Abstract
The germination of Arabidopsis seeds is promoted by gibberellin (GA). Arabidopsis GAI, and RGA are genes encoding key GA signal-transduction components (GAI and RGA) that mediate GA regulation of stem elongation. The Arabidopsis genome contains two further genes, RGL1 and RGL2, that encode proteins (RGL1 and RGL2) that are closely related to GAI and RGA. Here, we show that RGL2 regulates seed germination in response to GA, and that RGL1, GAI, and RGA do not. In addition, we show that RGL2 transcript levels rise rapidly following seed imbibition, and then decline rapidly as germination proceeds. In situ GUS staining revealed that RGL2 expression in imbibed seeds is restricted to elongating regions of pre-emergent and recently emerged radicles. These observations indicate that RGL2 is a negative regulator of GA responses that acts specifically to control seed germination rather than stem elongation. Furthermore, as RGL2 expression is imbibition inducible, RGL2 may function as an integrator of environmental and endogenous cues to control seed germination.
Keywords: RGL2/RGL1/GAI/RGA, gibberellin, imbibition, seed germination, Arabidopsis thaliana
Seeds contain embryonic plants that are arrested in their development, and that await the appropriate environmental conditions to continue with their life cycles (Bewley 1997). The transition of a seed from dormancy to germination is controlled by external environmental cues (including light quality, moisture, and transient exposure to cold) and by the internal growth regulators GA and ABA (abscisic acid). ABA establishes and maintains dormancy, whereas GA breaks dormancy and induces germination (Koornneef and Karssen 1994; McCarty 1995; Bewley 1997; Steber et al. 1998). Under favorable environmental conditions, GA biosynthesis is induced and, in turn, the de novo-synthesized GA induces the expression of genes encoding enzymes such as endo-β-1,3 glucanase (Leubner-Metzger et al. 1995), β-1,4 mannan endohydrolase (Bewley 1997; Sánchez and Miguel 1997), and the extensin-like protein AtEPR1 (Dubreucq et al. 2000). These enzymes hydrolyze the endosperm and release the inhibitory effects of ABA on embryo growth potential (Koornneef and Karssen 1994; McCarty 1995; Bewley 1997). The importance of the balance between ABA and GA signaling for seed germination is perhaps best shown by studies of ABA response mutants and GA-deficient mutants. In Arabidopsis, ABA-insensitive mutations (e.g., abi1, abi2, and abi3) reduce seed dormancy and allow germination at ABA concentrations that are normally inhibitory to wild-type germination (Koornneef et al. 1984). On the other hand, GA-deficient mutants (e.g., ga1-3 and ga2) fail to germinate in the absence of exogenous GA (Koornneef and van der Veen 1980). Although it has long been known that GA promotes seed germination, the molecular mechanisms by which GA signaling regulates germination, and the relationship between GA signaling and the environmental cues to germination are poorly understood.
In recent years, genes encoding several GA-signaling components have been identified and cloned (Harberd et al. 1998; Thornton et al. 1999; Sun 2000; Richards et al. 2001). Loss-of-function mutations of SPINDLY (spy mutant alleles) confer a phenotype that mimics that of GA-treated wild-type plants. spy mutants are slender and early flowering and have pale green foliage, indicating that SPINDLY is a negative regulator of GA responses (Jacobsen and Olszewski 1993; Jacobsen et al. 1996). In addition, spy alleles suppress the nongerminating phenotype conferred by ga1-2 (an allele of ga1-3) as follows: spy ga1-2 double-mutant seeds germinate in the absence of exogenous GA (Jacobsen and Olszewski 1993), suggesting that SPY plays an important role in the GA-mediated control of both stem elongation and seed germination. GAI and RGA are important negative regulators of the GA-signaling pathway for stem elongation, because the stem elongation of plants homozygous for null alleles of GAI or RGA (e.g., gai-t6 or rga-24) requires less GA than that of wild-type plants (Peng and Harberd 1993; Peng et al. 1997; Silverstone et al. 1997, 1998). Furthermore, the combination of gai-t6 and rga-24 (in a gai-t6 rga-24 double mutant) completely suppresses the dwarfing phenotype conferred by the ga1-3 mutation (Dill and Sun 2001; King et al. 2001). The GAI/RGA genes were originally defined by the cloning of the mutant gai allele (Peng et al. 1997). gai encodes a mutant protein that lacks a region of 17 amino acids from close to the N terminus and confers a dominant dwarf, reduced GA-response phenotype. Functional GAI/RGA orthologs have been identified in wheat (Rht), maize (d8), and rice (SLR1), and mutations that cause alterations in the N termini of these proteins also confer dominant dwarfism combined with reduced GA responses (Peng et al. 1999a; Ikeda et al. 2001). Recently, PHOR1 (photoperiod-response 1), initially identified as a photoperiod response gene in potato (Solanum. tuberosum ssp. Andigena), was also found to be a GA-signaling factor, suggesting that PHOR1 links photoperiod and GA signaling (Amador et al. 2001).
The plant height regulating factors GAI/RGA/RHT/d8/SLR1 (Peng et al. 1997, 1999a; Silverstone et al. 1998; Ikeda et al. 2001) belong to a family of putative transcription factors known as the GRAS family. GRAS proteins appear to be unique to plants (Peng et al. 1999a; Pysh et al. 1999; Richards et al. 2000), and there are ∼30 candidate GRAS ORFs in the Arabidopsis genome. Alignment of the GRAS proteins reveals that they share close homology in approximately two-thirds of their C-terminal regions, contain features with characteristics of transcription factors, and contain an SH2-like domain (Peng et al. 1999a; Pysh et al. 1999; Richards et al. 2000). In contrast, the N-terminal approximately one-third of the GRAS proteins are highly diversified (Peng et al. 1999a; Pysh et al. 1999; Richards et al. 2000). Mutant analysis has revealed that GRAS proteins regulate several different aspects of plant growth and development in addition to GA signaling, including root development (Di Laurenzio et al. 1996; Helariutta et al. 2000; Nakajima et al. 2001), lateral branch initiation (Schumacher et al. 1999), and phytochrome A signal transduction (Bolle et al. 2000). The specificity of the GRAS proteins is thought to reside in the diverse approximately one-third N-terminal regions, because they differ from each other mainly in this region. In fact, two highly conserved N-terminal regions (I and II; Peng et al. 1999a) are found in GAI and RGA, in GAI/RGA functional orthologs in other species, and in the ORFs of three related Arabidopsis genes of hitherto unknown function: RGA-like 1 (RGL1, chromosome 1), RGA-like 2 (RGL2, chromosome 3), and RGA-like 3 (RGL3, chromosome 5) (Peng et al. 1999a; Richards et al. 2000; Dill and Sun 2001). Regions I and II are not found in other Arabidopsis GRAS proteins. Analysis of the Arabidopsis mutant gai gene, of mutant forms of GAI/RGA orthologs in wheat and maize, and of the effects of in vitro-synthesized RGA and SLR1 mutant alleles that encode proteins lacking region I, have shown that regions I and II are critical for GA signaling (Peng et al. 1999a; Dill et al. 2001; Ikeda et al. 2001).
