Abstract
Translational studies using genomic techniques in cardiovascular diseases are still in their infancy. Access to disease-associated cardiovascular tissues from patients has been a major impediment to progress in contrast to the diagnostic advances made by oncologists using gene expression on readily available tumor samples. Nonetheless, progress is being made for atherosclerosis by carefully designed experiments using diseased tissue or surrogate specimens. This review details the rationale and findings of a study using freshly isolated blood mononuclear cells from patients undergoing carotid endarterectomy due to atherosclerotic stenosis and from matched normal subjects. Using this cardiovascular tissue surrogate, the mRNA levels of the Finkel-Biskis-Jinkins osteosarcoma (FOS) gene in circulating monocytes were found to correlate with atherosclerosis severity in patients, and with HMG CoA reductase inhibitor (statin) therapy in normal subjects. The major finding of this investigation is discussed in relation to observations from other human atherosclerosis gene expression studies. These distinct studies converge to demonstrate the unequivocal importance of inflammation in atherosclerosis. Although the clinical utility of the specific findings remains open, the identification of similar genes by different investigations serves to validate their reports. They also provide us with insights into pathogenesis that may impact future translational applications.
Diagnostic markers of inflammation and atherosclerosis
Based on our current understanding of atherosclerosis, the pathogenesis of this leading cause of morbidity and mortality in westernized societies is best described by chronic vascular inflammation that starts early in life and exacerbates with aging (Glass and Witztum 2001, Hansson 2005). Multiple hereditary and environmental factors contribute to the initiation and progression of this disease (Lusis 2000). However, some well established clinical risk factors such as low density lipoprotein (LDL) and the metabolic syndrome (including obesity and insulin-resistance, diabetes) and basic factors such as angiotensin II all appear to promote oxidative stress and inflammation (Daugherty and Cassis 2004, Glass and Witztum 2001, Libby 2002). Though a number of molecules linked to inflammation have been proposed for use as atherosclerosis markers, C-reactive protein (CRP) has gained prominence because of its association with increased cardiovascular risk in clinical trials (Jain and Ridker 2005). Revealing insights into statins' mechanism of action, the recent clinical trial PROVE IT-TIMI 22 separated the cholesterol lowering and anti-inflammatory effects of statin therapy by demonstrating the independent and additive predictive values of serum cholesterol and CRP levels on cardiovascular events, respectively (Jain and Ridker 2005, Ridker et al. 2005). A question that follows such fundamental observations is whether there exist other predictive markers that have equal or greater sensitivity and specificity.
The issue of tissue samples
Molecular oncology is pioneering the translational use of genomic data. Gene expression profiling has been used for the molecular diagnosis and prognosis of various human cancers including hematologic and breast malignancies (Ebert and Golub 2004, van de Vijver et al. 2002). In parallel, cardiovascular genomic studies have examined the transcriptional profiles of cardiomyopathies and their response to treatment using explanted and biopsied myocardium, respectively (Hwang et al. 2002, Lowes et al. 2002). However, the validation and application of these studies for routine practice have been limited by lack of access to patient tissue samples.
The routine examination of cardiac and vascular biopsies by translational investigators is difficult to justify in the light of potential complications and unclear benefits. Furthermore, tissue heterogeneity and localization of the disease may decrease the sensitivity and specificity of the gene expression changes. Unlike the controlled surgical excision of cancers with normal tissue margins, obtaining diseased cardiovascular tissues with adjacent normal structures are often not an option. Thus, the difficulty in obtaining appropriate human samples represents a significant barrier to the advancement of genomic cardiovascular research.
