Abstract
A group of evolutionarily conserved pleiotropic COP/DET/FUS proteins was initially defined by their ability to repress photomorphogenesis in Arabidopsis. It was proposed that this regulation be mediated by targeting degradation of key cellular regulators that promote photomorphogenesis. Among them, COP1 and the COP9 signalosome have been hypothesized to fulfill the roles as an ubiquitin ligase (E3) and an essential E3 modulator. Here we report that COP10 encodes a protein similar to ubiquitin-conjugating enzyme (E2) variant proteins (UEV). COP10 is part of a nuclear protein complex and capable of directly interacting with both COP1 and the COP9 signalosome. Our data indicates that COP10 defines a possible E2 activity, thus validating the working hypothesis that the pleiotropic COP/DET/FUS group of proteins defined a protein ubiquitination pathway.
Keywords: EUV, ubiquitination, proteasome, Arabidopsis, photomorphogenesis, protein degradation
Increasing evidence has suggested a prominent role of the regulated protein degradation through the ubiquitin-proteasome system in the control of multicellular organism development (Weissman 2001). Targeted protein degradation via the 26S proteasome is one of the most important means utilized by all eukaryotic organisms to control transcription, signal transduction, cell cycle progression, and metabolic activities. To use this proteolytic device effectively, the activity and the substrate specificity of the 26S proteasome must be tightly regulated in coordination with cellular signaling activities and homeostasis. For many substrates, becoming tagged by ubiquitin attachment is one important signal for proteasome-mediated degradation. Therefore, elaborate mechanisms have been employed by cells to selectively ubiquitinate substrates (Varshavasky 1996; Hershko and Ciechanover 1998; Laney and Hochstrasser 1999; Vierstra and Callis 1999). In most cases, three sequential enzymatic steps are involved in ubiquitination of substrates (Deshaies 1999; Weissman 2001): the ubiquitin activation enzyme (E1), ubiquitin conjugation enzyme (E2), and the ubiquitin protein ligase (E3).
A group of pleiotropic CONSTITUTIVE PHOTOMORPHOGENIC (COP/DET/FUS) proteins has been initially identified as repressors of photomorphogenic development and later shown to be conserved among multicellular eukaryotes (Osterlund et al. 1999; Wei and Deng 1999). It was recently hypothesized that those proteins may define a ubiquitin-proteasome system that mediates degradation of a key transcription factor, HY5, responsible for promoting photomorphogenesis within the nucleus (Hardtke et al. 2000; Osterlund et al. 2000; Schwechheimer and Deng 2001; Schwechheimer et al. 2001). Among them, COP1 was hypothesized to fulfill a role as a specific ubiquitin ligase (E3) for HY5 (3). The COP9 signalosome on the other side has been shown recently to interact directly with E3 ubiquitin ligases of the SCF-type and is suggested to act as an essential modulator of E3 complex activity (Wei and Deng 1999; Lyapina et al. 2001; Schwechheimer et al. 2001).
Arabidopsis COP10 was defined as one of the pleiotropic COP/DET/FUS loci that act to repress photomorphogenic seedling development in the absence of light (Wei and Deng 1999; Osterlund et al. 2000). Similar to other pleiotropic cop/det/fus mutations, mutations in COP10 also resulted in a defect in COP1-mediated degradation of the photomorphogenesis-promoting transcription factor HY5 (Osterlund et al. 2000). However, the identity and molecular basis of COP10 action is not known. Here we report the molecular characterization of COP10 and propose its possible mode of action in regulating protein degradation and development.
Results
Molecular cloning of the COP10 locus
A total of four mutant alleles of the COP10 locus have been described and are available (Fig. 1a; Materials and Methods). Three of those alleles (cop10-1, cop10-2, and cop10-3) showed a strong photomorphogenic phenotype in darkness and typically result in lethality after the seedling stage even grown under light conditions (Castle and Meinke 1994; Misera et al. 1994; Wei et al. 1994; Kwok et al. 1996). Occasionally, a strong cop10 mutant survived on growth medium and developed small and compact rosettes without elongation of leaf petioles, and with sterile flowers (Fig. 1b). The fourth allele, cop10-4, displayed the characteristic open cotyledon phenotype only after extended growth in the dark and was identified in a screen for cytokinin-insensitive mutants (Vogel et al. 1998). There is no observable adult phenotype for light-grown cop1-4 mutant plants.