Because RGL1 and RGL2 contain regions I and II, it seemed likely that they also function as GA-signaling factors. As described above, the absence of GAI and RGA suppresses the dwarf stem phenotype of ga1-3, indicating that GAI and RGA act as negative regulators of GA-mediated stem elongation. However, absence of GAI and RGA function is not sufficient to restore normal germination or normal floral development to ga1-3 in the absence of exogenous GA, indicating that other signaling components or pathways are involved in controlling these responses (Dill and Sun 2001; King et al. 2001). Here, we present genetic and molecular data showing that RGL2 is a GA-signaling component, and that it functions as a negative regulator of seed germination. In addition, we show that RGL2 expression in both wild-type and ga1-3 seeds is induced to high levels following the onset of imbibition. The levels of RGL2 transcripts in germinating wild-type seeds decrease rapidly once the radicle begins to protrude, but remain at a high level in imbibed, nongerminating ga1-3 mutant seeds. This suggests that the role of RGL2 is to prevent germination after imbibition and that GA promotes germination by down-regulating RGL2 expression. Thus, GA regulates stem elongation via GAI and RGA, and seed germination via RGL2. RGL2 acts as an integrating factor that links GA signaling and environmental cues in the regulation of seed germination.
Results
The N termini of RGL1 and RGL2 contain regions I and II, the conserved GA-signaling regions in GAI/RGA
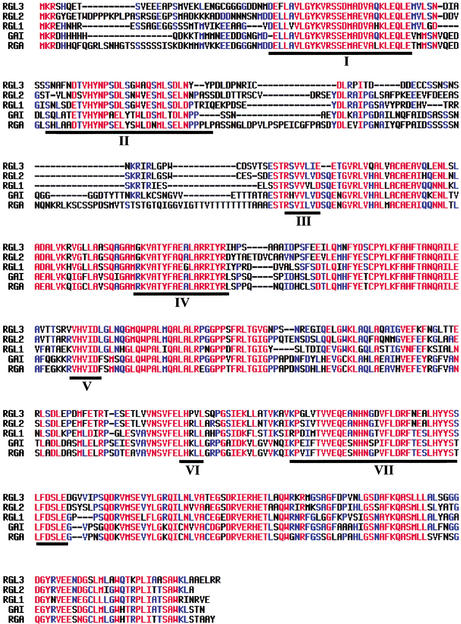
The Arabidopsis genome contains three additional genes, RGL1, RGL2, and RGL3 that encode proteins (RGL1, RGL2, and RGL3) whose amino acid sequences are closely related to those of GAI and RGA (Dill and Sun 2001). Overall, these sequences share 59% (RGL1 vs. RGL2), 58% (RGL1 vs. RGL3), and 68% (RGL2 vs. RGL3) homology. RGL1, RGL2, and RGL3 show, respectively, 58%, 55%, and 54% homology to GAI, and 55%, 57%, and 53% homology to RGA. RGL1, RGL2, and RGL3 differ from other GRAS proteins by showing close similarity, not only with the C-terminal regions of GAI and RGA but also to the N-terminal regions I and II (GA-response regions; Fig. 1). Previous reports have shown that mutant alleles with alterations in regions I and II confer greatly reduced responses to either exogenous or endogenous GA (Peng et al. 1997,1999a; Dill et al. 2001; Ikeda et al. 2001). The fact that RGL1, RGL2, and RGL3 display strong amino acid sequence conservation of regions I and II implies that these three proteins are also GA-response regulators.
Figure 1.
Amino-acid sequence alignment comparing the predicted RGL2, GAI, RGA, RGL1, and RGL3 proteins of Arabidopsis (for nomenclature, see Dill and Sun 2001). Gaps are introduced to maximize alignment. Regions I to VII are as defined previously (Peng et al. 1999a). Regions I and II are GA-response regions, which, when deleted from GAI, Rht (wheat), d8 (maize), SLR1 (rice), or RGA, confer reduced GA responses and dwarfism (Peng et al. 1997, 1999a; Dill et al. 2001; Ikeda et al. 2001).
Molecular characterization of three RGL2 Ds-GUS insertion lines
Because gai-t6 rga-24 ga1-3 mutant seeds fail to germinate, it has been suggested that signal components other than GAI or RGA are involved in controlling seed germination in response to GA (Dill and Sun 2001; King et al. 2001). The above described amino acid sequence similarities suggested that RGL1, RGL2, and RGL3 might act as GA-signaling components that control germination. We therefore screened a Ds-transposant collection (Sundaresan et al. 1995; Parinov et al. 1999) for Ds-GUS insertions within the RGL1, RGL2, and RGL3 ORFs.
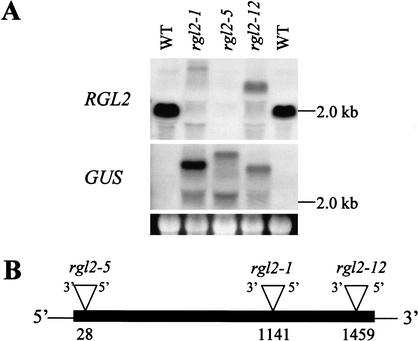
Initially, the RGL2 ORF was used to screen a database containing the sequences of the sites of insertion of Ds elements in transposants from within the collection (the SGT database; Parinov et al. 1999; V. Sundaresan, pers. comm.). Three lines (SGT625, SGT11937, and SGT13975) with Ds-GUS insertions in RGL2 were identified. DNA gel-blot analysis, using a hybridization probe derived from RGL2, revealed that the RGL2 ORF is interrupted in all three lines (S.C. Lee and J. Peng, unpubl.). In RNA gel-blot experiments (Fig. 2A), an RGL2 probe identified a 2.0-kb RGL2 transcript in wild type. In SGT625, SGT11937, and SGT13975, this 2.0-kb RGL2 transcript was no longer present and was replaced by larger transcripts that were thought likely to be fusion transcript products containing both RGL2 and Ds-GUS (Fig. 2A) similar to the pattern observed previously for GAI transcripts in the Ds- insertion gai-t6 mutant (Peng et al. 2001). As expected, the larger transcripts in SGT625, SGT11937, and SGT13975 also hybridized to a probe derived from the GUS gene (Fig. 2A). The GUS gene probe also identified additional, smaller, hybridization bands in SGT625, SGT11937, and SGT13975, which possibly represent alternative splicing products of RGL2-Ds-GUS fusion transcripts (Fig. 2A).
Figure 2.
Molecular characterization of three independent RGL2 Ds-insertion mutant lines. (A) RNA gel-blot hybridizations using DIG-labeled RGL2 and GUS probes (see Materials and Methods) and total RNA from inflorescences (28 days old). The RGL2 probe identified a ∼2-kb hybridization band in RNA from the wild-type control. This band was absent in RNA from all three mutant lines. The GUS probe identified hybridization bands in RNA from all three mutant lines, but failed to hybridize to RNA from wild-type controls. Below the GUS-hybridization panel, the gel fluorescence serves as a control for equal RNA sample loading on the gel. (B) Schematic diagram showing the Ds-insertion sites in the rgl2-1, rgl2-5, and rgl2-12 mutant alleles (thin line, noncoding region; filled thick line, RGL2-coding region, inverted triangle, Ds insertion). The Ds element insertion in all three alleles orientated 3′ to 5′ as shown. Numbers represent the site of insertion in the RGL2 ORF (in nucleotides, the A of the start ATG = 1).
By use of further information from the SGT database (V. Sundaresan, pers. comm.), RGL2-specific primers were designed and paired with primers derived from Ds-GUS, thus permitting the amplification via PCR of DNA fragments containing Arabidopsis genomic DNA–Ds junction regions (for both ends of the Ds) from each of SGT625, SGT11937, and SGT13975. The obtained PCR products were sequenced and compared with wild-type RGL2 DNA sequence to locate the precise Ds insertion sites. Ds-insertions in RGL2 were confirmed for all three mutant lines, and the precise position of insertion of Ds in each line is shown in Figure 2B. These three lines were thus designated as rgl2-1 (SGT625), rgl2-5 (SGT11937), and rgl2-12 (SGT13975), respectively.
