Abstract
DNA-binding activators and repressors recruit histone acetylases and deacetylases to promoters, thereby generating localized domains of modified histones that influence transcriptional activity. At the end of a transcriptional response, alterations in histone acetylation status are reversed, but the dynamics of this process are poorly understood. Here, we recruit histone deacetylases and acetylases to a well-defined yeast promoter in a regulated manner. Following dissociation of the recruiting protein from the promoter, targeted deacetylation and acetylation are reversed with rapid, yet distinct, kinetics. Reversal of targeted deacetylation occurs within 5–8 min, whereas reversal of targeted acetylation is more rapid, taking 1.5 min. These findings imply that untargeted, globally acting enzymes generate a highly dynamic equilibrium of histone acetylation and deacetylation reactions across chromatin. Targeted acetylases and deacetylases can locally perturb this equilibrium, yet once they are removed, the global activities mediate a rapid return to the steady-state level of histone acetylation. Our results also indicate that TBP occupancy depends on the presence of the activator, not histone acetylation status.
Keywords: HATs, HDACs, transcription, histone acetylation, chromatin, gene regulation
Acetylation of lysine residues on histone tails has long been correlated with transcriptional activation (Allfrey et al. 1964; Pogo et al. 1966). Acetate addition and removal experiments involving bulk histones suggest that this histone acetylation is dynamic, although the fraction of total histones involved and the modification rates depend on the organism (Davie 1997; Waterborg 1998, 2001). In mammalian cells, it has been suggested that histone acetylation turnover is more rapid at actively transcribed genes than at inactive genes (Covault and Chalkley 1980; Ip et al. 1988; Boffa et al. 1990). However, these analyses of bulk histone populations do not measure acetylation in the context of a defined transcriptional response, and it is unclear whether the dynamic changes in acetylation occur during histone synthesis, histone deposition on DNA, or on histones already assembled into chromatin.
A mechanistic understanding of the linkage between histone acetylation and transcription arose from the identification of histone acetylases (HATs) and histone deacetylases (HDACs) as transcriptional coactivators and corepressors, respectively, which are targeted to promoters by sequence-specific DNA-binding proteins (Pazin and Kadonaga 1997; Struhl 1998). Once recruited to chromatin, HATs and HDACs generate a localized domain of modified histones, resulting in activation or repression, respectively, of their target genes (Struhl 1999; Wu and Grunstein 2000). The yeast SAGA complex, containing the Gcn5 HAT is targeted to promoters by activators, such as Gcn4 and Swi5, in which it acetylates histone H3 (Kuo et al. 1998, 2000; Cosma et al. 1999; Krebs et al. 1999). Another yeast HAT, Esa1, is recruited via Rap1 to promoters of ribosomal protein genes (Reid et al. 2000). In an analogous mechanism leading to transcriptional repression, the Rpd3 HDAC is targeted to yeast promoters by the DNA-bound repressor Ume6, in which it deacetylates histones H3 and H4 (Kadosh and Struhl 1997, 1998; Rundlett et al. 1998).
As many transcriptional responses are regulated and transient, one might expect the basal level of histone acetylation to be restored after the recruiting activator or repressor has ended its function. Furthermore, reversal of an induced acetylation state may be essential for timely termination of the transcriptional response, as well as maintenance of proper chromatin structure. Changes in histone acetylation have been observed in a variety of situations in which an induced hyperacetylation is later followed by a return to the initial acetylation level (Cavalli and Paro 1999; Chen et al. 1999; Cosma et al. 1999; Krebs et al. 1999, 2000; Agalioti et al. 2000; Beresford and Boss 2001; Bouchard et al. 2001; Frank et al. 2001). In yeast, transient cell cycle-regulated binding of Swi5 to the HO promoter results in recruitment of SAGA and histone acetylation; later on in the cell cycle, SAGA departs, and acetylation is reversed (Cosma et al. 1999; Krebs et al. 1999). At promoters targeted by the estrogen receptor, a HAT complex including p300/CBP and ACTR, is recruited upon hormone induction, leading to histone hyperacetylation and gene activation. ACTR is then acetylated by p300/CBP, resulting in dissociation of the HAT complex from the promoter, and thus self-attenuation of histone acetylation (Chen et al. 1999). At the interferon-β enhancer, an enhanceosome containing PCAF and CBP is assembled in response to virus infection, leading to histone acetylation (Parekh and Maniatis 1999; Agalioti et al. 2000). The enhanceosome later disassembles when its HMGI(Y) component is acetylated by CBP, and the basal histone acetylation level is restored (Munshi et al. 1998; Agalioti et al. 2000). Conversely, cell cycle-dependent dissociation of E2F-Rb-targeted HDAC1 from the DHFR promoter is accompanied by increased histone acetylation and transcription (Ferreira et al. 2001). In essentially all of these studies, the time interval between the departure of the targeted HAT/HDAC and the restoration of the initial histone acetylation state has not been determined.