Blood as a tissue surrogate
As an alternative, blood is an easy sample to obtain and can be used for helping diagnose disease processes. This concept has received increasing interest by those examining diverse disorders including autoimmunity, asthma, osteoporosis, Huntington's disease and breast cancer (Borovecki et al. 2005, Hakonarson et al. 2005, Liu et al. 2005, Olsen et al. 2004, Sharma et al. 2005). Blood is a particularly fitting surrogate for atherosclerotic tissue, because it contains the inflammatory cells such as monocytes that are involved in its pathogenesis (Hansson 2005). However, overlapping explanations for changes in monocyte gene expression include concurrent atherosclerosis-independent processes or reaction to diseased vessels thereby serving as a “reporter” (Figure 1A). Blood cells also have well defined cell surface markers such as the CD14 antigen on monocytes to facilitate their purification, improving gene expression sensitivity and specificity (Figure 1B). Finally, it is critical to have in vitro models for testing gene function to gain insights into pathogenesis. In this regard, well described systems of primary cultures and established lines of human monocytic cells are available for examining these inflammatory cells.
Figure 1.
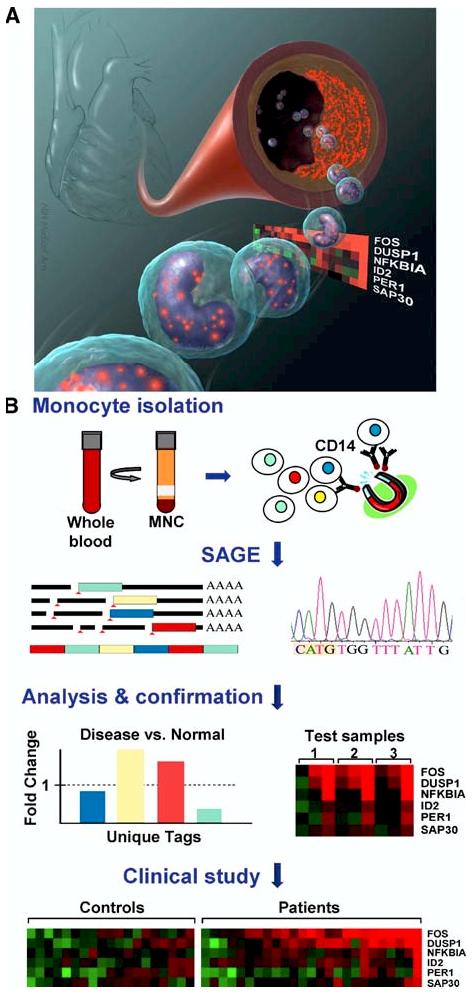
Circulating transcriptomes reveal makers of atherosclerosis. A) Illustration depicting the concept of genes by circulating cells in blood directly or indirectly reflecting atherosclerotic heart vessels. The red speckles represent the products of genes that may be important in heart attacks. By non-invasively querying circulating cells in patients with atherosclerosis, FOS gene was identified out of six candidates as being highly expressed in those with more extensive disease (color-coded panel: higher levels, red; lower levels, green). B) Schematic outline of purifying CD14+ monocytes with magnetic bead-conjugated antibodies from Disease and Normal blood samples followed by serial analysis of gene expression (SAGE) to identify atherosclerosis marker (Disease) genes. These are then confirmed by RT-PCR using known test samples before examining larger groups of subjects in a clinical study.
Genomic approach and technical considerations
Rather than investigate the levels of known inflammatory markers, we have recently explored the feasibility of using unbiased genomic techniques applied to in vivo human cells to identify disease markers and mediators. There are several well-established gene expression techniques for evaluating differences in gene expression (Fryer et al. 2002). However, the relative merits of the specific techniques have shifted with advancements in human genome annotation, marked improvements in gene expression technology, and decreased costs of high throughput experiments. Microarrays in particular have gained widespread popularity in translational medical research, both in oncology as referenced above and in cardiovascular medicine (Cook and Rosenzweig 2002).