While searching the SGT database, another line (SGT4763), with a Ds-GUS insertion 68 bp upstream of the ATG translation start codon of RGL1 was identified (V. Sundaresan, pers. comm.; S.C. Lee and J. Peng, unpubl.). RNA gel-blot analysis confirmed that this Ds-GUS insertion abolishes RGL1 gene expression (S.C. Lee and J. Peng, unpubl.). This mutant line was therefore designated rgl1-1.
The germination of rgl2 mutants is strongly resistant to the effects of paclobutrazol
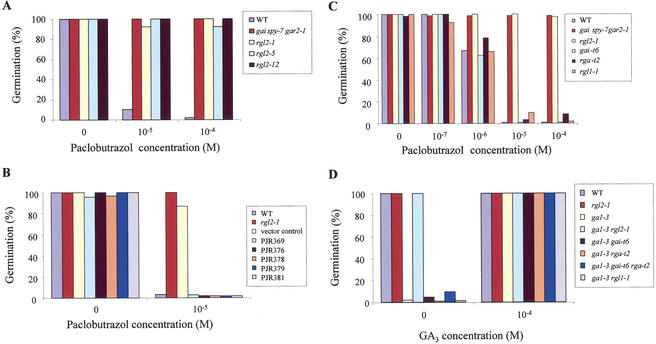
When grown in the greenhouse and in controlled-environment conditions (23°C, 16 h light/8 h dark), rgl2-1 plants were indistinguishable from wild type (data not shown). To determine whether disruption of RGL2 would affect seed germination, wild-type, rgl2-1, rgl2-5, and rgl2-12 seeds were placed on medium containing paclobutrazol (PAC) (Fig. 3A). PAC is a triazole derivative that inhibits GA biosynthesis at the kaurene oxidase reaction (Hedden and Graebe 1985). Seeds imbibed in the presence of PAC contain reduced or depleted endogenous GA levels, as a result of which their germination is inhibited. Previous experiments have shown that seeds of the gai spy-7 gar2-1 mutant line are strongly resistant to the inhibitory effects of PAC on seed germination (Peng et al. 1999b), and this line was used here as a positive control (Fig. 3A). The germination of wild-type seeds was inhibited completely by PAC at concentrations higher than 10−5 M. In contrast, seeds of the gai spy-7 gar2-1-positive control, and rgl2-1, rgl2-5, and rgl2-12 seeds, exhibited strong resistance to PAC and achieved high germination percentages (>90%) on 10−5 M and 10−4 M PAC medium (Fig. 3A).
Figure 3.
RGL2 is a GA-response negative regulator of seed germination. Germination begins when quiescent dry seeds are imbibed. Following this, the radicle begins to elongate. Germination becomes visible when the radicle protrudes outside of the seed coat. In these experiments, the frequency of radicle protrusion was used as a measure of germination. (A) Seed germination of RGL2 alleles on medium containing PAC. All three alleles (rgl2-1, rgl2-5, and rgl2-12) confer strong resistance to the inhibitory effects of PAC on seed germination. The gai spy-7 gar2-1 mutant line also displays PAC-resistant seed germination (Peng et al. 1999b) and was used here as a positive control. (B) A genomic DNA fragment containing RGL2 complements the rgl2-1 phenotype in five independent transgenic lines, PJR369, 376, 378, 379, and 381. Vector control, pCambia1300 (Cambia, Australia). (C) Comparison of the effects of increasing concentrations of PAC on the seed germination of wild-type and of the rgl2-1, rgl1-1, gai-t6, and rga-t2 mutants. At the highest PAC concentration, only the rgl2-1 mutant, and the gai spy-7 gar2-1-positive control seeds can germinate. (D) Suppression of the nongerminating phenotype of ga1-3 by the rgl2-1 mutation, but not by rgl1-1, gai-t6, rga-t2, or by gai-t6 rga-t2 in combination. Seeds were germinated on SM medium. All tests yielding germination rates between zero and <2% were given a value of 2% for the convenience of drawing the histogram. The seed germination experiments were repeated multiple times and the results shown are those of a single experiment and are clearly representative of what was seen in the repeat experiment.
To determine whether the PAC resistance of the rgl2-1, rgl2-5, and rgl2-12 mutants is truly conferred by insertional inactivation of RGL2, complementation experiments with an NdeI genomic DNA fragment containing the entire RGL2-coding region plus ∼3.3 kb 5′ noncoding and ∼2.0 kb 3′ noncoding sequences were performed. This DNA fragment was cloned into the binary vector pCambia1300 and transformed into rgl2-1 mutant plants. A number of independent transformants were obtained and seeds homozygous for the RGL2 transgene (from five independent transformant lines) were tested for their ability to germinate on medium containing PAC. As shown in Figure 3B, PAC sensitivity was restored in the seeds of all five transgenic lines (all of which display <5% germination at 10−5 M PAC). This result unequivocally shows that loss of RGL2 function confers PAC-resistant germination, demonstrating that RGL2 is a GA-response negative regulator of seed germination.
Experiments comparing the extent of the PAC resistance conferred by the rgl2 alleles with that conferred by loss-of-function mutations in other gene members of the GAI/RGA-like family were performed (Fig. 3C). In these experiments, seeds of the gai spy-7 gar2-positive control mutant achieved >95% germination on 10−4 M PAC medium. Although germinating slightly later than the positive control (data not shown), rgl2-1 seeds also achieved a high germination percentage (>95%) on this medium. However, the rgl1-1 (loss-of-function RGL1 allele), gai-t6 (loss-of-function GAI allele), and rga-t2 (loss-of-function RGA allele; see Materials and Methods) mutants all failed to germinate in these conditions, as did the wild-type control (Fig. 3C). These results show that rgl2 mutants, but not rgl1-1, gai-t6, or rga-t2 mutants, can germinate, regardless of the depleted endogenous GA levels conferred by PAC, suggesting that RGL2 and GAI/RGA, although closely related in amino acid sequence, might function in different GA-mediated signaling pathways.
rgl2-1 suppresses the nongerminating phenotype of ga1-3
GA1 encodes ent-CDP synthase, an enzyme that catalyzes a relatively early step in the biosynthesis of GA (Sun et al. 1992; Sun and Kamiya 1994). The ga1-3 loss-of-function allele causes GA defiency and abolishes seed germination (Koornneef and van der Veen 1980). Because rgl2-1 confers resistance to the inhibitory effect of PAC on seed germination, it seemed likely that rgl2-1 would also suppress the nongerminating phenotype conferred by ga1-3. To test this hypothesis, the seed germination of ga1-3 mutant lines carrying RGL2 or rgl2-1 was compared with that of other genotypes (Fig. 3D). As expected, ga1-3 mutants did not germinate, and absence of either or both of GAI and RGA (in ga1-3 gai-t6, ga1-3 rga-t2, and ga1-3 gai-t6 rga-t2) did not substantially suppress the nongermination phenotype of ga1-3 (Dill and Sun 2001; King et al. 2001). The ga1-3 rgl1-1 mutant also did not germinate, indicating that rgl1-1 germination is not GA independent. However, ga1-3 rgl2-1 achieved >95% germination, showing that the germination of rgl2-1 is largely independent of GA. As expected, high levels of germination were exhibited by all mutant lines in the presence of exogenous GA3 (Fig. 3D). These results, together with those obtained from the PAC test, clearly show that RGL2, but not RGL1, acts as a negative regulator of GA responses in the control of seed germination.