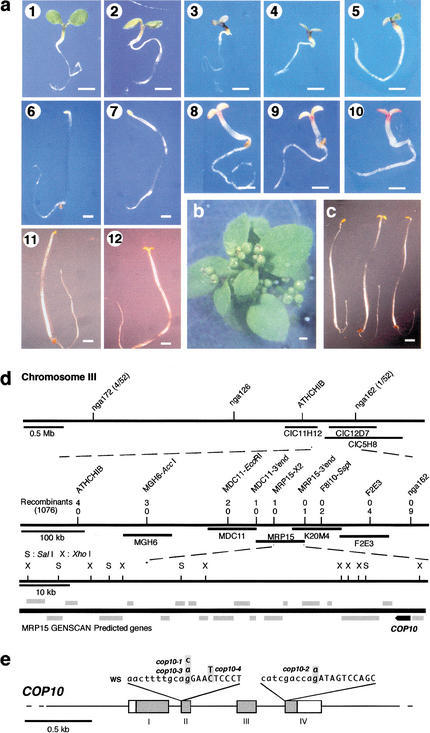
Figure 1.
Phenotype characteristics of cop10 mutants and the map-base cloning of COP10. (a) Morphology of the light- or dark-grown cop10 mutant seedlings. (Panels 1–5) Six-day-old light-grown wild- type, cop10-4, cop10-1, cop10-2, and cop10-3 seedlings, respectively. (Panels 6–10) Six-day-old dark-grown wild-type, cop10-4, cop10-1, cop10-2, and cop10-3 seedlings, respectively. (Panels 11,12) Eight-day-old dark-grown wild-type and cop10-4 seedlings, respectively. Bars, 1 mm. (b) A 5-week-old cop10-2 mutant plant. Bar, 1 mm. (c) Complementation of the cop10-4 mutation by a genomic fragment of the COP10 gene. From left to right, dark-grown 8-day-old wild-type seedling (left), cop10-4 (center), and cop10-4 transformed with COP10 genomic fragment (right). Bar, 1 mm. (d) Summary for the positional cloning of the COP10 gene. Partial genetic map of Arabidopsis chromosome III between the markers ATHCHIB and nga162 with several P1, TAC, and BAC clones are shown. (Gray lines) Open reading frames (ORFs) predicted by GENSCAN analysis. (e) Structure of COP10 and molecular nature of the mutations found in the four cop10 alleles. Boxes with Roman numerals indicate exons, whereas the introns are shown as lines between the exons. Shadowed area of exons indicates ORF coding region. The two splicing junctions where all four cop10 mutations are located are shown as inserts. The intron and exon sequences are shown as small and capital letters.
Based on the analysis of 1064 selected F2 plants from the mapping cross (see Materials and Methods), we located the COP10 locus within a 60-kb region of a P1 clone (Fig. 1d), MRP15, which was completely sequenced by the Arabidopsis Genome Initiative (GenBank accession no. AP000603). A GENSCAN analysis (genes.mit.edu/GENSCAN.html) of MRP15 predicted a number of open reading frames (ORFs). Database searches revealed that one of them (GenBank protein ID BAB01762), a 546-bp ORF encoding 182 amino acids, has high homology with ubiquitin-conjugating enzymes (E2). An expressed sequence tag (EST) containing a portion of the predicted ORF was also found in the database (GenBank accession no. AI999239). RT–PCR and 5′-RACE were performed to obtain the full-length cDNA for COP10 (GenBank accession no. AY034618), which was sequenced to confirm the ORF prediction. Our previous studies have indicated that both COP1 and the COP9 signalosome are involved in ubiquitin-proteasome pathway-mediated protein degradation (Osterlund et al. 2000; Schwechheimer et al. 2001). Thus this E2-like ORF was a prime candidate for COP10. To test whether this candidate gene corresponds to COP10, the genomic sequences of all four cop10 alleles were determined for this region by direct PCR followed by sequencing. Indeed, all four cop10 alleles had single base-pair mutations in the same gene-coding region (Fig. 1e). All three severe alleles contain splicing junction mutations, whereas the weak cop1-4 allele had a nucleotide transition that changed threonine 78 to isoleucine.
