Abstract
Specific recruitment of corepressor complexes containing histone deacetylases (HDAC) by transcription factors is believed to play an essential role in transcriptional repression. Recent studies indicate that repression by unliganded nuclear hormone receptors and by the Mad family of repressors requires distinct HDAC-containing corepressor complexes. In this work, we show that unliganded TR specifically recruits only the closely related N-CoR and SMRT–HDAC3 complexes, whereas the Mad1 recruits only the Sin3–HDAC1/2 complex. Significantly, both the Sin3 and Mi-2/NURD complexes also exhibit constitutive association with chromatin and contribute to chromatin deacetylation in a nontargeted fashion. These results suggest that HDAC complexes can contribute to gene repression by two distinct mechanisms as follows: (1) specific targeting by repressors and (2) constitutive association with chromatin.
Keywords: Nuclear hormone receptor, chromatin, corepressor, histone deacetylase, chromatin immunoprecipitation assay
Nuclear hormone receptors (NR) are a large group of structurally related transcription factors whose transcriptional activities are generally regulated by lipophilic ligands (Mangelsdorf et al. 1995). As members of the NR superfamily, the thyroid hormone receptor (TR) and retinoic acid receptor (RAR) have the capacity to alternately repress or activate transcription dependent on the absence or presence of their cognate hormones (Glass and Rosenfeld 2000). Studies over the last several years indicate that TR and RAR make use of distinct cofactors and chromatin modification to accomplish their dual functions in transcription (Wolffe et al. 1997; Urnov and Wolffe 2001). Whereas activation by liganded receptors is believed to associate with the recruitment of coactivators and targeted acetylation of chromatin, recent studies indicate that repression by unliganded receptors require corepressors SMRT and N-CoR and recruitment of HDAC activities (Glass and Rosenfeld 2000; Hu and Lazar 2000; Ordentlich et al. 2001; Urnov and Wolffe 2001).
Three well-characterized class I HDAC-containing complexes have been described as follows: the HDAC1/2-containing Sin3 (Zhang et al. 1997) and Mi-2/NURD/NuRD complexes (Wade et al. 1998; Xue et al. 1998; Zhang et al. 1998) (referred to as Sin3 and Mi-2/NURD complexes hereafter) and the HDAC3-containing SMRT and N-CoR complexes (Guenther et al. 2000; Li et al. 2000; Wen et al. 2000). Each of these complexes has been implicated in repression by unliganded TR and RAR. The identification of the Sin3 complex and the demonstration that the corepressors SMRT and N-CoR physically interact with Sin3 provided the first working model implicating HDACs in repression by unliganded receptors (Heinzel et al. 1997; Nagy et al. 1997). The importance of the Sin3 complex in repression by unliganded receptors is underscored by the ability of microinjected Sin3 and HDAC1/2 antibodies to block repression (Heinzel et al. 1997). However, the purified mammalian Sin3 complex contains neither SMRT nor N-CoR (Zhang et al. 1997). Furthermore, neither Sin3 nor HDAC1/2 are present in the purified SMRT complex (Guenther et al. 2000; Li et al. 2000), implying that the involvement of the Sin3 complex is not mediated through direct interaction with SMRT and N-CoR. In addition to the Sin3 complex, the Mi-2/NURD complex also appears to be involved in repression by unliganded TR, because the repression can be partially relieved through microinjection of Mi-2β/CHD4 antibodies (Xue et al. 1998). Whereas the functional evidence clearly suggests the involvement of both the Sin3 and Mi-2/NURD complexes in repression by unliganded receptors, the mechanisms by which these complexes are recruited is not yet clear.
The recent biochemical purification of SMRT and N-CoR indicates that both SMRT and N-CoR are associated with HDAC3 and a transducin β-like protein (TBL1) in large protein complexes 1.5–2 MD in size (Guenther et al. 2000; Li et al. 2000). Thus, SMRT and N-CoR define a new functional category of class I HDAC-containing complex. The interaction of SMRT and N-CoR with HDAC3 stimulates its histone deacetylase activity (Wen et al. 2000; Guenther et al. 2001), highlighting its functional significance. Recent studies also indicate that class II HDACs including HDAC4, HDAC5, HDAC6, and HDAC7 can interact directly with N-CoR and SMRT (Huang et al. 2000; Kao et al. 2000), although only a small fraction of endogenous N-CoR or SMRT appears to associate with the class II HDACs (Huang et al. 2000).