Although such regulated dissociation of the targeted HATs or HDACs from promoters would prevent further modification of histone tails, the mechanism by which the existing hyperacetylation or hypoacetylation is erased is less obvious. One possibility is that an enzyme with the reverse activity, recruited to the same promoter, can restore the initial acetylation state. In support of this idea, some transcriptional regulators can associate with both HATs and HDACs while functioning as activators and repressors, respectively (Kadosh and Struhl 1997; Xu et al. 1999). In a related mechanism, the reverse activity might be constitutively recruited to a promoter via a distinct DNA-binding protein, thereby restoring the initial histone acetylation level upon regulated dissociation of a HAT/HDAC. Such a mechanism might apply to the yeast HO promoter, which appears to be subject to multiple mechanisms of repression (Sternberg et al. 1987; Yu et al. 2000). However, sequential or simultaneous targeting of HATs and HDACs by DNA-bound proteins, although likely to occur at particular promoters under specific conditions, is unlikely to be a general mechanism for restoring histone acetylation levels after termination of a transcriptional response.
Yeast HATs (Gcn5 and Esa1) and HDACs (Rpd3 and Hda1) are not only targeted to specific loci, but they also function globally throughout the genome (Krebs et al. 1999; Kuo et al. 2000; Reid et al. 2000; Vogelauer et al. 2000). These global activities provide a possible general mechanism for resetting acetylation levels, with no need for promoter-specific targeting. However, the kinetics by which untargeted, global HATs and HDACs alter acetylation levels, and whether their action can account for rapid changes in histone acetylation, are not known. Here we address this issue using fusion constructs based on the tetracycline repressor (TetR) (Gossen and Bujard 1992; Hillen and Berens 1994), which target HATs and HDACs to a well-defined promoter in vivo, in a regulated manner. Our results, showing rapid reversal of targeted histone acetylation and deacetylation, reveal the highly dynamic nature of the global HDAC and HAT activities. Furthermore, they define a general mechanism by which histone acetylation status is rapidly restored to the steady-state level upon removal or inactivation of a transcriptional regulator in response to environmental or developmental signals.
Results
Experimental design
To monitor changes in histone acetylation at promoters following the removal of a targeted HAT or HDAC, we needed a DNA-binding protein that could be rapidly dissociated from the promoter in a regulated manner. The DNA-binding properties of TetR, encoded by the Escherichia coli Tn10 transposon, are optimal for such regulation, as this protein interacts with its target DNA site, the tet operator (tetO) in the absence of tetracycline and dissociates upon tetracycline addition (Hillen and Berens 1994). TetR fusions have been used as regulated activators and repressors in a variety of eukaryotic organisms including yeast (Gossen and Bujard 1992; Hillen and Berens 1994; Gari et al. 1997; Belli et al. 1998). Another advantage of TetR regulation, in contrast to most natural induction systems, is the absence of significant physiological effects that might indirectly affect the gene of interest. The potent tetracycline analog doxycycline (Dox) was reported not to affect viability or growth rate in yeast (Gari et al. 1997), and we did not detect any obvious change in viability, growth rate, or transcription in general during Dox treatment. We utilized HA-tagged TetR fusions with the Ume6 repression domain (Kadosh and Struhl 1997) and the VP16 activation domain (Sadowski et al. 1988) to target HDAC and HAT activities, respectively, to a tetO-driven promoter. Thus, the function of both HATs and HDACs could be monitored with the same experimental design.
We used a modified HIS3 promoter at its natural genomic locus as a reporter gene, because HIS3 promoter structure (Struhl et al. 1985) and histone acetylation properties (Kuo et al. 2000) have been studied in detail. Apart from the Gcn4-binding site, there are no known targeting sites for HATs or HDACs in the HIS3 promoter region. We replaced most of the HIS3 upstream region, including the Gcn4-binding site, by two copies of tetO; the core promoter region, including the TC and TR TATA elements, remains intact. Thus, changes in histone acetylation levels (other than those caused by the tetO-recruited activities) will presumably be due to the action of the untargeted, global enzymes. To monitor the transcriptional repression activity of the TetR–Ume6 fusion, another strain was constructed, in which the HIS3 locus contains a binding site for the Rap1 activator between the tetO sites and the TATA elements.
Reversal of targeted histone deacetylation takes 5–8 min
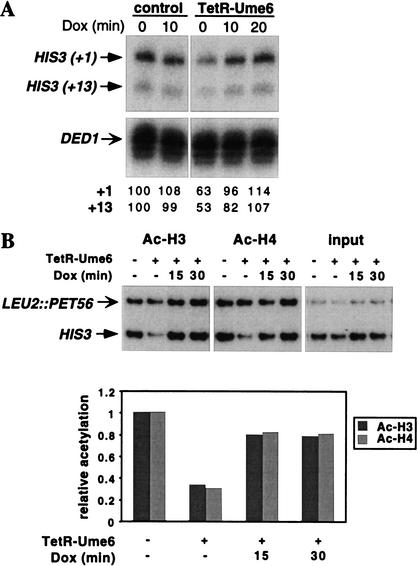
Targeted recruitment of the HDAC Rpd3 to yeast promoters is mediated by its interaction with the DNA-binding repressor Ume6 (Kadosh and Struhl 1997). Rpd3 then generates a local domain of histone H3 and H4 deacetylation, resulting in transcriptional repression (Kadosh and Struhl 1998; Rundlett et al. 1998). To target Rpd3 to the tetO sites in the HIS3 promoter, a portion of Ume6, which includes the repression domain (Kadosh and Struhl 1997), was fused to TetR. The TetR–Ume6 fusion represses transcription of the Rap1-activated HIS3 gene (Fig. 1A). Repression is rapidly relieved upon addition of Dox, within 10–20 min.