rgl2-1 does not confer PAC-resistant stem elongation or leaf expansion
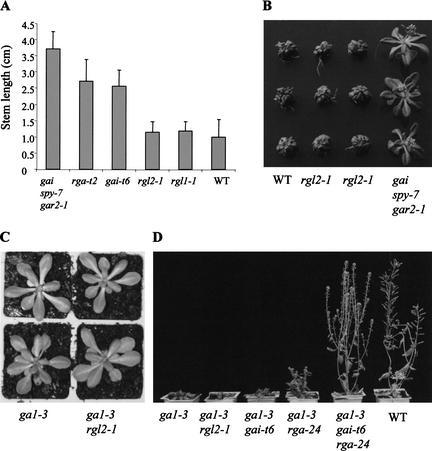
As shown above, wild-type seeds can germinate on medium containing 10−7 M PAC (Fig. 3C). However, continued growth on this medium causes dwarfism, with the reduced stem elongation and leaf expansion characteristic of GA deficiency. We tested the effects of long-term growth on PAC-containing medium on the stem elongation and leaf expansion of various genotypes. Growth of the gai spy-7 gar2-1 mutant line is relatively resistant to PAC, thus serving as a positive control for these experiments (Peng et al. 1999b; Fig. 4A,B). As expected, the absence of RGA had a marked effect on stem growth under these conditions (Silverstone et al. 1997). The rga-t2 mutant plants displayed strong PAC resistance and grew significantly taller than all of the other genotypes tested. The gai-t6 mutant plants also grew taller than wild type (Peng et al. 1997), but were shorter than the rga-t2 plants. However, no obvious differences were observed between the stem lengths of rgl1-1, rgl2-1, or wild-type plants grown in these conditions (Fig. 4A). The rosette sizes (used here as a measure of leaf expansion) of plants grown on 10−6 M PAC medium were also compared (Fig. 4B). The leaf expansion of wild-type and rgl2-1 plants was severely retarded by PAC. Wild-type and rgl2-1 rosettes were indistinguishable from one another, and markedly smaller than those of the gai spy-7 gar2-1 control. These results indicate that RGL2 is not involved in controlling stem elongation or leaf expansion growth or plays only a minor role in these processes.
Figure 4.
RGL2 plays a relatively minor role in the regulation of stem elongation and leaf expansion. (A) Comparison of stem lengths of wild-type and of rgl2-1, rgl1-1, gai-t6, and rga-t2 mutants grown on medium containing 10−7 M PAC (30 days old; n = 15–20). (B) Comparison of rosette size (a measure of leaf expansion) of two rgl2-1 lines, gai spy-7 gar2-1 (positive control) and wild-type (negative) control grown on medium containing 10−6 M PAC (30 days old). Each genotype is represented by three typical plants arranged in a column. (C) Rosette size of the rgl2-1 ga1-3 double mutant is indistinguishable from that of the ga1-3 single mutant (30 days old). Each genotype is represented by two typical plants arranged in a column. (D) rgl2-1 does not suppress the dwarf phenotype of ga1-3. All plants shown (except for the wild-type control) are homozygous for ga1-3. rgl2-1 and gai-t6 do not appreciably suppress ga1-3 phenotype, whereas rga-t2 partially, and the gai-t6 rga-24 combination fully, suppresses the dwarfing phenotype conferred by ga1-3 (adult plants, 45 days old, are shown).
rgl2-1 does not suppress the dwarf phenotype of ga1-3
To further investigate whether RGL2 is involved in controlling stem elongation growth, the growth of ga1-3 mutants carrying RGL2 or rgl2-1 was compared with the growth of additional mutant lines. When ga1-3 and ga1-3 rgl2-1 plants were compared at the late rosette stage, no significant phenotypic differences between the two lines could be discerned (Fig. 4C). In further experiments, the growth of ga1-3 rgl2-1 plants was compared with that of plants lacking GAI and/or RGA (Fig. 4D). Consistent with previous reports (Silverstone et al. 1998; Dill and Sun 2001; King et al. 2001), the dwarfism conferred by ga1-3 is partially suppressed by the absence of RGA, and completely suppressed by the absence of both GAI and RGA (Fig. 4D). In contrast, the growth of ga1-3 rgl2-1 plants was not significantly different from that of ga1-3 mutant plants (Fig. 4C,D). Thus, as shown above, rgl2-1 suppresses the effect of GA deficiency on seed germination, but does not suppress the effect of GA deficiency on stem elongation.
RGL2 expression is up-regulated during imbibition of seeds
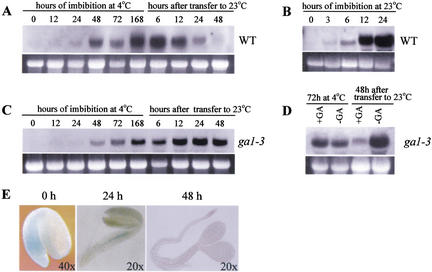
Imbibition is the process of water uptake by dry seeds, in the course of which the seed beomes hydrated. To further study how RGL2 is involved in seed germination, RNA gel-blot experiments were used to compare RGL2 transcript levels in dry seeds and in seeds imbibed at either 23 or 4°C. In dry wild-type seeds, RGL2 transcripts are present at a relatively low level. RGL2 transcript levels rise following onset of imbibition, are clearly visible after 12 h (at 23°C) and 48 h (at 4°C), and reach a high level at around 24 h (at 23°C) and 72 h (at 4°C) (Fig. 5A,B). The high levels of RGL2 transcripts are maintained if seeds are kept at 4°C (Fig. 5A, 168 h). When seeds imbibed at 4°C for 168 h are moved to 23°C, they germinate rapidly, with seedling emergence completed, and with hypocotyl and radicle elongation and cotyledon expansion clearly in progress within 48 h (data not shown). During this time, RGL2 transcript levels fall rapidly, and RGL2 transcripts are barely detectable 48 h following the move to 23°C (Fig. 5A).
Figure 5.
RGL2 expression in seeds is imbibition inducible. (A) RNA gel-blot analysis of RGL2 expression in wild-type seeds at different stages of imbibition and germination. RGL2 transcripts first become detectable at 24 h after the onset of imbibition at 4°C, then rise to a peak that is maintained for as long as the seeds are kept at 4°C. When wild-type seeds are subsequently moved to 23°C, RGL2 transcript levels begin to fall rapidly, and are barely detectable 48 h after the seeds were moved into the warmth, by which time germination had been completed. (B) RGL2 expression in wild-type seeds is also induced by imbibition at 23°C. Transcript levels rise more rapidly than they did at 4°C, with high levels being achieved by 24 h. (C) RGL2 transcripts remain at high levels in ga1-3 mutant seeds. Unlike RGL2 transcript levels in wild-type seeds, RGL2 transcript levels remained high in ga1-3 seeds following transfer from 4 to 23°C. (D) RGL2 expression is down-regulated in ga1-3 seeds grown at 23°C and treated with GA (10−4 M GA3). (E) In situ GUS expression patterns in seeds of rgl2-5 heterozygotes during seed germination. GUS staining is observed in the radicle of seeds that have been imbibed at 4°C (seed coat was removed after staining for observation). Following transfer to 23°C, GUS staining rapidly disappears, being still faintly detectable in the radicle at 24 h and barely detectable at 48 h, by which time germination is relatively advanced. In A, C, D, and E, seeds were chilled at 4°C for 7 d and then moved to a 23°C growth chamber with a 24 hr photoperiod.