We have utilized chromatin immunoprecipitation (ChIP) to assay active recruitment of three class I HDAC-containing complexes by unliganded TR. Repression by unliganded TR correlated with the recruitment of SMRT/N-CoR/HDAC3 complexes, but not with the recruitment of the Sin3 and Mi-2/NURD complexes. In contrast, repression by Mad1 featured recruitment of the Sin3/HDAC1/2 complex. Notably, both the Sin3 and NURD complexes displayed constitutive association with chromatin consistent with roles in global deacetylation. We propose that both the Sin3 and NURD complexes contribute to the repression by unliganded nuclear receptors, not because they are actively recruited by unliganded nuclear receptors, but because they contribute to the global deacetylation of chromatin.
Results and Discussion
Repression by unliganded TR is correlated with histone deacetylation
Using Xenopus oocytes as a model system for the study of transcriptional regulation by TR in the context of chromatin, we have reported previously that repression by unliganded TR requires HDAC activity (Wong et al. 1998). Recent experiments indicate that the known class I HDAC-containing complexes, including Sin3, Mi-2/NURD, and SMRT/N-CoR/HDAC3 are highly conserved between Xenopus and humans (Wade et al. 1998; Vermaak et al. 1999; Li et al. 2000). Thus, we wished to identify and compare the HDAC complex(es) recruited specifically by unliganded TR as well as by Mad1 using ChIP assays, as previous studies indicated that repression by Mad repressors required the Sin3-HDAC1/2 complex as well as N-CoR (Alland et al. 1997; Laherty et al. 1997). For this purpose, we constructed a Xenopus TRβA promoter-based reporter containing four Gal4-binding sites (4xUAS) (Fig. 1A). The reporter plasmid was assembled into chromatin with regularly spaced nucleosomes via the replication-coupled pathway (Almouzni and Wolffe 1993), as confirmed by Southern blot hybridization following partial micrococcal nuclease digestion (Fig. 1B). Expression of a Gal4–TR fusion protein in Xenopus oocytes led to repression of transcription in the absence of T3 and activation of transcription in the presence of T3 (Fig. 1C), resembling the reported result for TR/RXR heterodimers (Wong et al. 1995). The repression by Gal4–TR requires HDAC activity, as it can be blocked by addition of an HDAC inhibitor, trichostatin A (TSA) (data not shown; see Fig. 4D, below). To assess whether the repression by unliganded Gal4–TR is associated with chromatin deacetylation, we next carried out ChIP assays using an antibody specific for hyperacetylated histone H3 or H4, respectively. Subsequent PCR analysis (Fig. 1D) and quantification by PhosphorImaging (in each ChIP assay, the number in each lane was the relative value in comparison to 1 set for the control) revealed that expression of Gal4–TR led to a substantial decrease in levels of acetylation of histones H3 and H4 over the promoter proximal region only in the absence of T3. These results indicate that the repression by unliganded Gal4–TR is associated with chromatin deacetylation. The observed chromatin deacetylation was targeted specifically by the Gal4–TR to the 4xUAS proximal region, as no alteration in levels of acetylation of both H3 and H4 was detected in a region distal to the 4xUAS (∼2.5 kb) (data not shown; see Fig. 3B, below).
Figure 1.
Repression by unliganded TR is coupled to chromatin deacetylation and recruitment of the HDAC3-containing SMRT/N-CoR complexes, but not the Sin3 and Mi-2/NURD complexes. (A) The diagram illustrates the structure of the 4xUAS-TRβA reporter. Also indicated is the region for PCR amplification in the ChIP assay. (B) Partial MNase digestion confirmed the assembly of the injected 4xUAS-TRβA reporter DNA into chromatin. (C) Gal4–TR repressed transcription from the 4xUAS-TRβA reporter in the absence of T3 and activated in the presence of T3. The expression of Gal–TR was accomplished by injection of in vitro synthesized mRNA. (Ctrl) The primer extension product from the oocyte endogenous histone H4 mRNA, which serves as a loading control. Also shown was the expression of Gal4–TR by Western analysis using a Gal4–DBD-specific antibody. (D) ChIP assay using antibodies as indicated. The injection of oocytes was as in C. Input DNA used for PCR reaction was 5% of total input DNA. Negative control (−) for ChIP assay was performed as others but without addition of antibody. (E) ChIP assay using antibodies (serum) and corresponding preimmune serum (pre-) revealed the specific association of Sin3 and Mi-2/NURD complexes with chromatin.
Figure 4.