Figure 1.
TetR–Ume6-mediated transcriptional repression and histone deacetylation are reversed by Dox. (A) RNAs from YKT1R cells (contain a Rap1 site at the HIS3 promoter) transformed with a TetR–Ume6 or control plasmid grown in the presence of Dox for 0–30 min were analyzed by S1 nuclease protection, using probes to HIS3 and DED1. Quantitation of the signal for the +1 and +13 HIS3 transcripts, normalized to the DED1 signal, is shown at bottom, with the control untreated sample set as 100%. (B) Cross-linked chromatin from control or TetR–Ume6-expressing YKT1R cells treated with Dox for 0–30 min was immunoprecipitated with antibodies to acetylated histones H3 and H4. The smaller PCR product corresponds to the tetO-containing HIS3 locus, and the larger product corresponds to the LEU2∷PET56 locus, serving as internal control. After normalizing each immunoprecipitation (IP) signal to the input signal, the HIS3 results were divided by the LEU2∷PET56 results, and expressed relative to the control sample.
Changes in histone acetylation levels at the HIS3 locus were monitored by chromatin immunoprecipitation by use of antibodies to diacetylated histone H3 and tetra-acetylated H4. The strains contain a second copy of the HIS3 promoter, located, together with the adjacent PET56 gene, at the LEU2 locus. The primer pair used for the quantitative PCR analysis simultaneously amplifies both HIS3 alleles, thereby permitting the assessment of histone acetylation status at the tetO-containing promoter with a true internal control. The use of a true internal control eliminates many sources of experimental error, and hence, permits much more accurate measurements of small quantitative differences.
TetR–Ume6 causes a specific decrease in the level of acetylated histones at the HIS3 locus (Fig. 1B). Both histones H3 and H4 are affected, as observed for natural Ume6-repressed promoters (Kadosh and Struhl 1998; Rundlett et al. 1998; Deckert and Struhl 2001). Treatment with Dox for 15 or 30 min restores the level of histone acetylation to ∼80% of that observed in the absence of TetR–Ume6. Thus, Dox causes a rapid dissociation of TetR–Ume6 from the promoter, resulting in near basal levels of transcription and histone acetylation within 15–20 min.
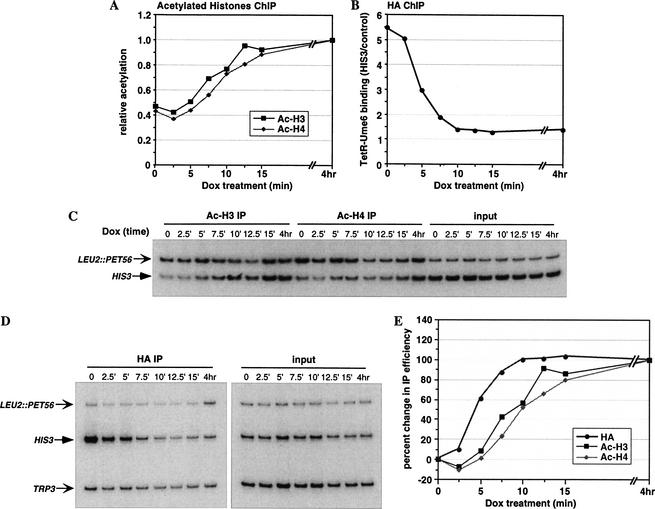
As the above results are suggestive of rapid changes in histone acetylation, a more detailed time-course experiment was performed with 2.5-min intervals of Dox treatment, up to 15 min. A culture treated with Dox for 4 h represented the situation of little or no TetR–Ume6 binding at HIS3. As shown in Figure 2 (A and C), the changes in histone H3 and H4 acetylation are continuous and reproducible, and they follow similar kinetics. A clear increase in acetylation level is detected 7.5 min after Dox addition, and by 12.5–15 min, the maximal level has been nearly reached.
Figure 2.
Reversal of targeted histone deacetylation upon dissociation of TetR–Ume6 from the HIS3 promoter. Cross-linked chromatin from TetR–Ume6-expressing YKT1R cells (contain a Rap1-activated HIS3 promoter) treated with Dox for 0–15 min or 4 h was immunoprecipitated with antibodies to acetylated histones H3 and H4 (A,C), or the HA epitope for measuring TetR–Ume6 occupancy (B,D), and analyzed by quantitative PCR. Histone acetylation (A,C) was analyzed as described in Figure 1. After normalizing each IP signal to the input signal, the HIS3 results were divided by the LEU2∷PET56 results, and expressed relative to the 4-hour-treated sample. TetR–Ume6 binding, as determined by HA IP (B,D), was analyzed using the HIS3 promoter primers described above in combination with primers amplifying TRP3 as internal control. After normalizing each IP signal to the input signal, the specific binding of TetR–Ume6 at HIS3 was determined by dividing the HIS3 results by the control TRP3 results. (E) The kinetics of changes in histone acetylation were compared with those of TetR–Ume6 dissociation from the promoter by quantitating the changes in these values throughout the time course. At each time-point, the change in normalized IP value from the time 0 value was determined, and expressed relative to the change at 4 h set as 100%
The continuous and reproducible nature of the time-course curves over 2.5-min intervals is notable, because formaldehyde treatment at each time point lasts for 20 min. Given that formaldehyde covalently binds lysine residues, we presume that cellular enzymes are inactivated almost immediately upon addition to the growing cells and that the 20-min incubation time merely increases the cross-linking in fixed and metabolically inert cells. As such, formaldehyde cross-linking provides a snapshot of protein–protein and protein–DNA interactions at the particular time-point.