The Ds insertion in the rgl2 alleles contains an intact but promoterless GUS reporter gene, with intron sequences and three alternative splice donor and acceptor sequences fused upstream of the GUS ATG codon (Sundaresan et al. 1995). If Ds is inserted in a transcribed region (5′-noncoding, intron, or exon), and providing that the GUS gene is in the correct orientation, a GUS fusion under control of the chromosomal gene promoter will result. Thus, GUS-staining patterns conferred by alleles containing such insertions can serve as markers for studying the tissue specificity of gene expression. The Ds insertion in rgl2-5 is between bases encoding G28 and A29 of RGL2 (Fig. 2B), and the GUS reporter gene is in the correct orientation. In addition, a 3.3-kb DNA fragment upstream of the start ATG in rgl2-5 was cloned via PCR. The DNA sequence of this fragment is identical to that obtained from a wild-type control (data not shown). Because we had already shown that the RGL2-coding region together with this same 3.3 kb upstream DNA sequence is sufficient to restore a RGL2 germination phenotype to rgl2-1 (Fig. 3B), we assumed that the RGL2 promoter in rgl2-5 is still fully functional. This is further supported by the detection of GUS fusion transcripts in rgl2-5 when a probe derived from GUS was used in RNA gel-blot experiments (Fig. 2B). In situ GUS staining of germinating seedlings heterozygous for rgl2-5 revealed staining in the pre-emergent radicle (embryonic axis; Bewley 1997) of seeds that had been imbibed for 168 h at 4°C (Fig. 5E, 0 h). This staining disappeared rapidly and was faintly detectable 24 h (Fig. 5E, 24 h), and barely detectable 48 h after the seeds were moved to 23°C (Fig. 5E, 48 h).
Imbibed ga1-3 mutant seeds maintain a high level of RGL2 transcripts
As shown above, rgl2-1 suppresses the nongerminating phenotype of ga1-3. We therefore reasoned that RGL2, as a repressor of seed germination, might be present at high levels in imbibed ga1-3 seeds, thus preventing their germination. To test this hypothesis, we assayed RGL2 transcript levels in ga1-3 mutant seeds. As for wild-type seeds, RGL2 transcript levels are relatively low in dry ga1-3 seeds, accumulate to much higher levels upon imbibition, and stay at high levels while imbibed seeds are maintained at 4°C (Fig. 5C). However, RGL2 transcript levels do not decline following transfer of ga1-3 seeds from 4 to 23°C. In marked contrast to what is seen in wild-type seeds, RGL2 transcripts are still at high levels in ga1-3 seeds after 48 h at 23°C (Fig. 5C; refer to 5A for wild-type control). In further experiments, the effect of GA3 on RGL2 transcript levels in ga1-3 seeds was studied. No detectable differences were observed between RGL2 transcript levels in GA-treated and GA-untreated ga1-3 seeds imbibed for 72 h at 4°C (Fig. 5D). However, RGL2 transcript levels were dramatically reduced by GA treatment when imbibed ga1-3 seeds were transferred to 23°C (Fig. 5D). These results are consistent with our genetic analysis, and with the hypothesis that RGL2 likely functions as a repressor of seed germination. High levels of RGL2 transcripts are maintained in ga1-3 seeds, and reduced by GA treatments. Germination of normal seeds presumably requires down-regulation of RGL2 expression by GA.
RGL2 is highly expressed in young inflorescences
We also investigated the expression of RGL2 in stages of the plant life cycle following seed germination. RNA gel-blot hybridization was used to measure the steady-state levels of RGL2, GAI, and RGA transcripts in tissues including young flower buds, siliques, bolting stem, cauline leaves, rosette leaves, and roots. With the exception of siliques, in which both RGA and GAI were expressed at low, but detectable levels, RGA and GAI were ubiquitously expressed in all tissues tested, with young flower buds showing the highest levels of transcript accumulation (Fig. 6A). In contrast, RGL2 transcripts are only detectable in the inflorescence, with high levels in young flower buds and significant levels in siliques, but not detectable in leaves, bolting stems, or roots (Fig. 6A).
Figure 6.
RGL2 is highly expressed in the inflorescences of bolting plants. (A) RNA gel-blot analysis of RGL2, GAI, and RGA transcripts in different plant tissues (28 days old). (B) GUS staining was detected mainly in the inflorescences and siliques of bolting plants heterozygous for rgl2-5. No GUS activity was detected at the seedling stage from day 4 to 12 after sowing (data not shown). (C) GUS staining was displayed in almost all flower organs. (D) Close up of a silique showing GUS staining at the base of the young developing seeds.
Following the reasoning outlined above, we also studied in situ GUS staining patterns in rgl2-5 heterozygotes, beginning at the seedling stage and assaying at 2-day intervals until the plants were 40 days old. GUS expression was not detected in young seedlings (6–12 d after sowing; data not shown). At day 14, GUS staining began to appear in young flower buds (clearly visible by day 20, Fig. 6B), and is maintained in inflorescences, but is not detectable in stem, leaves, or roots of bolting plants (Fig. 6B, 24 and 28 d). Clearly, the GUS expression pattern in rgl2-5 is consistent with the results obtained via RNA gel-blot analysis of RGL2 expression (Fig. 6A).
In a more detailed analysis of the GUS staining patterns in floral organs of rgl2-5 heterozygotes, young flower buds were examined microscopically. GUS staining was observed in almost all floral organs with particularly strong staining in stamen filaments, the top of the style (just below the stigma), and sepals (Fig. 6C). In young siliques, GUS staining was observed at the base of the young developing seed (Fig. 6D).
Discussion
The GAI/RGA/RHT/d8/SLR1 proteins mediate GA signals in a wide range of vascular plants (Peng et al. 1997, 1999a; Silverstone et al. 1998; Ikeda et al. 2001). Studies of loss-of-function alleles of GAI and RGA in Arabidopsis have shown that GAI and RGA function additively as negative regulators of GA responses controlling stem elongation growth (Peng et al. 1997; Silverstone et al. 1997, 1998; Dill and Sun 2001; King et al, 2001). The conserved N-terminal regions I and II of GAI/RGA/RHT/d8/SLR1 have been shown to be crucial for mediating GA signals. Alterations in regions I and II confer greatly reduced responses to both exogenous and endogenous GA (Peng et al. 1997, 1999a; Ikeda et al. 2001; Dill et al. 2001). RGL1 and RGL2 are related in amino acid sequence to GAI/RGA/RHT/d8/SLR1 throughout their entire lengths. The high homology shared by RGL1, RGL2, and GAI/RGA in N-terminal regions I and II (Fig. 1) implies that RGL1 and RGL2 are also regulators of GA responses.
In this study, we report our analyses of loss-of-function alleles of RGL1 (rgl1-1) and RGL2 (rgl2-1, rgl2-5, and rgl2-12). Under normal growth conditions, rgl1 and rgl2 mutant plants are indistinguishable from wild-type plants. However, rgl2 alleles confer resistance to the inhibitory effect of PAC on germination, whereas rgl1-1, gai-t6, and rga-t2 (loss-of-function alleles of RGL1, GAI, and RGA, respectively) do not. Furthermore, rgl2-1 suppresses the nongerminating phenotype of the GA-deficient ga1-3 mutant, whereas rgl1-1, gai-t6, or rga-t2 alone do not. Even the combined absence of GAI and RGA (in ga1-3 gai-t6 rga-t2) fails to suppress the nongerminating phenotype of ga1-3, as reported previously (Dill and Sun 2001; King et al. 2001). These observations suggest that RGL2 plays a major role in mediating the GA-regulated control of seed germination, whereas RGL1, GAI, and RGA play relatively minor roles.
On the other hand, rgl2-1 does not confer obvious resistance to the inhibitory effects of PAC on rosette leaf expansion and stem elongation. gai-t6 and rga-t2 mutants have significantly larger rosettes and display longer floral bolt stems than wild-type control plants when grown on medium containing PAC. rgl2-1 also fails to suppress the leaf expansion and stem elongation phenotypes of ga1-3. rgl2-1 ga1-3 double mutant plants are indistinguishable from the ga1-3 single mutant. This is in marked contrast to the significant suppression of the ga1-3 dwarfing phenotype conferred by rga alleles and to the complete suppression of the ga1-3 dwarfing phenotype conferred by the gai-t6 rga-24 double knockout (Dill and Sun 2001; King et al. 2001).