For our studies, we used the Serial Analysis of Gene Expression technique (SAGE) for its ease of data analyses and comparisons to previous or future SAGE library databases (Patino et al. 2002). Facile comparisons, internet-based exchange of quantitative transcript data without need for normalization and ability to “re-mine” SAGE databases with progressive improvements in genomic annotation continue to remain key attributes of this technique. For more details on its principles and methodology, the reader is referred to selected reviews (Harbers and Carninci 2005, Polyak and Riggins 2001) and resource websites (www.sagenet.org; www.ncbi.nlm.nih.gov/SAGE/). Additionally, there was an impetus to pursue in vivo disease studies using this technique as previously published in vitro human monocyte SAGE transcriptomes revealed promising results (Hashimoto et al. 1999).
Differentially expressed monocyte genes in atherosclerosis
Owing to the high resource requirements of the SAGE technique, we first performed global gene expression studies on a limited number of subjects to identify candidate marker genes followed by confirmation with RT-PCR in larger numbers of subjects (Figure 1B). Patients with atherosclerosis were selected from a pool of eligible subjects undergoing carotid artery endarterectomy and were matched closely with normal volunteers (Patino et al. 2005). We created SAGE libraries using both CD14+ monocytes purified from blood mononuclear cells (MNC) to increase sensitivity and CD14-depleted MNCs to enable in silico selection of monocytes-specific genes. Though positive selection using antibodies against CD14 antigen efficiently separated this specific subpopulation of cells, it has the potential to stimulate or alter monocyte gene expression. We optimized the purification scheme to minimize changes, but more importantly, we measured the candidate monocyte gene mRNA levels in the purified and starting mononuclear fractions to confirm that they were similar as described in the next section.
The correlation between our SAGE transcriptomes and previously published in vitro data was low, emphasizing the importance of examining in vivo cells (Hashimoto et al. 1999, Patino et al. 2005). We found that the top two genes differentially expressed between patient and control monocyte libraries were FOS, a proto-oncogene known to be involved in monocyte activation and differentiation, and DUSP1, a stress response phosphatase important for mitogen-activated protein kinase (MAPK) regulation (Farooq and Zhou 2004, Shaulian and Karin 2002).
FOS as reactive marker and mediator of atherosclerosis
We next took our SAGE findings obtained from relatively few subjects with or without atherosclerosis and asked whether the genes identified were helpful in discriminating disease states in a larger cohort of patients. To simplify sample preparation and to test whether the CD14 monocyte purification process may affect levels of FOS and DUSP1 transcripts, we compared the relative transcript levels in total unprocessed mononuclear cell (MNC, composed of ∼20% monocytes) and in CD14+ monocyte fractions by RT-PCR. The relative mRNA ratios between patient and controls (P/C) in both the MNC and CD14+ monocyte fractions were similar, indicating the feasibility of this approach and, importantly, confirming that FOS and DUSP1 expression were not altered by the monocyte purification process (Figure 2) (Patino et al., 2005).
Figure 2.
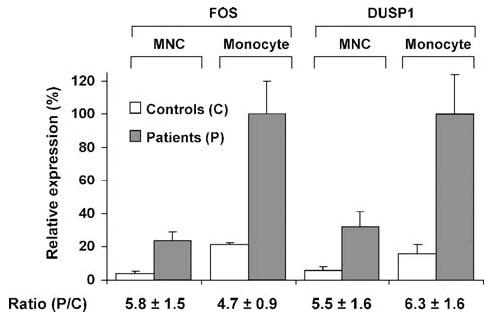
Finkel-Biskis-Jinkins osteosarcoma (FOS) and dual specificity phosphatase 1 (DUSP1) relative mRNA expression levels determined by RT-PCR. The fold change ratios in patients compared with controls [Ratio (P/C)] were preserved whether whole mononuclear cells (MNC) or purified monocytes (Monocyte) were used for RT-PCR. Values shown as mean ± SE, n = 6, for patients and controls. Reproduced with permission from Patino et al. 2005.