The changes in histone acetylation levels depicted in Figure 2A reflect two processes, namely Dox-induced dissociation of TetR–Ume6 (with the associated Rpd3) from DNA, followed by restoration of the basal acetylation level by globally acting HATs. To separate these processes, we monitored occupancy of the HA-tagged TetR–Ume6 at HIS3 during this time course. PCR was done with primers spanning the HIS3 promoter together with an additional primer set, which amplifies the TRP3 promoter as an internal control. Figure 2 (B and D) shows a rapid dissociation of TetR–Ume6 from HIS3, starting 2.5–5 min and being nearly completed 7.5–10 min after Dox addition.
The time interval between TetR–Ume6 dissociation (Fig. 2B) and the changes in histone acetylation levels (Fig. 2A) should reflect the actual HAT-mediated acetylation process. To facilitate the comparison between these time-course curves, we calculated the change that occurred at every time point, relative to the value at time 0, with the change at the 4-hour end-point being set as 100% (Fig. 2E). These data clearly show a time interval, in a range of minutes, between the initial event of TetR–Ume6 dissociation, and the subsequent changes in histones H3 and H4 acetylation levels. The time difference between these curves, here determined at the point of 70% change (Δt70), represents the time required to restore the steady-state histone acetylation level. For both H3 and H4 acetylation, this time interval is ∼6 min. Thus, following the dissociation of a DNA-bound repressor, the basal level of histone acetylation is rapidly, though not instantly, restored.
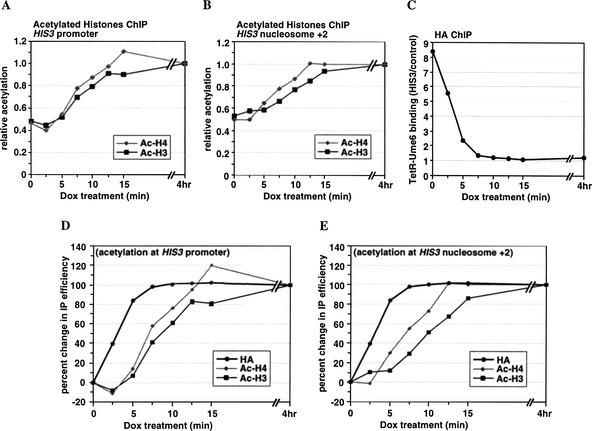
As a Rap1-associated HAT might contribute to the observed changes in acetylation levels, we performed the same experiment in a strain that contains no activator-binding site at the HIS3 locus. As shown in Figure 3 (A,C,D), the kinetics of histone acetylation are similar to those observed at the Rap1-activated promoter. The changes in histone acetylation levels were also monitored by use of primers encompassing nucleosome +2, within the HIS3 ORF, and the PGK1 promoter as an internal control. The level of histone deacetylation caused by TetR–Ume6 is slightly lower at HIS3 nucleosome +2, yet similar kinetics of changes in acetylation are observed (Fig. 3B,C,E). Altogether, the analyses of TetR–Ume6-mediated histone deacetylation show a time interval of 6.9 ± 1.7 min and 5.6 ± 1.8 min for H3 and H4, respectively, at the point of 70% change (Δt70 from three independent experiments). Thus, global HATs rapidly restore the basal level of histone acetylation.
Figure 3.
Reversal of targeted histone deacetylation upon dissociation of TetR–Ume6 from the HIS3 promoter. Cross-linked chromatin from TetR–Ume6-expressing YKT2 cells (contain a nonactivated HIS3 promoter) treated with Dox for 0–15 min or 4 h was immunoprecipitated with antibodies to acetylated histones H3 and H4 or the HA epitope for measuring TetR–Ume6 occupancy, and analyzed by quantitative PCR. (A) Histone acetylation at the HIS3 promoter was analyzed as described in Figure 1. (B) Histone acetylation at the nucleosome +2 region within the HIS3 ORF was determined using the PGK1 promoter as an internal control. After normalizing each IP signal to the input signal, the HIS3 results were divided by those of the control locus, and expressed relative to the 4-hour-treated sample. (C) TetR–Ume6 binding at the HIS3 promoter was determined using the PGK1 promoter as an internal control. After normalizing each IP signal to the input signal, the specific binding of TetR–Ume6 at HIS3 was determined by dividing the HIS3 results by the control PGK1 results. (D,E) Kinetics of changes in histone acetylation and TetR–Ume6 dissociation from the promoter throughout the time course. At each time-point, the change in IP value from the time 0 value was determined, and expressed relative to the change at 4 h set as 100%.