Both the Sin3 and Mi-2/NURD complexes are likely to contribute to a global deacetylation of chromatin. (A) ChIP assays revealed that both the Sin3 and Mi-2/NURD complexes were constitutively associated with various chromatin templates. The structure of each reporter and the relative position of PCR products of ChIP assay were shown at right. (Lane 1) ChIP assay without antibody (beads only, negative control); (lanes 2,4,6) ChIP assays using preimmune serum (pre-); (lanes 3,5,7) ChIP assays using serum against CHD4, Sin3, and HDAC1, respectively. (B) TSA treatment led to increased histone acetylation. Groups of oocytes as in A were treated with (+) or without (−) TSA (0.3 μM) overnight, and acetylation of histones H3 and H4 were analyzed by ChIP assays. Input DNA used for PCR reaction in lanes 1 and 2 was 5% of total input DNA, whereas lanes 3 and 4 were control ChIP assays without addition of antibody (beads only) (C). TSA treatment revealed the repression of transcription by no-targeted deacetylation. The oocyte injection and TSA treatment were as in B. After overnight incubation, the levels of transcription were assayed by primer extension. (*) The expected product from each reporter. (D) The nontargeted deacetylation also contributes to repression by both unliganded TR and Mad1. Groups of oocytes were injected with the 4xUAS-TRβA reporter and mRNA encoding Gal–TR or Gal–Mad1 and treated with (+) or without (−) TSA as indicated. After overnight incubation, levels of transcription were determined by primer extension assay (top), and the levels of histone H3 acetylation were determined by ChIP assay (bottom).
Figure 3.
The SMRT/N-CoR complexes but not the Sin3 complexes are recruited by unliganded TR/RXR heterodimers and tethering of either HDAC3 or HDAC1 is sufficient for repression. (A) The expression of TR/RXR heterodimers repressed transcription in the absence of T3 and activated transcription in the presence of T3. (B) ChIP assays examining the TRE proximal region (PCR A) and a distal region (∼2 kb) (PCR B). The ChIP assay conditions were as in Figure 1D, except different sets of primers were used for PCR. Note that the association of CHD4, Sin3A, and HDAC1 with both the TRE proximal and the distal region was detected, whereas the association of N-CoR, SMRT, and HDAC3 was detected only in the TRE vicinity with unliganded TR/RXR. (C) Tethering of HDAC1 and HDAC3 to chromatin is sufficient for repression. The transcription assay was as in Figure 1C except that the mRNA encoding Gal4–HDAC1 or Gal4–HDAC3 was used. Three dilutions of mRNA corresponding to 3, 1, and 0.3 ng/oocyte were injected, respectively, for Gal4–HDAC1 (lanes 2–4) and Gal4–HDAC3 (lanes 4–6). Also shown was the Western analysis showing the expression of Gal4–HDAC1 and Gal4–HDAC3.
Unliganded TR recruits SMRT/N-CoR/HDAC3
Next, we carried out ChIP assays to determine which of the HDAC-containing complexes could be recruited by Gal4–TR. As shown in Figure 1D, the association of N-CoR with chromatin was only detected in the presence of Gal4–TR and in the absence of T3 (with a 12-fold increase). Similar results were observed for SMRT and HDAC3. These results indicate that Gal4–TR actively recruits the corepressors SMRT, N-CoR, and HDAC3 in the absence of T3. These ChIP results are consistent with the recent biochemical purification of SMRT and N-CoR complexes from HeLa cells, showing that HDAC3 is a subunit of the SMRT and N-CoR complexes (Guenther et al. 2000; Li et al. 2000).
Both Sin3 and Mi-2/NURD complexes exhibited constitutive chromatin association and were not recruited by unliganded TR
We next carried out ChIP assays to determine whether the Sin3 and Mi-2/NURD complexes would also be recruited by unliganded Gal4–TR. ChIP assays revealed the constitutive association of the Mi-2/NURD complex with chromatin in the absence of Gal4–TR (Fig. 1D). No increased association of Mi-2/CHD4 with chromatin was observed in the presence of Gal4–TR, indicating that unliganded Gal4–TR did not recruit the Mi-2/NURD complex. Similarly, Xenopus Sin3A and HDAC1/2 also associated with chromatin in a Gal4–TR independent manner (Fig. 1D). Thus, in contrast to the clear evidence for the active recruitment of SMRT, N-CoR, and HDAC3, neither the Mi-2/NURD nor the Sin3 complex was recruited by Gal4–TR in the absence of T3.
As controls, we performed parallel ChIP assays comparing Sin3A, HDAC1, and Mi-2/CHD4 with their corresponding preimmune serum. In each case (Fig. 1E), much less DNA was pulled down by the corresponding preimmune serum (indicated as pre-). The constitutive association of the Mi-2/NURD complex with chromatin was further supported by ChIP assays with two different affinity purified CHD4 antibodies (data not shown) and with an antibody against MBD3, a different subunit of the Mi-2/NURD complex (Wade et al. 1999; Fig. 1E). We thus concluded that both the Sin3 and Mi-2/NURD complexes show constitutive association with chromatin and that unliganded TR recruits neither complex.