To further support the conclusion that this gene corresponds to COP10, RNA blot analysis was performed using a 3-kb genomic fragment containing the predicted gene. As shown in Figure 2a, no significant difference was detected in the level of a 0.9-kb transcript between wild-type and cop10-4seedlings. Conversely, the transcripts from cop10-1, cop10-2, and cop10-3 alleles, all having single base-pair changes in splicing junctions, were reduced in abundance and varied in size compared to transcripts in wild-type seedlings, possibly due to improper splicing. To confirm further the COP10 gene identity, a 3-kb genomic fragment that contains the entire predicted coding region and regulatory sequences was introduced into homozygous cop10-4 mutants and rescued the defects of dark-grown cop10-4 seedlings (Fig. 1c). Taken together, these data suggested that the E2-like ORF encode the COP10 protein.
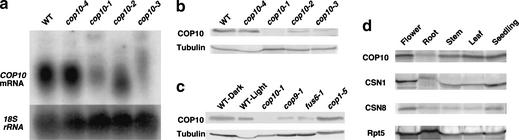
Figure 2.
The COP10 mRNA and protein expression profiles. (a) RNA blot analysis. RNAs were prepared from 5-day-old wild-type and cop10-4, cop10-1, cop10-2 and cop10-3 mutant seedlings grown under continuous white light. Total RNA from equal numbers of seedlings was used. The band in the wild-type lane corresponds to a size of 0.9 kb. As a loading control, the blot was reprobed with the 18S ribosomal RNA gene. (b) Immunoblot of light-grown wild-type (WT), cop10-4, cop10-1, cop10-2, and cop10-3 seedling extracts using anti-COP10 antibodies. (c) COP10 immunoblot from light- and dark-grown wild-type, light-grown cop10-1, cop9-1, fus6-1 and cop1-5 seedlings. (d) COP10 immunoblot of protein extracts from different tissue types. Flower, root, stem, or leaf protein extracts were obtained from 4-week-old light-grown mature plants. The seedlings were 5-day-old under continuous white light. A tubulin immunoblot was used as control for b and c and a Rpt5 immunoblot was used as a loading control in d.
The COP10 protein accumulation requires functional COP9 signalosome
To analyze the COP10 protein, polyclonal antibodies specific for COP10 were raised and used for immunoblot analysis (see Materials and Methods). Immunoblot analysis showed that COP10 migrates as an expected 21-kD protein regardless of light conditions (Fig. 2b,c). Except for the weak cop10-4 allele, which has normal levels of COP10, the abundance of COP10 in the other three cop10 alleles was significantly reduced or undetectable as in the case of cop10-1. This further confirmed the COP10 gene identity. Interestingly, expression of COP10 in cop9-1 and fus6-1 mutants, which lack COP9 signalosome, was reduced significantly compared to wild type, suggesting that the COP9 signalosome be involved in the regulation of COP10 stability (Fig. 2c). Conversely, there was no significant difference in the levels of COP10 in the cop1-5 and wild-type seedlings. In Arabidopsis adult plants, COP10 levels in flowers and leaves were similar to that in seedlings, but roots had reduced levels of COP10 (Fig. 2d). Thus the COP10 protein is present in all tissues examined albeit at variable levels. Although light or COP1 does not affect the accumulation of COP10, its accumulation depends on the COP9 signalosome.