Reversal of targeted histone acetylation and TBP occupancy takes ∼1.5 min
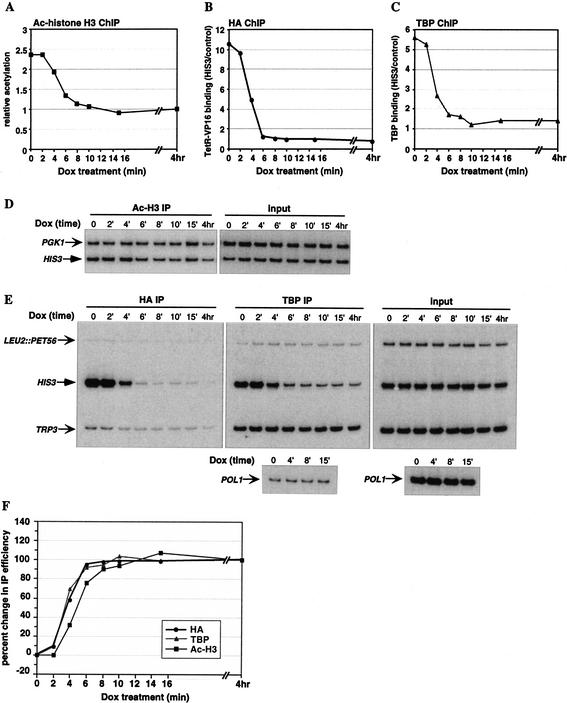
The acidic activation domain of VP16 recruits the SAGA complex, which contains the Gcn5 HAT, to promoters in vivo (Tumbar et al. 1999; Larschan and Winston 2001), leading to localized histone hyperacetylation and transcriptional activation. We therefore targeted SAGA to the HIS3 promoter using a HA-tagged TetR–VP16 fusion. We observe an increase in histone H3 acetylation at the HIS3 locus upon introduction of TetR–VP16, with maximal acetylation at nucleosome +2 (data not shown). Treatment with Dox for 4 h eliminates this hyperacetylation. A time-course experiment was performed to determine the kinetics of this process (Fig. 4). In addition to histone acetylation and TetR–VP16 occupancy levels, we also analyzed the samples with a TBP antibody, to determine the presence of the basal transcription machinery at HIS3 upon TetR–VP16 dissociation. Histone acetylation levels were determined by duplex PCR using primers to HIS3 nucleosome +2 and the PGK1 promoter as an internal control. For TetR–VP16 and TBP occupancy, primers spanning the HIS3 promoter (with TRP3 as internal control) were used.
Figure 4.
Reversal of targeted histone acetylation and TBP binding upon dissociation of TetR–VP16 from the HIS3 promoter. Cross-linked chromatin from TetR–VP16-expressing YKT2 cells (contain a nonactivated HIS3 promoter) treated with Dox for 0–15 min or 4 h was immunoprecipitated with antibodies to acetylated histone H3, the HA epitope for measuring TetR–VP16 occupancy, and TBP, and analyzed by quantitative duplex PCR. (A,D) Histone acetylation at the nucleosome +2 region of HIS3 was analyzed as described in Figure 3. (B,E) TetR–VP16 binding to the HIS3 promoter was determined by normalizing each IP signal to its corresponding input signal, and dividing the HIS3 results by the control TRP3 results. (C,E) TBP binding to the HIS3 promoter was determined by normalizing each IP signal to its corresponding input signal, and dividing the HIS3 results by the control TRP3 results. The POLI ORF signal represented background binding by TBP, and the results are expressed relative to the average normalized POL1 background signal. (F) Kinetics of changes in histone acetylation, and occupancy by TetR–VP16 and TBP at the HIS3 promoter throughout the time course. At each time point, the change in IP value from the time 0 value was determined, and expressed relative to the change at 4 h set as 100%.
This time-course experiment shows a rapid decline in all three parameters upon Dox treatment (Fig. 4), TetR–VP16 occupancy (B and E), histone acetylation (A and D), and TBP occupancy (C and E). In all cases, near minimal levels are reached within 6–8 min. To compare between the different processes, we determined the change from the initial (time 0) level, occurring at each time point, as described above for the TetR–Ume6 experiment (Fig. 4F). As in the case of TetR–Ume6, a time interval exists between TetR–VP16 dissociation and the change in acetylation level, yet, here the two graphs differ by only ∼1.5 min at the point of 70% change (Δt70 = 1.5 ± 0.25 from three independent experiments). The change in TBP occupancy, however, follows almost the exact time course as TetR–VP16 occupancy, with no evident time interval between the two curves.
Collectively, the TetR-based recruitment experiments show that for both targeted histone acetylation and deacetylation, a rapid return to the basal acetylation level occurs within a range of minutes, with reversal of targeted acetylation showing a significantly faster response. In addition, TBP occupancy depends on the presence of the activator, not histone acetylation status per se.
Reversal of targeted histone acetylation from the natural UGT51 promoter
To further examine whether the rapid reversal of targeted histone acetylation applies to natural promoters, we identified additional TetR-binding sites within yeast promoter regions. The tetO site consists of 19 bp, most of which contribute to DNA-binding affinity (Wissmann et al. 1988; Sizemore et al. 1990; Hillen and Berens 1994). We found a putative imperfect TetR-binding site 77 bp upstream of the translation start site of UGT51, the gene for sterol glucosyltransferase (Warnecke et al. 1999). TetR–VP16 occupancy and TetR–VP16-mediated histone H3 hyperacetylation at the UGT51 promoter were detected, although both values were significantly lower than that seen with the modified HIS3 locus (data not shown). TetR–Ume6-mediated histone deacetylation was weak at UGT51, and, therefore, was not further analyzed.