We compared the levels of FOS and DUSP1 mRNAs in MNCs to plasma CRP protein in 25 patients and 19 controls. We observed that both FOS and DUSP1 were more differentially expressed between patients and controls than CRP, a clinical marker of increased cardiac risk, and FOS appeared to identify patients with more extensive atherosclerosis with greater sensitivity and specificity than CRP (Patino et al. 2005). Among the control subjects those on statin treatment clustered together with lower levels of FOS, possibly reflecting the anti-inflammatory effects of statins. By immunohistochemistry we observed high expression and co-localization of FOS with macrophage CD14+ immunoreactivity in human carotid and coronary artery plaque suggesting its potential involvement in pathogenesis (Figure 3) (Patino et al. 2005). Higher levels of FOS have also been detected by RT-PCR in atherectomy samples obtained from acute coronary syndrome patients versus those with stable angina, giving additional clinical significance to our observation (Rossi et al. 2004).
Figure 3.
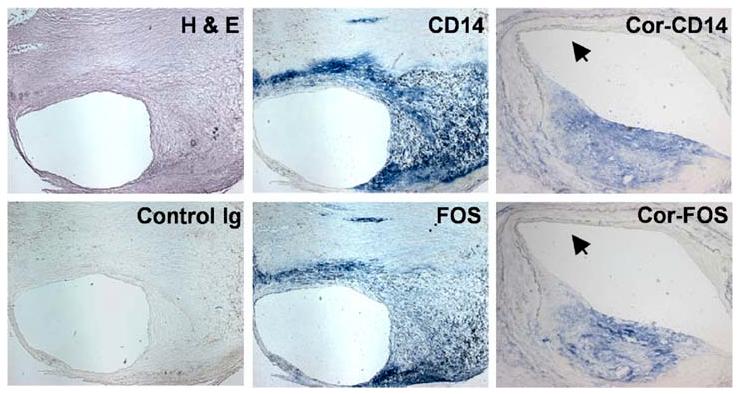
Expression of FOS in human carotid and coronary artery plaques. Fresh frozen sections of human carotid and coronary (Cor) artery plaques stained with hematoxylin and eosin (H&E), negative control immunoglobulin (Control Ig) and antibodies against CD14 or FOS. CD14+ staining of macrophages colocalized with FOS immunoreactivity. Similar colocalization of CD14 and FOS can be seen in the coronary artery plaque (Cor-CD14, Cor-FOS), while in the more “normal” appearing segment (black arrowhead) there is virtual absence of staining. (Carotid plaque, 25X magnification; coronary plaque, 50X magnification.) Reproduced with permission from Patino et al. 2005.
Concordance of transcriptional studies of human atherosclerosis using blood
Exploring the utility of cardiovascular disease gene expression studies for translational purposes has previously been examined. In particular, the concept of using blood gene expression as indicator of atherosclerosis has been explored by us and others as summarized in Table 1. Interestingly, the reported results and conclusions appear to be generally concordant amongst the studies helping validate the individual findings.
Table 1.
Blood mononuclear cell gene expression studies in human atherosclerosis
| Samples | Techniques | Findings | Reference |
|---|---|---|---|
| Purified CD14+ monocytes from CEA and normal subjects | SAGE, RT-PCR | FOS was the most differentially expressed gene in patients with atherosclerosis and may mediate pathogenesis | (Patino et al. 2005) |
| Blood MNCs from CEA patients and controls | Microarray: Affymetrix U133A, RT-PCR | EGR1 gene was identified as the most highly expressed marker in patients | (Gui et al. 2004) |
| Blood MNCs from acute ischemic stroke and normal subjects | Microarray: Affymetrix U133A, RT-PCR | A panel of differentially expressed genes including FOS was able to identify stroke subjects with 78% sensititivity and 80% specificity | (Moore et al. 2005) |
| Blood MNCs from hyperlipidemic patients treated with atorvastatin over 4 weeks | Microarray: Affymetrix U95Av2, RT-PCR | 31% of the statin responsive genes were observed at 36h, prior to any significant decrease in serum lipid levels | (Wibaut-Berlaimont et al. 2005) |
| Purified CD14+ monocytes from nondiabetic, well- or poorly-controlled diabetic subjects | RT-PCR | The degree of diabetic control and activated monocyte/macrophage marker gene expression were found to be correlated | (Cipolletta et al. 2005) |
| The CardioGene Study: blood MNCs from 350 consecutive patients undergoing coronary artery stent placement | Microarray: Affymetrix U133A | One of the genomic components of this multicenter study examines the relationship between MNC gene expression at pre-stent, 2 weeks and 6 months after stent placement and restenosis at 6 and 12 months | (Ganesh et al. 2004) |
Abbreviations: MNCs, mononuclear cells; RT-PCR, real-time polymerase chain reaction; CEA, carotid endarterectomy; FOS, Finkel-Biskis-Jinkins osteosarcoma; EGR1, early growth response gene 1
One of the studies examined gene expression in un-fractionated blood MNCs isolated from carotid endarterectomy patients and matched healthy subjects with the use of the Affymetrix genechip (Gui et al. 2004). Similar to observations in our study, EGR1, an early response transcriptional factor previously associated with atherosclerosis (McCaffrey et al. 2000), was one of the most up-regulated of the 267 genes identified in their screen. In retrospective analysis, EGR1 was also expressed at higher levels in our patient versus control monocytes though it was not significantly increased in our SAGE libraries (Patino et al. 2005).