COP10 encodes an E2 variant protein conserved in higher plants
Data base search with predicted COP10 protein sequence revealed EST clones from tomato (Lycopersicon esculentum) and soybean (Glycine max) that encode proteins highly similar to Arabidopsis COP10 (Fig. 3a). The deduced amino acid sequence of COP10 exhibited a high homology with E2 enzymes such as UBC4/UBC5 (52% amino acid identity) from Saccharomyces cerevisiae and UBC8/UBC9 (48% amino acid identity) from Arabidopsis (Fig. 3a,c). However, the invariant cysteine residue of canonical E2 catalytic domains is replaced by a serine residue in COP10 (Fig. 3b), even though COP10 has all the other conserved amino acids of the catalytic domain of E2 enzymes (Fig. 3a,b). This feature of COP10 resembles those of the ubiquitin-conjugating E2 enzyme variant (UEV) proteins defined by TSG101 and MMS2 (or called UEV1) (Koonin and Abagyan 1997; Sancho et al. 1998; Thomson et al. 1998). However, COP10 is phylogenetically closer to yeast UBC4/UBC5 and Arabidopsis UBC8/UBC9, than to the TSG101 or MMS2 (UEV1) families of UEVs, suggesting that COP10 defines a new group of UEV proteins. As opposed to E2 ubiquitin conjugation enzymes, UEV proteins are themselves inactive E2 variant enzymes that appear to function together with bona fide E2 enzymes (Hofmann and Pickart 1999). For example, UEV1 forms a heterodimer with UBC13, and this complex catalyzes the elongation of unusual polyubiquitin chain in yeast and mammals (Hofmann and Pickart 1999; Deng et al. 2000; VanDemark et al. 2001). Although COP10 is closely related to UBC4 and UBC5 family of E2 enzymes, it fails to rescue the yeast ubc4/ubc5 double mutant (data not shown). This is consistent with the hypothesis that COP10 itself is not an active E2 enzyme and likely works together with other partner(s).
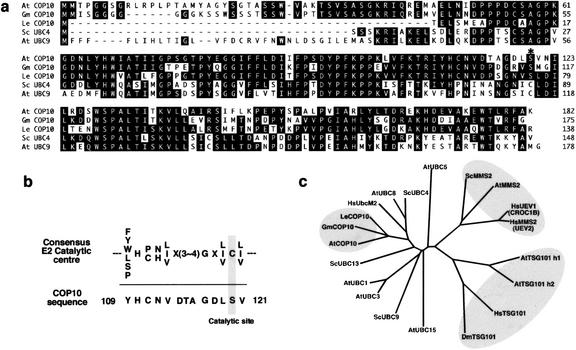
Figure 3.
Comparison of COP10 and ubiquitin conjugation enzyme E2 related proteins. (a) Sequence alignment of COP10 from Arabidopsis (AtCOP10), soybean (GmCOP10, GenBank AI460798), and tomato (LeCOP10, GenBank AW223526), and E2 enzymes from Saccharomyces (ScUBC4, GenBank S22857) and Arabidopsis UBC9 (AtUBC9, GenBank AAG40371). (Black boxes) Identical amino acids. Asterisk denotes active-site cysteine of E2 enzymes. (b) Sequence comparison between consensus catalytic center of UBC (PROSITE: PDOC00163) and Arabidopsis COP10. The catalytic cysteine and its corresponding serine in COP10 were highlighted. (c) Phylogenetic relationships of selected UBCs, COP10s, Ubc domain-like regions of TSG101, and UEVs. The gene families lacking catalytic cysteine are clustered in three gray boxes. The tree was constructed by phylogenetic analysis using ClustalW (Thompson et al. 1994). The accession numbers for the proteins are given below in parentheses: Homo sapiens (Hs)UbcM2 (AAD40197), HsUEV1 (AAB72016), HsMMS2 (CAA66717), HsTSG101 (AAC52083); Arabidopsis thaliana (At)UBC1 (P25865), AtUBC3 (P42746), AtUBC5 (P42749), AtUBC8 (P35131), AtUBC15 (AAC39324), AtMMS2 (AAK68786), AtTSG101-h1 (AAG51025), AtTSG101-h2 (BAB11114); Drosophila melanogaster (Dm)TSG101 (AAG29564); Saccharomyces cerevisiae (Sc)UBC9 (S52414), ScUBC13 (NP010377), and ScMMS2 (AAC24241). Note that Arabidopsis contains members in all three groups of E2V protein families.
COP10 is part of a large protein complex whose integrity and stability depend on the COP9 signalosome
To examine whether COP10 functions as a complex in vivo, gel filtration analysis was performed (Fig. 4a). The majority of COP10 fractionates with an ∼300-kD complex, whereas only small amounts are detected in the fractions corresponding to the 21-kD COP10 monomeric form. The high molecular mass fractions of COP10 were clearly different from that of the COP9 signalosome (Fig. 4a). However, the amount of COP10 complex is greatly reduced in the COP9 signalosome mutants, such as cop9-1 and fus6-1. Furthermore, the molecular mass of the COP10 complex in cop9-1 and fus6-1 mutants was smaller (∼250 kD) compared to the wild type. This suggests that lack of the COP9 signalosome not only reduced the stability of the COP10 complex, but also affected its integrity.
