome Center, funded by the NIH, for providing strains; the Stern lab for advice on scoring SMs; Liz Ryder for advice on scoring HSNs; Wendy Hanna-Rose for help with vulval anatomy; Shinya Kuroda and The TEAM for helpful discussions; and Michel Labousse, James McGhee, David Miller, and Rob- ert Waterston for providing antibodies. H.Q. and K.K. were supported by grants-in-aid for Scientific Research from the Ministry of Education, Science, and Culture, Japan; by the Japan Society of the Promotion of Science Research for the Future; and by the Human Frontier Science Program. C.M. and M.S. were supported by grants from the NIH, by the HHMI, by a postdoctoral grant from the American Cancer Society, and an NIH training grant (5T32HD07312).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL craig.mello@umassmed.edu; FAX (508) 856-4289.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.955702.
References
- Baumgartner S, Martin D, Chiquet-Ehrismann R, Sutton J, Desai A, Huang I, Kato K, Hromas R. The HEM proteins: A novel family of tissue-specific transmembrane proteins expressed from invertebrates through mammals with an essential function in oogenesis. J Mol Biol. 1995;251:41–49. doi: 10.1006/jmbi.1995.0414. [DOI] [PubMed] [Google Scholar]
- Blelloch R, Newman C, Kimble J. Control of cell migration during Caenorhabditis elegans development. Curr Opin Cell Biol. 1999;11:608–613. doi: 10.1016/s0955-0674(99)00028-9. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EB, Stern MJ. Understanding cell migration guidance: Lessons from sex myoblast migration in C. elegans. Trends Genet. 1998;14:322–327. doi: 10.1016/s0168-9525(98)01507-8. [DOI] [PubMed] [Google Scholar]
- Chen W, Lim HH, Lim L. A new member of the ras superfamily, the Rac1 homologue from Caenorhabditis elegans. J Biol Chem. 1993;268:320–324. [PubMed] [Google Scholar]
- Chen W, Chen S, Yap SF, Lim L. The Caenorhabditis elegans p21-activated kinase (CePAK) colocalizes with CeRac1 and CDC42Ce at hypodermal cell boundaries during embryo elongation. J Biol Chem. 1996;271:26362–26368. doi: 10.1074/jbc.271.42.26362. [DOI] [PubMed] [Google Scholar]
- Chin-Sang ID, Chisholm AD. Form of the worm: Genetics of epidermal morphogenesis in C. elegans. Trends Genet. 2000;16:544–551. doi: 10.1016/s0168-9525(00)02143-0. [DOI] [PubMed] [Google Scholar]
- Chin-Sang ID, George SE, Ding M, Moseley SL, Lynch AS, Chisholm AD. The ephrin VAV-2/EFN-1 function in neuronal signaling to regulate epidermal morphogenesis in C. elegans. Cell. 1999;99:781–790. doi: 10.1016/s0092-8674(00)81675-x. [DOI] [PubMed] [Google Scholar]
- Costa M, Draper BW, Priess JR. The role of actin filaments in patterning the Caenorhabditis elegans cuticle. Dev Biol. 1997;184:373–384. doi: 10.1006/dbio.1997.8530. [DOI] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotti JG, Merz DC. DCC and netrins. Curr Opin Cell Biol. 1998;10:609–613. doi: 10.1016/s0955-0674(98)80036-7. [DOI] [PubMed] [Google Scholar]
- Desai C, Garriga G, McIntire SL, Horvitz HR. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature. 1988;336:638–646. doi: 10.1038/336638a0. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Frohli Hoier E, Mohler WA, Kim SK, Hajnal A. The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes & Dev. 2000;12:874–886. [PMC free article] [PubMed] [Google Scholar]
- Garriga G, Desai C, Horvitz HR. Cell interactions control the direction of outgrowth, branching and fasciculation of the HSN axons of Caenorhabditis elegans. Development. 1993;117:1071–1087. doi: 10.1242/dev.117.3.1071. [DOI] [PubMed] [Google Scholar]
- George SE, Simokat K, Hardin J, Chisholm AD. The VAB-1 Eph receptor tyrosine kinase functions in neural and epithelial morphogenesis in C. elegans. Cell. 1998;92:633–643. doi: 10.1016/s0092-8674(00)81131-9. [DOI] [PubMed] [Google Scholar]