Repression by Mad1 correlates with recruitment of Sin3–HDAC1/2 complex
As repression by Mad1 was reported to involve both Sin3 and SMRT/N-CoR (Alland et al. 1997; Laherty et al. 1997), we next wished to identify the complexes recruited by the repressor Mad1. For this purpose, we constructed a Gal4 fusion protein containing the amino acids 1–47 of Mad1, which is known to contain a repression domain and be sufficient for interaction with Sin3 (Eilers et al. 1999). Expression of the Gal4–Mad1 fusion in Xenopus oocytes was sufficient to repress transcription from the 4xUAS-TRβA reporter (Fig. 2A). As a control, the expression of Gal4–VP16 activated transcription (Fig. 2A). ChIP assays showed that, similar to unliganded Gal4–TR, expression of Gal4–Mad1 also led to a targeted deacetylation of both H3 and H4 (Fig. 2B). Consistent with previous findings that Mad1 interacts with Sin3A, the ChIP assay revealed a substantially increased association of Sin3A with chromatin (6.5-fold). Moreover, the association of HDAC1 was also increased (6.2-fold), consistent with the notion that HDAC1 is a component of the Sin3 complex (Zhang et al. 1997). In contrast to the results with Gal4–TR (Fig. 1D), expression of Gal4–Mad1 did not lead to an increase in association of SMRT, N-CoR, or HDAC3 with chromatin (Fig. 2B). Furthermore, recruitment of Sin3 and HDAC1 was not observed with Gal4–VP16 under the same experimental conditions (Fig. 2B) or with the Gal–DBD control (data not shown), indicating that the Sin3 complex was actively recruited by Gal4–Mad1. Together with the results that Gal4–TR actively recruits the HDAC3-containing SMRT and N-CoR complexes, we conclude that Sin3 and SMRT/N-CoR demarcate distinct HDAC-containing complexes that can be actively recruited for local chromatin deacetylation and repression by distinct repressors.
Figure 2.
The repressor Mad1 recruits the HDAC1/2-containing Sin3 complex, but not the SMRT/N-CoR complexes. (A) Expression of Gal4–Mad1 through microinjection of Gal4–Mad1 mRNA synthesized in vitro led to a dose-dependent repression. The experimental conditions were as in Figure 1C and the expression of Gal4–Mad1 and Gal4–VP16 was detected by Western analysis using a Gal4–DBD-specific antibody. (B) ChIP assay revealed that the repression by Mad1 is coupled to the chromatin deacetylation and recruitment of the Sin3 complex. The ChIP assay and PCR reactions were performed as in Figure 1D.
Unliganded TR/RXR also recruit only the SMRT/N-CoR/HDAC3
The Gal4–TR proteins used in the above experiments contain the ligand-binding domain but not the full-length TR. Whereas these results indicate that recruitment of the SMRT/N-CoR complexes does not result in the recruitment of the Sin3 complex, and vice versa, we cannot exclude the possibility that the TR/RXR heterodimer may recruit the Sin3 complex. To test this possibility, we used a reporter containing the Xenopus TRβA promoter, which contains a native T3 response element (TRE) located downstream of the transcriptional start site. Expression of TR/RXR heterodimers in Xenopus oocytes repressed transcription in the absence of T3, and activated transcription in the presence of T3 (Fig. 3A). ChIP assays confirmed that the repression by unliganded TR/RXR was associated with the deacetylation of both H3 and H4 and with the recruitment of N-CoR, SMRT, and HDAC3 (Fig. 3B). Deacetylation of histones and recruitment of N-CoR, SMRT, and HDAC3 were targeted by unliganded TR/RXR to the TRE region, because neither were detected in a region ∼2 kb downstream of the TRE site (PCR B). Importantly, we found no evidence for the recruitment of Sin3A and HDAC1 by unliganded TR/RXR under the same conditions (Fig. 3B). However, the association of the Sin3 and NURD complexes with chromatin was detected in both the TRE (PCR A) and the control region (PCR B) (Fig. 3B), further supporting the idea that both the Sin3 and NURD complexes exhibit constitutive association with chromatin.