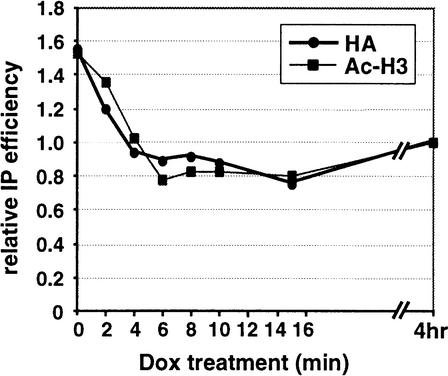
To monitor the time course of histone hyperacetylation reversal at UGT51, samples from the TetR–VP16 experiments described above were analyzed using PCR primers spanning the UGT51 promoter. Both TetR–VP16 occupancy and histone H3 acetylation rapidly decline, reaching basal values by 4 min following Dox addition (Fig. 5). Despite the relatively low binding and hyperacetylation at UGT51, the reproducibly short time interval between the two curves, similar to that observed at the HIS3 locus, indicates rapid reversal of targeted histone acetylation.
Figure 5.
Reversal of targeted histone acetylation upon dissociation of TetR–VP16 from the UGT51promoter . Samples used in Figure 4 were analyzed by quantitative duplex PCR using UGT51 primers instead of HIS3 primers. After normalizing each IP signal to the input signal, the UGT51 results were divided by the those of the control locus, and expressed relative to the 4-hour-treated sample.
Discussion
Highly dynamic acetylation and deacetylation reactions maintain, and can rapidly restore the steady-state acetylation status of nucleosomal histones
At many eukaryotic promoters, the level of histone acetylation undergoes dynamic changes in response to external stimulation or as a function of cell cycle progression. Although previous work has focused on the establishment of altered histone acetylation states by DNA-bound transcription factors recruiting HATs and HDACs, restoration of the initial acetylation level may be no less important. In line with this notion, our results suggest a general mechanism by which the basal state of histone acetylation is rapidly restored, involving untargeted, global HAT and HDAC activities.
We monitored the changes in histone acetylation upon removal of a VP16-derived activator and a Ume6-derived repressor targeted to the HIS3 promoter. As the HIS3 sequence present in our strains appears to include no recruitment sites for HATs or HDACs, the restoration of basal acetylation levels is due to untargeted, global enzymes. Our results show that following the dissociation of the targeting protein, the basal level of histone acetylation is restored rapidly, although not instantly, in a time frame of minutes. In the case of Ume6-mediated histone H3 and H4 hypoacetylation, most of the change in acetylation was achieved by 5–8 min (Δt70 of 6.9 ± 1.7 and 5.6 ± 1.8, respectively), whereas VP16-mediated H3 hyperacetylation was removed within ∼1.5 min (Δt70 of 1.5 ± 0.25). Histone H3 hyperacetylation at the UGT51 promoter was reversed with similar kinetics. Thus, localized alterations in the histone acetylation state, mediated by recruited HATs and HDACs, appear to be rapidly erased when recruitment ends by the action of globally acting HDACs and HATs.
Our results show that untargeted HAT and HDAC activities are highly dynamic. Such rapid processes occurring throughout chromatin can reset the basal acetylation state upon termination of the various transcriptional responses, within a short time frame, ensuring complete and timely reversal of each response. Although altered states of histone acetylation are often linked to transcriptional regulation, the level of histone acetylation is likely to be important also for other processes occurring on chromatin, such as recombination, DNA repair, and replication. The rapid kinetics of the global acetylation and deacetylation processes indicate that maintenance of a proper acetylation state is an important aspect of chromatin dynamics. Thus, chromatin structure, with its globally acting HATs and HDACs, is in a dynamic equilibrium, in which acetylation and deacetylation reactions occur continuously, generating a steady-state level of histone acetylation. Localized perturbations of this equilibrium can occur by promoter-specific targeting of HATs and HDACs; yet, once targeting terminates, the cell rapidly restores its initial acetylation state
Global histone deacetylation is more efficient than global histone acetylation
Global histone acetylation and deacetylation are largely mediated by the same enzymes catalyzing targeted histone modifications (Krebs et al. 1999; Kuo et al. 2000; Reid et al. 2000; Vogelauer et al. 2000). Our results show a clear difference between the kinetics by which targeted hyperacetylation and hypoacetylation are reversed, as VP16-mediated histone H3 hyperacetylation declines more rapidly than Ume6-mediated hypoacetylation on the same promoter. In contrast, no obvious difference was observed between the reversal of histone H3 and H4 hypoacetylation, although distinct HATs are probably involved. These observations suggest that in vivo globally acting HAT and HDAC complexes may differ in some basic property, such as their interaction with chromatin or their ability to access and modify histone tails. For example, global HDAC complexes may be more widely distributed along chromatin, or may be able to move faster, and thus, would gain better access to the histone tails, whereas global HAT complexes may be more restricted in their distribution or movement. An intriguing possibility is that a mechanistic difference between global HDAC and HAT complexes reflects distinct kinetic requirements related to their roles in reversing activator-targeted acetylation versus repressor-targeted deacetylation. Alternatively, such different properties may be important for their function in maintaining distinct steady-state levels of histone acetylation across chromosomal regions. Lastly, the observation that global histone deacetylation is more efficient than global histone acetylation is consistent with the fact that most lysine residues on histone tails are nonacetylated in yeast cells (Waterborg 2000).