- Hanna-Rose W, Han M. COG-2, a Sox domain protein necessary for establishing a functional vulval–uterine connection in Caenorhabditis elegans. Development. 1999;126:169–179. doi: 10.1242/dev.126.1.169. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Gomes AV, Kenyon C, Liu J, Krause M, Fire A. An analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes & Dev. 1998;12:2623–2635. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Hall DH. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis of C. elegans. Neuron. 1990;4:61–85. doi: 10.1016/0896-6273(90)90444-k. [DOI] [PubMed] [Google Scholar]
- Heid PJ, Raich WB, Smith R, Mohler WA, Simokat K, Gendreau SB, Rothman JH, Hardin J. The zinc finger protein DIE-1 is required for late events during epithelial cell rearrangement in C. elegans. Dev Biol. 2001;236:165–180. doi: 10.1006/dbio.2001.0315. [DOI] [PubMed] [Google Scholar]
- Hummel T, Leifker K, Klambt C. The Drosophila HEM-2/NAP1 homolog KETTE controls axonal pathfinding and cytoskeletal organization. Genes & Dev. 2000;14:863–873. [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu Rev Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Kitamura Y, Kitamura T, Sakaue H, Maeda T, Ueno H, Nishio S, Ohno S, Osada S, Sakaue M, Ogawa W, et al. Interaction of Nick-associated protein 1 with activated GTP-binding protein Rac. Biochem J. 1997;322:873–878. doi: 10.1042/bj3220873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, Matsuura Y, Yoshida-Kubomura N, Iwamatsu A, Kaibuchi K. p140Sra-1 (Specifically Rac1– associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem. 1998;273:291–295. doi: 10.1074/jbc.273.1.291. [DOI] [PubMed] [Google Scholar]
- Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3:983–991. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- Labouesse M, Hartwieg E, Horvitz HR. The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development. 1996;122:2579–2588. doi: 10.1242/dev.122.9.2579. [DOI] [PubMed] [Google Scholar]
- Legouis R, Gansmuller A, Sookharea S, Bosher JM, Baillie DL, Labouesse M. LET-413 is a basolateral protein required for the assembly of adherens junctions in Caenorhabditis elegans. Nat Cell Biol. 2000;2:415–422. doi: 10.1038/35017046. [DOI] [PubMed] [Google Scholar]
- Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–4488. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb DT, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Priess JR. The maternal genes apx-1 and glp-1 and establishment of dorsal–ventral polarity in the early C. elegans embryo. Cell. 1994;77:95–106. doi: 10.1016/0092-8674(94)90238-0. [DOI] [PubMed] [Google Scholar]
- Mohler WA, Simske JS, Williams-Masson EM, Hardin JD, White JG. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr Biol. 1998;8:1087–1090. doi: 10.1016/s0960-9822(98)70447-6. [DOI] [PubMed] [Google Scholar]
- Montell DJ. The genetics of cell migration in Drosophila melanogaster and Caenorhabditis elegans development. Development. 1999;126:3035–3046. doi: 10.1242/dev.126.14.3035. [DOI] [PubMed] [Google Scholar]
- Newman AP, White JG, Sternberg PW. Morphogenesis of the C. elegans hermaphrodite uterus. Development. 1996;122:3617–3626. doi: 10.1242/dev.122.11.3617. [DOI] [PubMed] [Google Scholar]