Taken together, our results show that loss-of-function mutations in RGL2 suppress the seed germination, but not the leaf expansion and stem elongation components of the GA-deficient phenotype conferred either by treatments with PAC, or by ga1-3. Our observations indicate that RGL2 (but not RGL1, GAI, or RGA) acts as a negative regulator of GA-promoted seed germination, whereas GAI and RGA act as negative regulators of GA-promoted stem elongation and leaf expansion.
After they have been stored for a while (normally >1 mo post harvesting), wild-type seeds will germinate ∼3 d following imbibition at room temperature and ∼2 wk following imbibition at 4°C (H. Cheng and J. Peng, unpubl.; Koornneef and Karssen 1994). However, seeds of the severely GA-deficient mutant ga1-3 do not germinate in either of these conditions, implying that GA is necessary for germination to proceed (Koornneef and van der Veen 1980; this work). Here, we have shown that RGL2 transcripts accumulate following imbibition of both wild-type and ga1-3 mutant seeds, either at 23 or 4°C. However, whereas RGL2 transcript levels decline rapidly in wild-type seeds during germination, high levels of RGL2 transcripts are maintained in the nongerminating ga1-3 seeds during the same period of time. These observations are significant and have three important implications. First, the expression of RGL2 is imbibition inducible. This induction may be a direct effect of imbibition, or it may simply be that the hydration of seeds sets in motion a developmental program, of which RGL2 up-regulation is a part. Second, down-regulation of RGL2 transcript levels is regulated via GA. Third, because loss-of-function RGL2 alleles suppress the nongerminating phenotype of ga1-3, it is likely that the elevated levels of RGL2 transcripts in ga1-3 prevent seed germination.
Previous analyses of GAI and RGA function have led to the concept that these proteins operate as repressors of GA responses, and that their repressive activities are opposed by GA (the GA-derepressible repressor hypothesis; Peng et al. 1997, 1999a; Harberd et al. 1998; Dill et al. 2001; Fu et al. 2001; King et al. 2001; Richards et al. 2001). Here, we have shown that RGL2 represses germination in the absence of GA (in ga1-3 or in the presence of PAC), and that this repression is released by GA. Thus, RGL2 operates as a GA-derepressible repressor of seed germination. RNA gel-blot analyses have indicated that GA treatments have little effect on the abundance of GAI and RGA transcripts (Silverstone et al. 1998), However, recent experiments have shown that GA treatments cause rapid disappearance of the RGA protein from the nuclei of plant cells. One interpretation of these observations is that GA counteracts the repressive effects of RGA by targeting the protein for destruction, rather than by altering the levels of transcripts that encode it (Dill et al. 2001; Silverstone et al. 2001). Although it is possible that GA also alters the stability of RGL2, we have shown here that GA causes a rapid decline in the levels of RGL2 transcripts during seed germination. Thus, in the case of RGL2, GA counteracts RGL2-mediated repression of seed germination by down-regulating the abundance of RGL2-encoding transcripts. It is possible that there are several different mechanisms by which GA can counteract the action of the GAI/RGA family of GA-derepressible repressors.
Seed germination is complete when embryo growth overcomes the mechanical restraint imposed by the testa, and the radicle protrudes from the seed coat (Koornneef and Karssen 1994; Bewley 1997). Bioactive GAs promote seed germination by inducing the expression of genes encoding enzymes that hydrolyze the endosperm and loosen cell walls, thus facilitating the protrusion of the radicle (Bewley 1997). As discussed above, RGL2 is an imbibition-inducible GA-repressible negative regulator of seed germination. However, the exact mechanism of RGL2 function is still not clear. The GAI/RGA/RHT/d8/SLR1 proteins are putative transcription factors (Ikeda et al. 2001; Richards et al. 2001). RGL2 may therefore act as a negative regulator that represses the expression of genes encoding hydrolyzing enzymes, such that this repression can be released by GA. Alternatively, RGL2 may act as negative regulator that prevents cell expansion by other means. We have shown, via in situ GUS staining, that RGL2 is expressed early during germination, and especially in the pre-emergent radicle. It is therefore reasonable to suggest that RGL2 functions as a restraint on radicle cell expansion, a restraint that can be counteracted by GA.
rgl2-1 strongly suppresses the nongerminating phenotype of ga1-3. However, seed germination can be rendered even more GA independent, because germination of the gai spy-7 gar2-1 triple mutant is even more resistant to PAC than is rgl2-1. Thus, GA-response mediators additional to RGL2 are also involved in controlling seed germination. On the basis of sequence alignments, RGL3 (68% identity to RGL2) is a likely candidate. Previous reports have shown that, whereas the absence of GAI or RGA alone can partially suppresses GA-deficiency phenotypes (Peng et al. 1997; Silverstone et al. 1998), it is necessary to remove both GAI and RGA to achieve full suppression of the dwarfed stem phenotype of ga1-3 (Dill and Sun 2001; King et al. 2001). RGL2 and RGL3 might be coupled in a similar way in the control of seed germination. Analysis of the effects of combined loss-of-function alleles of both RGL2 and RGL3 on ga1-3 seed germination would allow us an improved understanding of the relative roles of RGL2 and RGL3.
The absence of an obvious role in controlling stem elongation for RGL2 is consistent with RGL2 expression patterns in the adult plant. RNA gel-blot analysis and in situ GUS staining showed that RGL2 is specifically expressed in the young inflorescence and is not detectable in expanding leaves and internodes. GAI and RGA are ubiquitously expressed in all of these tissues, and are known to play key roles in internode and leaf expansion. RGL2, GAI, and RGA are all highly expressed in inflorescences. Intriguingly, ga1-3 mutants exhibit clear floral phenotypes, with retarded stamen and petal development, suggesting an important role for GA signaling in the regulation of floral organ growth. However, loss of GAI and RGA, either singly or in combination, does not suppress the floral organ phenotypes of ga1-3 (Dill and Sun 2001; King et al. 2001). Loss of RGL2 or RGL1, in a ga1-3 rgl2-1 or ga1-3 rgl1-1 mutant, also fails to suppress the ga1-3 floral phenotype (J. Peng, unpubl.). These observations suggest either that there is substantial redundancy of function between RGL2, RGL1, GAI, and RGA in the regulation of floral organ growth, or that these processes are largely regulated by RGL3. Analysis of ga1-3 gai-t6 rga-t2 rgl2-1 mutant plants in the future will enable us to understand more fully the relative roles of RGL2, GAI, and RGA in regulating the growth of stamens and petals.
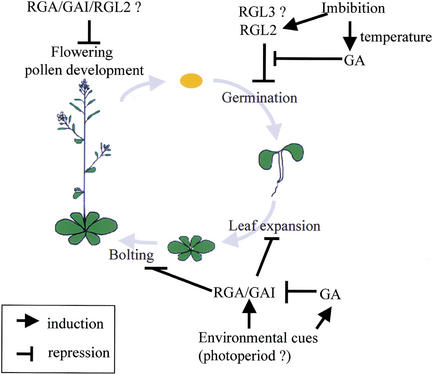
The Arabidopsis genome contains five members (GAI, RGA, RGL1, RGL2, and RGL3) of the GAI/RGA gene family (Dill and Sun 2001). Genetic and physiological studies of mutant alleles of four of these genes (RGL1, RGL2, GAI, and RGA) have shown, perhaps not surprisingly, that they act, sometimes alone, sometimes collectively, to control different plant developmental processes. Figure 7 provides a summary of our current understanding of the relative roles of the proteins encoded by RGL2, GAI, and RGA. Signaling through RGL2 is the main pathway for the GA-mediated control of seed germination. Signaling through GAI/RGA is the main pathway for GA-mediated control of stem elongation and leaf expansion. It is also possible that RGL2, GAI, and RGA are redundantly involved in the regulation of floral organ growth.