Continued TBP association depends on the presence of the activator, not histone hyperacetylation
An interesting event observed upon TetR–VP16 dissociation from the promoter is the immediate departure of TBP, which precedes the reversal of histone hyperacetylation. This finding suggests that the presence of the activator is critical for continued association of TBP with the promoter; shortly after the activator departs, the persisting histone hyperacetylation is not in itself sufficient to stabilize full TBP binding. As reversal of other chromatin modifications (e.g., nucleosome remodeling) is likely to take some time after dissociation of the activator, it is possible that activator-dependent changes in chromatin structure might not be sufficient for continued TBP association.
Although highlighting the importance of the activator for the stable association of TBP with the promoter, this result does not contrast with the notion that maintaining a proper steady-state level of histone acetylation, and restoring basal acetylation levels at the end of a transcriptional response, can be important for transcriptional regulation (Vogelauer et al. 2000). Under conditions when TBP binding is stabilized by contacts with a DNA-bound activator, or possibly by some alteration of chromatin structure, an abnormally high acetylation level may further stabilize the transcription complex and enhance transcription. It is also possible that although full TBP occupancy requires the presence of the activator at the promoter, persisting histone hyperacetylation could maintain a certain level of promoter-associated TBP after the activator has departed. Our results thus support on one hand the crucial role of the activator in continuously tethering TBP to the promoter during the activation process, and on the other hand, as suggested by the rapid kinetics of global acetylation and deacetylation, the importance of restoring and maintaining a steady-state level of histone acetylation.
Materials and methods
DNAs and yeast strains
The constructs used to create the tetO-driven HIS3 allele are based on a previously described plasmid, in which the HIS3 upstream region has been deleted and replaced by a Rap1-binding site (Iyer and Struhl 1995). YIp211HisTetRap was generated by cloning a 150-bp PCR fragment from pCM172 (Gari et al. 1997) harboring two copies of tetO, into the XhoI site, upstream of the Rap1 site. Deletion of the Rap1 site by EcoRI digestion of YIp211HisTetRap and self ligation produced YIp211HisTet. The HIS3 loci of YIp211HisTetRap and YIp211HisTet were introduced into FT5 (α ura3-52 trp1-Δ63 his3-Δ200 leu2∷PET56) by two-step gene replacement to generate YKT1R (with a Rap1 site at HIS3) and YKT2 (lacking activator sites upstream of the HIS3 core region), respectively. These strains were transformed with TRP1 centromeric plasmids expressing TetR-derived chimeras, or empty vectors as control. The TetR–Ume6 expression construct p414ADH-Tet3HAUN596, cloned in p414ADH (Mumberg et al. 1995), includes the following (in this order): TetR, a nuclear localization signal, three copies of the HA epitope, and amino acids 1–596 of Ume6. The TetR–VP16 expression construct p414CYC1-TetLVP-3HA, cloned in p414CYC1 (Mumberg et al. 1995), includes the following: TetR, a nuclear localization signal, a linker from the λ bacteriophage cI gene (amino acids 92–132), the VP16 activation domain (amino acids 367–490), and three copies of the HA epitope. The linker and VP16 portion was cloned from pCM172. Yeast strains were grown under standard conditions in glucose medium containing casamino acids.
Chromatin immunoprecipitation
The association of TetR fusions with tetO DNA was regulated by adding doxycycline (Dox) (10 μg/mL) to logarithmically growing cultures at various times before formaldehyde treatment. Immunoprecipitation (IP) of chromatin from cross-linked cells and quantitative PCR were performed essentially as described (Kuras and Struhl 1999), except that IPs were done in 150 mM NaCl. For quantitation of histone acetylation levels, antibodies to diacetylated histone H3 and tetra-acetylated histone H4 (Upstate Biotechnology) were used. A 24-cycle PCR was performed with ∼1/100 of the immunoprecipitated DNA and 1/10,000 of the input DNA. IPs with monoclonal anti-HA (F7, Santa Cruz) and polyclonal anti-TBP were followed by a 26-cycle PCR using ∼1/40 of the immunoprecipitated DNA and 1/40,000 of the input DNA. Each PCR reaction from the IPs for acetylated histones and TBP generated two products of the tested locus (HIS3 or UGT51) and an irrelevant locus serving as internal control. For the HA IPs, the tested locus and control locus were amplified either in the same PCR reaction (in the TetR–VP16 experiments and some TetR–Ume6 experiments) or in different reactions. IP efficiency for a given locus was calculated as the ratio between the amount of IP PCR product and input PCR product. The IP efficiency value of the tested locus (HIS3 or UGT51) was then divided by that of the control locus to correct for variation between different samples. PCR reactions were done in duplicates or triplicates, and the individual values reported in each figure have an error of ±10%. For each TetR fusion, histone acetylation and occupancy at HIS3 (by HA IP) were determined by use of samples from three independent time-course experiments, which gave similar results. Analyses of TBP occupancy and the UGT51 promoter were done for two independent experiments.
Transcriptional analysis
Logarithmically growing cultures were treated with Dox (10 μg/mL) for different time periods. Total RNA was subjected to S1 nuclease protection analysis to determine HIS3 RNA levels, as described previously (Iyer and Struhl 1996).