- Podbilewicz B, White JG. Cell fusion in the developing epithelia of C. elegans. Dev Biol. 1994;161:408–424. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- Priess JR, Hirsh DI. Caenorhabditis elegans morphogenesis: The role of the cytoskeleton in elongation of the embryo. Dev Biol. 1986;117:156–173. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol. 2000;2:131–136. doi: 10.1038/35004000. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha Y-H, Ali M, Priess JR, Mello CC. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Yasuda J, Shin TH, Lin R, Sawa H, Okano H, Priess JR, Davis RJ, Mello CC. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell. 1999;97:717–726. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- Roy PJ, Zheng H, Warren CE, Culotti JG. mab-20 encodes Semaphorin-2a and is required to prevent ectopic cell contacts during epidermal morphogenesis in Caenorhabditis elegans. Development. 2000;127:755–767. doi: 10.1242/dev.127.4.755. [DOI] [PubMed] [Google Scholar]
- Settleman J. Rho GTPases in development. Prog Mol Subcell Biol. 1999;22:201–229. doi: 10.1007/978-3-642-58591-3_10. [DOI] [PubMed] [Google Scholar]
- Shelton CA, Clayton CJ, Ellis GC, Bowerman B. The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior–posterior polarity, and body morphogenesis during Caenorhabditis elegans embryogenesis. J Cell Biol. 1999;146:439–451. doi: 10.1083/jcb.146.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mello C. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes & Dev. 1998;12:943–955. doi: 10.1101/gad.12.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simske JS, Hardin J. Getting into shape: Epidermal morphogenesis in Caenorhabditis elegans embryos. BioEssays. 2001;22:12–23. doi: 10.1002/1521-1878(200101)23:1<12::AID-BIES1003>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev Biol. 1981;82:41–55. doi: 10.1016/0012-1606(81)90427-9. [DOI] [PubMed] [Google Scholar]
- Sulston J, Schierenberg E, White J, Thomson N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:67–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Nishiyama K, Yamamoto A, Inazawa J, Iwaki T, Yamada T, Kanazawa I, Sakaki Y. Molecular cloning of a novel apoptosis-related gene, human Nap1 (NCKAP1), and its possible relation to Alzheimer Disease. Genomics. 2000;63:246–254. doi: 10.1006/geno.1999.6053. [DOI] [PubMed] [Google Scholar]
- Tabara H, Grishok A, Mello CC. RNAi in C. elegans: Soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- Trent C, Tsung N, Horvitz HR. Egg-laying defective mutants of the nematode C. elegans. Genetics. 1983;104:610–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshenker D, Garriga G, Thomas JH. Genetic and pharmacological analysis of neurotransmitters controlling egg laying in C. elegans. J Neurosci. 1995;15:6975–6985. doi: 10.1523/JNEUROSCI.15-10-06975.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Masson EM, Malik AN, Hardin J. An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development. 1997;124:2889–2901. doi: 10.1242/dev.124.15.2889. [DOI] [PubMed] [Google Scholar]
- Williams-Masson EM, Heid PJ, Lavin CA, Hardin J. The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev Biol. 1998;204:263–276. doi: 10.1006/dbio.1998.9048. [DOI] [PubMed] [Google Scholar]
- Wissmann A, Ingles J, McGhee JD, Mains PE. Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinases and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes & Dev. 1997;11:409–422. doi: 10.1101/gad.11.4.409. [DOI] [PubMed] [Google Scholar]
- Wissmann A, Ingles J, Mains PE. The Caenorhabditis elegans mel-11 myosin phosphatase regulatory subunit affects tissue contraction in the somatic gonad and the embryonic epidermis and genetically interacts with the Rac signaling pathway. Dev Biol. 1999;209:111–127. doi: 10.1006/dbio.1999.9242. [DOI] [PubMed] [Google Scholar]
- Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 β-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- Zipkin ID, Kindt RM, Kenyon CJ. Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell. 1997;90:883–894. doi: 10.1016/s0092-8674(00)80353-0. [DOI] [PubMed] [Google Scholar]