Figure 7.
Model outlining the roles of GAI/RGA-like GA-signaling components in the plant life cycle. In this study, we have shown that RGL2 links an environmental cue (moisture) with the GA-signaling pathway during the regulation of seed germination. Sequence similarity suggests that RGL3 may also regulate seed germination in response to GA. Signaling through GAI and RGA mediates GA-promoted stem elongation and leaf expansion (Peng et al 1997, 1999a; Silverstone et al. 1998; Dill and Sun 2001; King et al. 2001; this work). Signaling through GAI, RGA, and RGL2 may mediate GA-promoted flower and pollen development (Dill and Sun 2001; S.C. Lee, H. Cheng, and J. Peng, unpubl.). The activities of GAI, RGA, and RGL1 may well be modulated in response to environmental variables at stages other than seed germination, and possible variables are shown.
Plant growth hormones, such as GA, are often held to be endogenous factors that regulate plant growth and development in response to environmental change. How the GA-signaling system interacts with environmental signals is likely to be an important topic of research in the coming years. Recent observations have identified PHOR1, a possible link between photoperiod sensing and GA signaling in plants (Amador et al. 2001). Here, we have shown that, while absorbing water from the environment in the process of hydrating, Arabidopsis seeds accumulate relatively high levels of RGL2-encoding transcripts. RGL2 appears to act as an inhibitor of seed germination, and GA promotes germination by counteracting RGL2.
Materials and methods
Genetic nomenclature and plant materials
In this study, genotypes are written in italics; the wild-type genotype is in capitals (e.g., RGL2), and the mutant genotype is in lowercase letters (e.g., rgl2-1). The polypeptide products of genes are written in nonitalic capitals (e.g., RGL2).
All mutants described here were derived from Landsberg erecta (wild type). Single mutant (gai-t6 and ga1-3), double mutant (gai-t6 ga1-3; rga-24 ga1-3), and triple mutant (gai spy-7 gar2-1; gai-t6 rga-24 ga1-3) lines were obtained as described previously (Peng et al. 1997; Silverstone et al. 1998; King et al. 2001). Five Ds-insertion lines (rgl1-1, rgl2-1, rgl2-5, rgl2-12, and rga-t2) were obtained from a previously described Ds-tagging population (Sundaresan et al. 1995; Parinov et al. 1999). During segregation analysis, a semidwarfing mutation was found to be closely linked (∼2.3 cM; data not shown) to rgl2-1 and rgl2-5. This semidwarfing mutation was separated from rgl2-1 and rlg2-5 via genetic recombination, and rgl2-1, rgl2-5, rgl2-12, rgl1-1, and rga-t2 were then backcrossed twice with wild type to further purify the genetic background. These backcrossed lines were used for all experiments described in this work. Double mutants (ga1-3 rgl2-1, ga1-3 rgl1-1, and ga1-3 rga-t2) were obtained from crosses between the relevant single mutant and ga1-3. The triple mutant gai-t6 rga-t2 ga1-3 was obtained from a gai-t6 ga1-3 × rga-t2 ga1-3 cross.
For plants grown in soil, seeds were allowed to imbibe on water-moistened filter paper (in the case of ga1-3, using GA3 solution-moistened filter paper) at 4°C for 7 d (to break dormancy), and then planted on Arabidospsis mix (three parts Florobella potting compost/1 sand). The plants were then grown in a growth room (16 h light/8 h darkness photoperiod, 20–23°C), or in a greenhouse. For germination tests, seeds (stored more than 1 mo) were surface sterilized and sown on SM or GM medium supplemented (where appropriate) with GA3 or PAC. The seeds were then chilled in a cold room for 7 d. Germination was recorded 8 d after the plates were moved to the above-mentioned growth room.
To confirm the exact locations of the Ds-insertions in rgl1-1, rga-t2, rgl2-1, rgl2-5, and rgl2-12, DNA was amplified from genomic DNA using primers flanking the Ds region, and the products were cloned in the pGEMTeasy (Promega) vector and sequenced. For rga-t2, primers 906F (5′-GCCGGAGCTATGA GAAAAGTGG-3′) and DS3-2 (5′-CCGGTATATCCCGTTT TCG-3′) were used to amplify DNA adjacent to the 3′ end of the Ds, and primers 2076R (5′-AAGAATTTTAAACAAGTGA ACG-3′) and DS5-3 (5′-CGGTCGGTACGGGATTTTCC-3′) were used to amplify sequences adjacent to the 5′ end of Ds. Primers 906F and 2076R were derived from RGA sequence (Peng et al. 1997; Silverstone et al. 1998) and DS3-2 and DS5-3 were derived from Ds sequence (Parinov et al. 1999). The Ds insertion in rga-t2 was located at 1521 nucleotides from the ATG start codon of RGA. For rgl1-1, primers DS3-2 and 2295R (5′-CCACAGAGCGCGTAGAGGATAAC-3′) were used to amplify DNA adjacent to the 3′ end of the Ds, and primers Ds5-P1 (5′-CATGGGCTGGGCCTCAGTG-3′) and 1670F (5′-AAGC TAGCTCGAAACCCAAAT-3′) were used to amplify DNA adjacent to the 5′ end of Ds. Primers 2295R and 1670F were derived from RGL1 sequence and Ds5-P1 is derived from Ds sequence. The Ds insertion in rgl1-1 was located at 68 nucleotides upstream of the ATG start codon of RGL1. For rgl2-1, primers 856F (5′-GCTGGTGAAACGCGTGGGAACA-3′) and DS3-2 were used to amplify DNA fragments adjacent to the 3′ end of Ds, whereas 1883R (5′-ACGCCGAGGTTGTGATGAGTG-3′) and DS5-3 were used to amplify DNA fragments adjacent to the 5′ end of Ds. For rgl2-5, primers 78F (5′-GTAACCAAATCA CAACAAAGA-3′) and DS3-2 were used to obtain the sequence adjacent to the 3′ end of Ds, whereas 700R (5′-GCTGC TAGCTTCCTCGTCAAA-3′) and DS5-3 were used to amplify DNA adjacent to the 5′ end of Ds. For rgl2-12, primer 1355F (5′-TTCGAAACCCGACCC-3′) and DS3-2 were used to obtain the sequence adjacent to the 3′ end of Ds, whereas RGL2(FL)Rv (5′-TCAGGCGAGTTTCCACGCCGAGGTT-3′) and DS5-3 were used to amplify DNA adjacent to the 5′ end of Ds. Primer sequences 856F, 1883R, 78F, 700R, 1355F, and RGL2(FL)Rv were all derived from the RGL2 sequence. For the rgl2 mutant alleles, Ds was inserted at 1141, 28, and 1459 nucleotides from the ATG start codon of RGL2 in rgl2-1, rgl2-5, and rgl2-12, respectively. The primers used to identify the ga1-3 mutation were as described (Silverstone et al. 1997).
RNA gel-blot hybridization, DNA sequencing
Total RNA was extracted from different tissues from 28-day-old plants using the Tri Reagent (Molecular Research Center) according to the manufacturer's protocols. RNA from seeds was extracted using the method of Vicient and Delseny (1999) and RNA from siliques was extracted following the method of Bekesiova et al. (1999). DNA–RNA hybridizations were carried out using procedures that are standard for the DIG system, as advised by the manufacturer (Roche). Gene-specific probes were used for the RNA gel-blot hybridizations as follows: RGL2 (−165–142 nucleotides), RGA (−97–402 nucleotides), GAI (−150–219 nucleotides) probes were labeled by use of the PCR DIG probe synthesis kit (Roche). DNA sequences were determined by use of the Big Dye terminator cycle sequencing kit (Perkin Elmer).