In another study, gene expression in blood MNCs from ischemic stroke and control subjects revealed a panel of genes with 78% sensitivity and 80% specificity at correctly identifying stroke subjects (Moore et al. 2005). Though not all patients had documented atherosclerosis, coronary artery disease history and other cardiac risk factors were more prevalent in the group of elderly patients with stroke. In their list of stroke “classifier genes”, the monocyte activation genes identified included FOS, DUSP1, CD14 and the scavenger receptor CD36 (Moore et al. 2005, Patino et al. 2005).
Just as the inflammatory marker CRP has been shown to be responsive to statins in clinical trials, FOS expression by monocytes would also be expected to show treatment responses as suggested by our observations (Patino et al. 2005). Using a unique experimental design with blood mononuclear cells, 10 hyperlipidemic male subjects were initiated on 20 mg of atorvastin per day for a total of four weeks, with blood samples drawn after 12 hours, 36 hours, 1 week and 4 weeks of treatment(Wibaut-Berlaimont et al. 2005). Notably, the mRNA levels of 31% of the 240 genes that responded to statin treatment changed within 36 hours, well before any significant effect on lipid level. Both FOS and another member of the activating protein (AP-1) complex JUNB decreased during this early response period, possibly representing one of the molecular pathways mediating the anti-inflammatory effects of statins (Jain and Ridker 2005, Wibaut-Berlaimont et al. 2005).
Additional insights into disease pathogenesis can be gleaned by studying a condition that is associated with but distinct from the disease itself. Diabetes is considered to be a cardiac risk factor equivalent to atherosclerotic coronary artery disease. In a study examining the activation state of circulating monocytes, there was a significant correlation between the expression levels of macrophage differentiation markers and the state of diabetes control with medication, independently supporting the concept of monocyte gene activation in atherosclerosis (Cipolletta et al. 2005).
Taken together, the studies discussed identify similar genes that collectively reflect circulating monocyte activity, corresponding to atherosclerotic disease burden and response to therapeutic intervention. The concordant nature of the results likely reflects the strong effector role of monocytes in mediating inflammation. Not surprisingly, these molecular studies also mirror a number of prior clinical investigations demonstrating a simple association between peripheral monocyte count and subclinical atherosclerosis, post-myocardial infarction ventricular remodeling and in-stent restenosis (Chapman et al. 2004, Fukuda et al. 2004, Maekawa et al. 2002). Though we have simply summarized the findings reported by the studies in Table 1, it will be useful in the future to perform a more systematic and complete analysis of all the primary data. This could be accomplished by using web-based bioinformatics tools such as List of Lists-Annotated (LOLA) and L2L that allow the comparison of data from multiple microarray studies or even different genomic techniques such as SAGE (Cahan et al. 2005)(Newman and Weiner 2005).