Acknowledgments
We thank Jutta Deckert, Mario Mencia, Juliet Reid, Elmar vom Baur, and Gerry Crabtree for fruitful discussions. This work was supported by a European Molecular Biology Organization postdoctoral fellowship to Y.K-K. and by research grants to K.S. from the National Institutes of Health (GM30186 and GM53720).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kevin@hms.harvard.edu; FAX (617) 432-2529.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.967302.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Allfrey V, Faulkner RM, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belli G, Gari E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beresford GW, Boss JM. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat Immunol. 2001;2:652–657. doi: 10.1038/89810. [DOI] [PubMed] [Google Scholar]
- Boffa LC, Walker J, Chen TA, Sterner R, Mariani MR, Allfrey VG. Factors affecting nucleosome structure in transcriptionally active chromatin. Histone acetylation, nascent RNA and inhibitors of RNA synthesis. Eur J Biochem. 1990;194:811–823. doi: 10.1111/j.1432-1033.1990.tb19474.x. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes & Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286:955–958. doi: 10.1126/science.286.5441.955. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Covault J, Chalkley R. The identification of distinct populations of acetylated histone. J Biol Chem. 1980;255:9110–9116. [PubMed] [Google Scholar]
- Davie JR. Nuclear matrix, dynamic histone acetylation and transcriptionally active chromatin. Mol Biol Rep. 1997;24:197–207. doi: 10.1023/a:1006811817247. [DOI] [PubMed] [Google Scholar]
- Deckert J, Struhl K. Histone acetylation at promoters is differentially affected by activators and repressors. Mol Cell Biol. 2001;21:2726–2735. doi: 10.1128/MCB.21.8.2726-2735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R, Naguibneva I, Mathieu M, Ait-Si-Ali S, Robin P, Pritchard LL, Harel-Bellan A. Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter. EMBO Rep. 2001;2:794–799. doi: 10.1093/embo-reports/kve173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes & Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen W, Berens C. Mechanisms underlying expression of Tn10-encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- Ip YT, Jackson V, Meier J, Chalkley R. The separation of transcriptionally engaged genes. J Biol Chem. 1988;263:14044–14052. [PubMed] [Google Scholar]
- Iyer V, Struhl K. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol Cell Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- ————— Targeted recruitment of the Sin3–Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs JE, Kuo M-H, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes & Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs JE, Fry CJ, Samuels ML, Peterson CL. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell. 2000;102:587–598. doi: 10.1016/s0092-8674(00)00081-7. [DOI] [PubMed] [Google Scholar]
- Kuo M-H, Zhou J, Jambeck P, Churchill MEA, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes & Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M-H, vom Baur E, Struhl K, Allis CD. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes & Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMGI(Y) by CBP turns off IFNβ expression by disrupting the enhancesome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- Parekh BS, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- Pogo BGT, Allfrey VG, Mirsky AE. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc Natl Acad Sci. 1966;55:805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4–VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sizemore C, Wissmann A, Gulland U, Hillen W. Quantitative analysis of Tn10 Tet repressor binding to a complete set of tet operator mutants. Nucleic Acids Res. 1990;18:2875–2880. doi: 10.1093/nar/18.10.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg PW, Stern MJ, Clark I, Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- ————— Fundamentally different logic of gene expression in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- Struhl K, Chen W, Hill DE, Hope IA, Oettinger MA. Constitutive and coordinately regulated transcription of yeast genes: Promoter elements, positive and negative regulatory sites, and DNA binding proteins. Cold Spring Harbor Symp Quant Biol. 1985;50:489–503. doi: 10.1101/sqb.1985.050.01.061. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- Warnecke D, Erdmann R, Fahl A, Hube B, Muller F, Zank T, Zahringer U, Heinz E. Cloning and functional expression of UGT genes encoding sterol glucosyltransferases from Saccharomyces cerevisiae, Candida albicans, Pichia pastoris, and Dictyostelium discoideum. J Biol Chem. 1999;274:13048–13059. doi: 10.1074/jbc.274.19.13048. [DOI] [PubMed] [Google Scholar]
- Waterborg JH. Dynamics of histone acetylation in Chlamydomonas reinhardtii. J Biol Chem. 1998;273:27602–27609. doi: 10.1074/jbc.273.42.27602. [DOI] [PubMed] [Google Scholar]
- ————— Steady-state levels of histone acetylation in Saccharomyces cerevisiae. J Biol Chem. 2000;275:13007–13011. doi: 10.1074/jbc.275.17.13007. [DOI] [PubMed] [Google Scholar]
- ————— Dynamics of histone acetylation in Saccharomyces cerevisiae. Biochemistry. 2001;40:2599–2605. doi: 10.1021/bi002480c. [DOI] [PubMed] [Google Scholar]
- Wissmann A, Meier I, Hillen W. Saturation mutagenesis of the Tn10-encoded tet operator O1. Identification of base-pairs involved in Tet repressor recognition. J Mol Biol. 1988;202:397–406. doi: 10.1016/0022-2836(88)90273-2. [DOI] [PubMed] [Google Scholar]
- Wu J, Grunstein M. 25 years after the nucleosome model: Chromatin modifications. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Yu Y, Eriksson P, Stillman DJ. Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription. Mol Cell Biol. 2000;20:2350–2357. doi: 10.1128/mcb.20.7.2350-2357.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]