Plant transformation and histochemical localization of β-glucuronidase (GUS) activity
An NdeI DNA fragment of 6.95 kb was obtained from the BAC clone T21P5 (Arabidopsis Biological Resource Center). This region encompasses the RGL2 ORF, 3.3 Kb upstream sequence, and 2.0 kb downstream sequence. The DNA fragment was cloned into the binary vector pCAMBIA 1300 (CAMBIA) and transformed into Agrobacterium EHA105. The construct was transformed into rgl2-1 mutant plants following the method of Clough and Bent (1998).
In situ GUS staining was performed using the method of Jefferson (1987). Whole plants were transferred to microfuge tubes or 15-mL falcon tubes and incubated at 37°C overnight in GUS-staining solution containing 100 mM Na Phosphate buffer (pH 7.0), 10 mM EDTA, 0.1% Triton X-100, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, and 1 mg/mL X-Gluc (Biosynth AG). Imbibed seeds were vacuum infiltrated together with GUS staining buffer for 2 d. The staining solution was then removed and replaced with several changes of 70% ethanol.
Acknowledgments
We thank Venkatesan Sundaresan for his generosity of providing tagging lines rgl1-1, rga-t2, rgl2-1, rgl-5, and rgl-12, which made everything possible. We thank De Ye, Weicai Yang, and Sergey Parinov for advice on techniques and useful discussions and Daoxin Xie, Zilong Wen, Alvin Eun, and Chen Jun for helpful discussions. This work is financially supported by National Science and Technology Board (NSTB) in Singapore. The work of K.K. and N.P.H. was supported by the Core Strategic Grant from the BBSRC to the John Innes Centre, and by BBSRC grant 208/P15108 to NPH. BAC clone T21P5 was obtained from Arabidopsis Biological Resource Center.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pengjr@ima.org.sg; FAX 65-8727007.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.969002.
References
- Amador V, Monte E, García-Martínez J-L, Prat S. Gibberellin signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell. 2001;106:343–354. doi: 10.1016/s0092-8674(01)00445-7. [DOI] [PubMed] [Google Scholar]
- Bekesiova I, Nap J-P, Mlynarova L. Isolation of high quality DNA and RNA from leaves of the carnivorous plant Drosera rotundifolia. Plant Mol Biol Reporter. 1999;17:269–277. [Google Scholar]
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua NH. PAT1, a new member of the RGAS family, is involved in phytochrome A signal transduction. Genes & Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organizatin of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T-P. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159:777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Jung H-S, Sun T-P. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreucq B, Berger N, Vincent E, Boisson M, Pelletier G, Caboche M, Lepiniec L. The Arabidopsis AtEPR1 extensin-like gene is specifically expressed in endosperm during seed germination. Plant J. 2000;23:643–652. doi: 10.1046/j.1365-313x.2000.00829.x. [DOI] [PubMed] [Google Scholar]
- Fu X, Sudhakar D, Peng J, Richards DE, Christou P, Harberd NP. Expression of Arabidopsis GAI in transgenic rice represses multiple gibberellin responses. Plant Cell. 2001;13:1791–1802. doi: 10.1105/TPC.010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd NP, King KE, Carol P, Cowling RJ, Peng JR, Richards DE. Gibberellin: inhibitor of an inhibitor of…? BioEssays. 1998;20:1001–1008. doi: 10.1002/(SICI)1521-1878(199812)20:12<1001::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Hedden P, Graebe JE. Inhibition of gibberellin biosynthesis by paclobutrazol in cell-free homogenates of Cucurbita maxima endosperm and Malus pumila embryos. J Plant Growth Regu. 1985;4:111–122. [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser M-T, Benfey PN. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopetide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- King KE, Moritz T, Harberd NP. Gibberellins are not required for stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics. 2001;159:767–776. doi: 10.1093/genetics/159.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Karssen CM. Seed dormancy and germination. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 313–334. [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61:377–383. [Google Scholar]
- Leubner-Metzger G, Fründt C, Vögeli-Lange R, Meins F., Jr Class 1 β-1,3-glucanase in the endosperm of tobacco during germination. Plant Physiol. 1995;109:751–759. doi: 10.1104/pp.109.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR. Genetic control and integration of maturation and germination pathways in seed development. Ann Rev Plant Physiol Pl Mol Biol. 1995;46:71–93. [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang W-C, Kumaran M, Sundaresan V. Analysis of flanking sequences from dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell. 1999;11:2263–2270. doi: 10.1105/tpc.11.12.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JR, Harberd NP. Derivative alleles of the Arabidopsis gibberellin-insensitive (gai) mutation confer a wild-type phenotype. Plant Cell. 1993;5:351–360. doi: 10.1105/tpc.5.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JR, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes & Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JR, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. “Green Revolution” genes encode mutant gibberellin response modulators. Nature. 1999a;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Peng JR, Richards DE, Moritz T, Cano-Delgado A, Harberd NP. Extragenic suppressors of the Arabidopsis gai mutation alter the dose-response relationship of diverse gibberellin responses. Plant Physiol. 1999b;119:1199–1207. doi: 10.1104/pp.119.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J.R., Richards, D.E., Moritz, T., Ezura, H., Carol, P., and Harberd, N.P. 2002. Molecular and physiological characterization of Arabidopsis GAI alleles obtained in targeted Ds-tagging experiments. Planta (published on-line: DOI 10.1007/s004250100643) (in press). [DOI] [PubMed]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Richards DE, Peng JR, Harberd NP. Plant GRAS and metazoan STATs: One family? Bioassays. 2000;22:573–577. doi: 10.1002/(SICI)1521-1878(200006)22:6<573::AID-BIES10>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-ali T, Harberd NP. How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Ann Rev Plant Physiol Pl Mol Biol. 2001;52:67–88. doi: 10.1146/annurev.arplant.52.1.67. [DOI] [PubMed] [Google Scholar]
- Sánchez RA, de Miguel L. Phytochrome promotion of mannan-degrading enzymes in the mycropylar endosperm of Datura ferox seeds requires the presence of the embryo and gibberellin synthesis. Seed Sci Res. 1997;7:27–33. [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci. 1999;96:290–295. doi: 10.1073/pnas.96.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Martinez EC, Sun TP. The RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics. 1997;146:1087–1099. doi: 10.1093/genetics/146.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun TP. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung H-S, Dill A, Kawaide H, Kamiya Y, Sun T-P. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1565. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, Cooney SE, McCourt P. Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics. 1998;149:509–521. doi: 10.1093/genetics/149.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. Gibberellin signal transduction. Curr Opin Plant Biol. 2000;3:374–380. doi: 10.1016/s1369-5266(00)00099-6. [DOI] [PubMed] [Google Scholar]
- Sun TP, Kamiya K. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Goodman HM, Ausubel FM. Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell. 1992;4:119–128. doi: 10.1105/tpc.4.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Ma H, Martienssen R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes & Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- Thornton T, Swain SM, Olszewski N. Gibberellin signal transduction presents: The SPY who O-GlcNAc'd me. Trends Plant Sci. 1999;4:424–428. doi: 10.1016/s1360-1385(99)01485-5. [DOI] [PubMed] [Google Scholar]
- Vicient CM, Delseny M. Isolation of total RNA from Arabidopsis thaliana seeds. Analyt Biochem. 1999;268:412–413. doi: 10.1006/abio.1998.3045. [DOI] [PubMed] [Google Scholar]