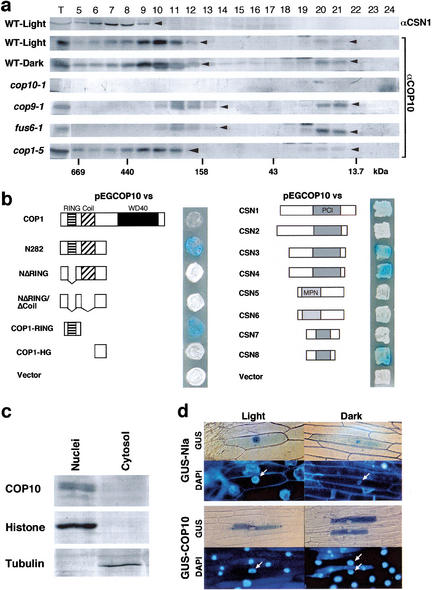
Figure 4.
The conformation, protein interaction, and nuclear localization of COP10. (a) Gel filtration analysis of the COP10 complex. Protein extracts were prepared from 5-day-old seedlings of continuous light or dark-grown wild-type, and cop10-1, cop9-1, fus6-1, and cop1-5 mutants in continuous light condition. Numbers below the panel indicate the approximate molecular mass. T represents total soluble protein extracts before gel filtration. The fraction numbers are indicated on the top of the panel. An anti-CSN1 immunoblot of light-grown wild type was performed for revealing the different peak position of the COP9 signalosome. (b) Interaction of COP10 with truncated COP1 proteins (Ang et al. 1998) and individual subunits (Serino et al. 1999) of the COP9 signalosome by a yeast two-hybrid assay (Ang et al. 1998). (c) COP10 immunoblot of protein extracts from nuclear or cytosolic fractions of cauliflower tissues. Anti-tubulin or anti-histone immunoblots were used as a cytosolic or nuclear protein controls, respectively. (d) Subcellular localization of the GUS–COP10 fusion protein in transiently transfected onion epidermal cells (von Arnim and Deng 1994). The entire COP10 coding region was fused to COOH end of GUS (von Arnim and Deng 1994). Cells stained for GUS and DAPI are shown next to each other. Results of GUS–NIa fusion protein and GUS alone were used as controls.
COP10 is capable of directly interacting with COP1RING-finger and the COP9 signalosome
To test for the potential direct interactions between COP10 and the COP9 signalosome or COP1, a yeast two-hybrid assay was performed. As shown in Figure 4b, there was a strong interaction between COP10 and the RING finger domain of COP1. This is consistent with previous observations that the RING-finger domain of E3 enzymes is the interacting site for E2 partner (Weissman 2001). The lack of interaction between COP10 and full-length COP1 in the yeast two-hybrid system could be caused by improper conformation of the COP1 in yeast and/or absence of required accessory factors. Furthermore, we have observed interactions between COP10 and three distinct COP9 signalosome subunits (CSN3, CSN4, and CSN8) in our yeast two-hybrid assay, suggesting a direct interaction between COP10 and the COP9 signalosome. Therefore, COP10 is potentially capable of direct interactions with both COP1 and the COP9 signalosome, suggesting they work together in mediating the repression of photomorphogenesis.
The COP10 complex is nuclear enriched
Because both COP1 and the COP9 signalosome act within the nucleus where they mediate the degradation of the photomorphogenesis-promoting transcription factor HY5, we examined whether COP10 also colocalized within the nucleus. To this end, nuclear and cytoplasmic fractions were isolated from florets of cauliflower and Western blot analysis was performed. As anticipated, COP10 was present abundantly in nuclear fractions and not detectable in cytoplasm (Fig. 4c). This result supports the notion that COP10 acts together with both COP1 and the COP9 signalosome in mediating light-regulated degradation of HY5 within the nucleus. It should be noted that COP10 lacks a recognizable nuclear targeting signal. Thus, the COP10 nuclear localization is likely mediated by other subunits within the COP10 complex. Indeed, a transient expression assay for GUS–COP10 fusion protein in onion epidermal cells resulted in only cytoplasm localization in both light and dark conditions (Fig. 4d). This result implies that COP10 itself does not contain a functional nuclear localization signal.