Conclusions and future directions
The future role of genomic translational studies in the clinical management of patients remains to be seen, but the pathogenetic insights derived from these studies may be more directly applicable for devising preventive and therapeutic strategies against atherosclerosis. For example, molecularly assessing sub-clinical therapeutic responses in surrogate tissues from human have the potential to complement the current approaches taken in drug development. It may facilitate screening and focusing on the most promising approaches based on disease mechanism with less dependence on empirical trials.
With a more balanced view of human atherosclerosis gene expression studies, studying surrogates such as blood cells has constituted only a small part of the work done to advance genomic atherosclerosis knowledge. Carefully designed studies have revealed genes capable of identifying different stages of atherosclerotic disease in human aortas, the SNPs for which may be useful for genomic classification of patients with clinical implications (Seo et al. 2004). Extending this type of approach, combined results from microarray experiments using mouse atherosclerosis models and human coronary gene expression datasets have also identified a shared group of genes for classifying disease stage (Tabibiazar et al. 2005). These classifier genes were able to distinguish between native coronary artery disease and in-stent restenosis.
The CD14+ monocyte gene expression profiles that we have reported are likely to be altered by their local tissue environment. There are no published studies to date comparing circulating monocytes in atherosclerosis to plaque tissue macrophages within the same patient. This may be a fruitful area to investigate by purifying macrophages from atherosclerotic lesions such as carotid endarterectomy plaques to determine the relative fidelity of monocytes as tissue surrogate in atherosclerosis. Though we focused on the CD14+ monocyte fraction, genomic investigations using sub-populations of lymphocytes or granulocytes may be equally useful, as they are known to be important in the pathogenesis of atherosclerosis (Vanderlaan and Reardon 2005). In fact, even the CD14+ monocytes are composed of different sub-populations that when further fractionated, may improve gene expression sensitivity and provide new insights. Finally, by combining genomic information from other studies examining plaque macrophages or peripheral mononuclear cells in distinct immunologic processes such as cardiac allograft rejection (Horwitz et al. 2004), we may uncover unique information that will assist us in genetically dissecting the complexities of human atherosclerosis.
Much work remains to be done for translating our genomic knowledge of human atherosclerosis for use in the clinics. Currently underway is the CardioGene Study, a multi-center trial examining the use of genomic and proteomic techniques for predicting in-stent restenosis in 350 prospectively enrolled patients undergoing coronary artery stent placement (Table 1) (Ganesh et al. 2004). One component of the study will investigate the relationship between peripheral mononuclear cell gene expression assessed at prestent, 2 weeks and 6 months after stent placement and restenosis at 6 and 12 months. The results of this comprehensive evaluation should be illuminating not only with regard to discovering genes important for inflammation and neointimal proliferation but also for determining the role of genotyping and proteomics in the management of a vast clinical problem.
Acknowledgements
The communicating author wishes to thank Michael N. Sack and Toren Finkel for helpful suggestions and critical reading of this manuscript. We would also like to acknowledge Omar Y. Mian for technical assistance and artwork by Lydia Kibiuk and Donald F. Bliss, II, NIH Medical Arts. This research was supported by the Intramural Program of the National Heart, Lung, and Blood Institute of the National Institutes of Health.