Discussion
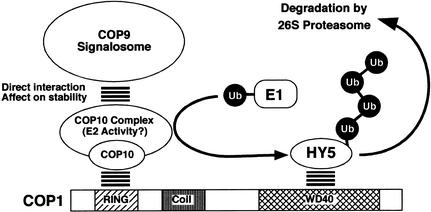
Taken together, our results have established that COP10 is another component in addition to the COP1 and the COP9 signalosome in the ubiquitin-proteasome pathway responsible for repression of photomorphogenesis in darkness. COP10 acts as a protein complex whose stability and integrity depend on a functional COP9 signalosome. COP10 is capable of directly interacting with both the COP9 signalosome and COP1 in the nucleus where these factors work together to mediate light regulated degradation of HY5 and thus the light control of photomorphogenic development. There are two possible biochemical activities for the COP10 complex. The COP10 complex may simply serve as a specific ubiquitin conjugation enzyme (E2) for the proposed E3 activity defined by COP1 (Osterlund et al. 2000). This model is illustrated in Figure 5. Alternatively, the COP10 complex may act as an E2 activity that promotes the formation of an unusual ubiquitin chain on COP1 and thus positively regulates COP1 activity. This would put COP10 function analogous to the E2V1 complex E2 activity reported for both yeast and mammalian systems (Hofmann and Pickart 1999; Deng et al. 2000; VanDemark et al. 2001). In both models, COP10 is part of a ubiquitin E2 activity that works together with COP1, a putative E3, and the COP9 signalosome, an essential and conserved E3 modulator. In either of the two cases, the COP9 signalosome would be a key participant in the process by directly associating with this E2–E3 complex. This role of the COP9 signalosome is in line with its reported role in directly interacting with SCF type E3 ubiquitin ligases (Schwechheimer and Deng 2001). This work thus supports the conclusion that the evolutionarily conserved pleiotropic COP/DET/DET group of proteins defines a ubiquitination pathway for regulated protein degradation through the 26S proteasome.
Figure 5.
A working model depicting possible functional relationships of COP1, COP9 signalosome, and COP10 complex in protein ubiquitination and proteasome-mediated degradation. The COP9 signalosome directly interacts with COP10 complex and regulates its accumulation and assembly, whereas COP10 complex acts as E2 activity for the COP1-mediated substrate ubiquitination. In repressing photomorphogenesis, those three components work together to target photomorphogenesis-promoting transcription factors (such as HY5) degradation via 26S proteasome. It is assumed that COP10 complex contains at least one more subunit that provides the E2 catalytic activity. Stacked parallel lines indicated the direct protein–protein interactions. The other abbreviations are: Ub, ubiquitin; RING, RING-finger motif; Coil, Coil–coil domain; WD40, WD-40 repeat domain.
Materials and methods
Plant materials and growth condition
All Arabidopsis growth conditions have been described previously (Wei et al. 1994) unless otherwise stated. The cop10-1(Wei et al. 1994) and cop10-4 (cin4) (Vogel et al. 1998) mutants were isolated from Wassilewskija (WS) and Columbia (Col-O) ecotypes, respectively. Both cop10-2 (fus9-T321) and cop10-3 (fus9-U401) were isolated from the Landsberg erecta (Ler) ecotype (Castle and Meinke 1994; Misera et al. 1994).
Cloning and analysis of COP10
For the fine-mapping of the COP10 gene, an F2 mapping population was generated by crossing heterozygous cop10-2(Ler ecotype) with wild type (Col-O). DNAs isolated from 1076 individual F2 plants were used for SSLP analysis (Bell and Ecker 1994). After scoring these 2152 chromosomes, cop10-2 was mapped to a region flanked by SSLP marker nga162 and ATHCHIB. Selected BAC or P1 clones in the region were used to generate RFLP markers for further fine mapping using the same F2 population. The TAIL–PCR (Liu and Whittier 1995) method was used to isolate both the right and left ends of several selected BAC clones for RFLP markers as well. To obtain the full-length cDNA for COP10, 5′-RACE were performed using 5′RACE System 2.0 (GIBCO BRL).
To complement the cop10-4 phenotype, a genomic fragment (from 76,235 bp to 79,840 bp of MRP15) that contains the whole genomic region of COP10 was amplified by PCR and subcloned into pPZP221 vector (Hajdukiewicz et al. 1994). The resulting construct was introduced into cop10-4 plants by Agrobacterium-mediated transformation.
For RNA blot analysis, total RNA from Arabidopsis seedlings was isolated using the RNeasy kit (QIAGEN). Blots were prepared using RNA from equal numbers of seedlings and were probed with radioactively labeled DNA fragments (Wei et al. 1994).
COP10 antibody, immunoblot analysis, and gel filtration chromatography
We generated a polyclonal COP10 antibody by cloning the COP10 cDNA into the pET28 expression vector (Novagen). The entire ORF of the COP10 cDNA was amplified by PCR with the primers 5′-TTCATAT GATGACACCTGGCGGAAG-3′ and 5′-GACTCGAGTCACTTGGCA AATCGCAATG-3′, thereby introducing a NdeI site at the 5′ terminus and an XhoI site at the 3′ terminus. This cDNA fragment was ligated into pET28c. The resulting plasmid pETCOP10 was transformed into BL21 (DE3) cells. The purified histidine-tagged COP10 protein was injected into rabbits as antigen. Polyclonal anti-COP10 antibodies were purified from rabbit serum using the purified GST–COP10 with glutathione sepharose 4B (Amersham Pharmacia). For immunoblot analysis, 5- to 7-day-old seedlings were homogenized with grinding buffer (25 mM Tris-HCl at pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 10μM leupeptin, 1μg/mL pepstatin A, 10% glycerol). For gel filtration chromatography, 7-day-old seedlings were homogenized in grinding buffer. The extract was centrifuged for 10 min at 4°C, and subsequently filtered though a 0.2 μm syringe filter. Total soluble protein was fractionated through a 25 mL Superdex-200 FPLC column (Amersham Pharmacia) with the grinding buffer. After a 7-mL void volume, consecutive fractions of 0.5 mL each were collected.
Cellular localization studies
Cauliflower floret tissue was fractionated according to a described procedure (Masuda et al. 1991). Briefly, 10 g of cauliflower floret tissue were homogenized in a grinding buffer containing 25 mM Mes-KOH (pH 5.6), 5 mM MgCl2, 10 mM KCl, 0.35 M sucrose, 30% glycerol, and Complete mini protease inhibitor set (Boehringer) and then centrifuged at 3500g for 10 min. Supernatant was collected as a cytoplasmic fraction. For nuclear fraction, the pellet was further subjected to a Percoll gradient (32%–48%) fractionation.
To generate in-frame fusion to the C-terminal end of the GUS protein, the entire COP10 coding sequence was subcloned into pRTL2–GUS (von Arnim and Deng 1994). The resulting construct for GUS–COP10 was introduced into onion epidermal cells using a particle bombardment system as described previously (von Arnim and Deng 1994) with pRTL2–GUS used as a control.
Yeast two-hybrid assays
The two-hybrid interaction assay in yeast was performed as described previously (Ang et al. 1998). Truncated versions of the COP1 and each subunit of COP9 signalosome expression vector in yeast were constructed as described using pEG202 and pJG4-5 (Ang et al. 1998; Serino et al. 1999). A β-galactosidase plate assay for yeast was achieved on selection agar plates containing galactose and X-gal in the medium (buffered to pH 7.0). Colonies were allowed to grow at 30°C for 4 d.
Acknowledgments
We thank G. Serino for her valuable help during this work, C. Schwechheimer and T. Nelson for critical commenting the manuscript, and Dr. S. Jentsch for providing yeast ubc4/ubc5 double mutant strain. This work was supported by a National Institutes of Health Grant GM47850 (to X.W.D.) and a Human Frontier Science Program Grant RG0043/97 (to M.M. and X.W.D.). X.W.D. is a National Science Foundation Presidential Faculty Fellow and Y.Y. is a postdoctoral fellow of Japan Society for the Promotion of Science (JSPS).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL xingwang.deng@yale.edu; FAX (203) 432-5726.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.964602.