REFERENCES
- Borovecki F, Lovrecic L, Zhou J, et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci U S A. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Ahmad AM, Burke H, et al. List of lists-annotated (LOLA): a database for annotation and comparison of published microarray gene lists. Gene. 2005;360:78–82. doi: 10.1016/j.gene.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Chapman CM, Beilby JP, McQuillan BM, et al. Monocyte count, but not C-reactive protein or interleukin-6, is an independent risk marker for subclinical carotid atherosclerosis. Stroke. 2004;35:1619–1624. doi: 10.1161/01.STR.0000130857.19423.ad. [DOI] [PubMed] [Google Scholar]
- Cipolletta C, Ryan KE, Hanna EV, Trimble ER. Activation of peripheral blood CD14+ monocytes occurs in diabetes. Diabetes. 2005;54:2779–2786. doi: 10.2337/diabetes.54.9.2779. [DOI] [PubMed] [Google Scholar]
- Cook SA, Rosenzweig A. DNA microarrays: implications for cardiovascular medicine. Circ Res. 2002;91:559–564. doi: 10.1161/01.res.0000036019.55901.62. [DOI] [PubMed] [Google Scholar]
- Daugherty A, Cassis L. Angiotensin II-mediated development of vascular diseases. Trends Cardiovasc Med. 2004;14:117–120. doi: 10.1016/j.tcm.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Golub TR. Genomic approaches to hematologic malignancies. Blood. 2004;104:923–932. doi: 10.1182/blood-2004-01-0274. [DOI] [PubMed] [Google Scholar]
- Farooq A, Zhou MM. Structure and regulation of MAPK phosphatases. Cell Signal. 2004;16:769–779. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Fryer RM, Randall J, Yoshida T, et al. Global analysis of gene expression: methods, interpretation, and pitfalls. Exp Nephrol. 2002;10:64–74. doi: 10.1159/000049901. [DOI] [PubMed] [Google Scholar]
- Fukuda D, Shimada K, Tanaka A, et al. Circulating monocytes and in-stent neointima after coronary stent implantation. J Am Coll Cardiol. 2004;43:18–23. doi: 10.1016/j.jacc.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Ganesh SK, Skelding KA, Mehta L, et al. Rationale and study design of the CardioGene Study: genomics of in-stent restenosis. Pharmacogenomics. 2004;5:952–1004. doi: 10.1517/14622416.5.7.949. [DOI] [PubMed] [Google Scholar]
- Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- Gui M, Fan H, Steplewski K, Carpenter J, et al. Gene expression profiling of peripheral blood mononuclear cells in humans yields potential biomarkers of accelerated atherosclerotic disease that are independent of known cardiovascular risk factors. Circulation. 2004;110(Suppl S):1309. [Google Scholar]
- Hakonarson H, Bjornsdottir US, Halapi E, et al. Profiling of genes expressed in peripheral blood mononuclear cells predicts glucocorticoid sensitivity in asthma patients. Proc Natl Acad Sci U S A. 2005;102:14789–14794. doi: 10.1073/pnas.0409904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- Harbers M, Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nat Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Suzuki T, Dong HY, et al. Serial analysis of gene expression in human monocytes and macrophages. Blood. 1999;94:837–844. [PubMed] [Google Scholar]
- Horwitz PA, Tsai EJ, Putt ME, et al. Detection of cardiac allograft rejection and response to immunosuppressive therapy with peripheral blood gene expression. Circulation. 2004;110:3815–3821. doi: 10.1161/01.CIR.0000150539.72783.BF. [DOI] [PubMed] [Google Scholar]
- Hwang JJ, Allen PD, Tseng GC, et al. Microarray gene expression profiles in dilated and hypertrophic cardiomyopathic end-stage heart failure. Physiol Genomics. 2002;10:31–44. doi: 10.1152/physiolgenomics.00122.2001. [DOI] [PubMed] [Google Scholar]
- Jain MK, Ridker PM. Anti-inflammatory effects of statins: clinical evidence and basic mechanisms. Nat Rev Drug Discov. 2005;4:977–987. doi: 10.1038/nrd1901. [DOI] [PubMed] [Google Scholar]
- Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- Liu YZ, Dvornyk V, Lu Y, et al. A novel pathophysiological mechanism for osteoporosis suggested by an in vivo gene expression study of circulating monocytes. J Biol Chem. 2005;280:29011–29016. doi: 10.1074/jbc.M501164200. [DOI] [PubMed] [Google Scholar]
- Lowes BD, Gilbert EM, Abraham WT, et al. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med. 2002;346:1357–1365. doi: 10.1056/NEJMoa012630. [DOI] [PubMed] [Google Scholar]
- Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa Y, Anzai T, Yoshikawa T, et al. Prognostic significance of peripheral monocytosis after reperfused acute myocardial infarction:a possible role for left ventricular remodeling. J Am Coll Cardiol. 2002;39:241–246. doi: 10.1016/s0735-1097(01)01721-1. [DOI] [PubMed] [Google Scholar]
- McCaffrey TA, Fu C, Du B, et al. High-level expression of Egr-1 and Egr-1-inducible genes in mouse and human atherosclerosis. J Clin Invest. 2000;105:653–662. doi: 10.1172/JCI8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DF, Li H, Jeffries N, Wright V, et al. Using peripheral blood mononuclear cells to determine a gene expression profile of acute ischemic stroke: a pilot investigation. Circulation. 2005;111:212–221. doi: 10.1161/01.CIR.0000152105.79665.C6. [DOI] [PubMed] [Google Scholar]
- Newman JC, Weiner AM. L2L: a simple tool for discovering the hidden significance in microarray expression data. Genome Biol. 2005;6:R81. doi: 10.1186/gb-2005-6-9-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen NJ, Moore JH, Aune TM. Gene expression signatures for autoimmune disease in peripheral blood mononuclear cells. Arthritis Res Ther. 2004;6:120–128. doi: 10.1186/ar1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino WD, Mian OY, Hwang PM. Serial analysis of gene expression: technical considerations and applications to cardiovascular biology. Circ Res. 2002;91:565–569. doi: 10.1161/01.res.0000036018.76903.18. [DOI] [PubMed] [Google Scholar]
- Patino WD, Mian OY, Kang JG, et al. Circulating transcriptome reveals markers of atherosclerosis. Proc Natl Acad Sci U S A. 2005;102:3423–3428. doi: 10.1073/pnas.0408032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Riggins GJ. Gene discovery using the serial analysis of gene expression technique: implications for cancer research. J Clin Oncol. 2001;19:2948–2958. doi: 10.1200/JCO.2001.19.11.2948. [DOI] [PubMed] [Google Scholar]
- Ridker PM, Cannon CP, Morrow D, et al. C-reactive protein levels and outcomes after statin therapy. N Engl J Med. 2005;352:20–28. doi: 10.1056/NEJMoa042378. [DOI] [PubMed] [Google Scholar]
- Rossi ML, Marziliano N, Merlini PA, et al. Different quantitative apoptotic traits in coronary atherosclerotic plaques from patients with stable angina pectoris and acute coronary syndromes. Circulation. 2004;110:1767–1773. doi: 10.1161/01.CIR.0000142865.04816.89. [DOI] [PubMed] [Google Scholar]
- Seo D, Wang T, Dressman H, et al. Gene expression phenotypes of atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:1922–1927. doi: 10.1161/01.ATV.0000141358.65242.1f. [DOI] [PubMed] [Google Scholar]
- Sharma P, Sahni NS, Tibshirani R, et al. Early detection of breast cancer based on gene-expression patterns in peripheral blood cells. Breast Cancer Res. 2005;7:R634–644. doi: 10.1186/bcr1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Tabibiazar R, Wagner RA, Ashley EA, et al. Signature patterns of gene expression in mouse atherosclerosis and their correlation to human coronary disease. Physiol Genomics. 2005;22:213–226. doi: 10.1152/physiolgenomics.00001.2005. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Vanderlaan PA, Reardon CA. Thematic review series: the immune system and atherogenesis. The unusual suspects:an overview of the minor leukocyte populations in atherosclerosis. J Lipid Res. 2005;46:829–838. doi: 10.1194/jlr.R500003-JLR200. [DOI] [PubMed] [Google Scholar]
- Wibaut-Berlaimont V, Randi AM, Mandryko V, et al. Atorvastatin affects leukocyte gene expression in dyslipidemia patients: in vivo regulation of hemostasis, inflammation and apoptosis. J Thromb Haemost. 2005;3:677–685. doi: 10.1111/j.1538-7836.2005.01211.x. [DOI] [PubMed] [Google Scholar]