References
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsisdevelopment. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Castle LA, Meinke DW. A FUSCA gene of Arabidopsisencodes a novel protein essential for plant development. Plant Cell. 1994;6:25–41. doi: 10.1105/tpc.6.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW. HY5 stability and activity in Arabidopsisis regulated by phosphorylation in its COP1 binding domain. EMBO J. 2000;19:4997–5006. doi: 10.1093/emboj/19.18.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Abagyan RA. TSG101 may be the prototype of a class of dominant negative ubiquitin regulators. Nat Genet. 1997;16:330–331. doi: 10.1038/ng0897-330. [DOI] [PubMed] [Google Scholar]
- Kwok SF, Piekos B, Misera S, Deng XW. A complement of ten essential and pleiotropic ArabidopsisCOP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 1996;110:731–742. doi: 10.1104/pp.110.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshiaes RJ. Promotion of NEDD8–CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Masuda K, Takahashi S, Nomura K, Inoue M. A simple procedure for the isolation of pure nuclei from carrot embryos in synchronized cultures. Plant Cell Rep. 1991;10:329–333. doi: 10.1007/BF00193152. [DOI] [PubMed] [Google Scholar]
- Misera S, Muller AJ, Weiland-Heidecker U, Jurgens G. The FUSCA genes of Arabidopsis: Negative regulators of light responses. Mol Gen Genet. 1994;244:242–252. doi: 10.1007/BF00285451. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Ang LH, Deng XW. The role of COP1 in repression of Arabidopsisphotomorphogenic development. Trends Cell Biol. 1999;9:113–118. doi: 10.1016/s0962-8924(99)01499-3. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Sancho E, Vila MR, Sanchez-Pulido L, Lozano JJ, Paciucci R, Nadal M, Fox M, Harvey C, Bercovich B, Loukili N, et al. Role of UEV-1, an inactive variant of the E2 ubiquitin-conjugating enzymes, in in vitro differentiation and cell cycle behavior of HT-29–M6 intestinal mucosecretory cells. Mol Cell Biol. 1998;18:576–589. doi: 10.1128/mcb.18.1.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Deng X. COP9 signalosome revisited: A novel mediator of protein degradation. Trends Cell Biol. 2001;11:420–426. doi: 10.1016/s0962-8924(01)02091-8. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292:1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- Serino G, Tsuge T, Kwok S, Matsui M, Wei N, Deng XW. Arabidopsiscop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell. 1999;11:1967–1980. doi: 10.1105/tpc.11.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson TM, Khalid H, Lozano JJ, Sancho E, Arino J. Role of UEV-1A, a homologue of the tumor suppressor protein TSG101, in protection from DNA damage. FEBS Lett. 1998;423:49–52. doi: 10.1016/s0014-5793(98)00060-x. [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Molecular insights into polyubiquitin chain assembly: Crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105:711–720. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule: Functions, mysteries, uses. Proc Natl Acad Sci. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD, Callis J. Polypeptide tags, ubiquitous modifiers for plant protein regulation. Plant Mol Biol. 1999;41:435–442. doi: 10.1023/a:1006323317890. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Schuerman P, Woeste K, Brandstatter I, Kieber JJ. Isolation and characterization of Arabidopsismutants defective in the induction of ethylene biosynthesis by cytokinin. Genetics. 1998;149:417–427. doi: 10.1093/genetics/149.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. Light inactivation of Arabidopsisphotomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW. Making sense of the COP9 signalosome. A regulatory protein complex conserved from Arabidopsisto human. Trends Genet. 1999;15:98–103. doi: 10.1016/s0168-9525(98)01670-9. [DOI] [PubMed] [Google Scholar]
- Wei N, Kwok SF, von Arnim AG, Lee A, McNellis TW, Piekos B, Deng XW. ArabidopsisCOP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell. 1994;6:629–643. doi: 10.1105/tpc.6.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]