Our results thus far indicate that the repression exerted by unliganded TR or Mad1 correlates with a targeted deacetylation of chromatin and the active recruitment of the HDAC3-containing SMRT/N-CoR complexes or the HDAC1/2-containing Sin3 complex, respectively. These results raise a question as to whether tethering of HDAC3 or HDAC1 alone to chromatin could be sufficient for repression in Xenopus oocytes. Expression of both Gal4–HDAC1 and Gal4–HDAC3 led to a dose-dependent repression of transcription from the 4xUAS-TRβA reporter (Fig. 3C). Thus, as shown previously in mammalian cells (Hassig et al. 1998), tethering a single HDAC to chromatin is sufficient for repression in Xenopus oocytes. As expected, this repression is sensitive to inhibition by TSA (data not shown).
The constitutive chromatin association of the Sin3 and Mi-2/NURD complexes likely contributes to global chromatin deacetylation and chromatin-mediated repression
The results that tethering a single HDAC to chromatin is sufficient to repress transcription (Fig. 3C) and that both TR and Mad1 recruit a single, unique HDAC complex, apparently contradict the observed requirement for multiple HDAC complexes for repression. The association of both the Sin3 and Mi-2/NURD complexes with chromatin in the absence of TR and Mad1 led us to hypothesize that these complexes could contribute to histone deacetylation and repression through their intrinsic association with chromatin in the absence of active recruitment by sequence-specific repressors. To test this idea, we first examined whether their observed chromatin association is unique to the TRβA reporter. Two additional reporters (TK- and MMTV-CAT) were assembled into chromatin via a replication-coupled pathway in Xenopus oocytes and the association of the Sin3 and Mi-2/NURD was determined by ChIP assay. Both complexes were found to be associated with all the regions tested (Fig. 4A). Thus, the observed chromatin association most likely reflects an intrinsic association of both the Sin3 and Mi-2/NURD complexes with chromatin.
Having established their likely intrinsic chromatin association, we next wished to show that the Sin3 and Mi-2/NURD complexes contribute to chromatin deacetylation and repression in the absence of specific repressors. Although it is clear that the HDAC1/2 present in the Sin3 and Mi-2/NURD complexes represents the major HDAC activity in Xenopus oocytes (Wade et al. 1998; Vermaak et al. 1999), no reagent that could block specifically the HDAC1/2 activity in these complexes is currently available. Therefore, we used TSA, a specific HDAC inhibitor, to evaluate the potential contribution of these enzymes to the properties of chromatin assembled via the replication-coupled pathway. We assembled three different reporters into chromatin via this pathway and tested the effect of TSA treatment on acetylation and transcription. ChIP assays revealed that TSA treatment led to a two- to threefold increase in acetylation of H3 and H4 over all regions that we have tested on the TRβA, TK, and MMTV promoters (Fig. 4B). These results indicate that chromatin is under dynamic acetylation and deacetylation and that the Sin3 and Mi-2/NURD complexes are likely the major contributors to such nontargeted, global deacetylation. Importantly, this nontargeted global deacetylation appears to be important for repression mediated by chromatin assembly, as TSA treatment substantially enhanced transcription from all three reporters assembled into chromatin via replication-coupled pathway (Fig. 4C).
The contribution of global deacetylation to the repression by TR and Mad1 could also be revealed by TSA treatment. Both unliganded Gal4–TR and Gal4–Mad1 repressed transcription from the 4xUAS-TRβA reporter (Fig. 4D, cf. lane 1 with lanes 3 and 5). The addition of TSA not only blocked this repression, but also elevated transcription beyond the basal level (Fig. 4C, cf. lane 1 with lanes 4 and 6). Consistent with its effect on transcription, TSA treatment blocked the deacetylation targeted by Gal4–TR or Gal4–Mad1 and elevated the levels of acetylation beyond that in the control (Fig. 4A, cf. lane 1 with lanes 4 and 6). Thus, the final levels of chromatin deacetylation, as well as repression by unliganded TR and Mad1, appears to be determined by the combined effect of an actively targeted HDAC complex (SMRT/N-CoR/HDAC3 by TR and Sin3 by Mad1) and the global association of the Sin3 and Mi-2/NURD complexes with chromatin.
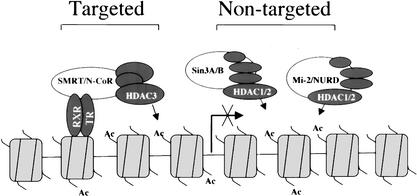
A working model for repression by unliganded receptors
Our results provide clear evidence that unliganded TR recruits the HDAC3-containing SMRT/N-CoR complexes but not the Sin3 and Mi-2/NURD complexes, and that Mad1 recruits the Sin3 complex but not the SMRT/N-CoR complexes. Both the Sin3 and Mi-2/NURD complexes exhibit constitutive association with chromatin and most likely contribute to chromatin deacetylation in a global fashion. On the basis of these observations, we propose a working model (Fig. 5), in which unliganded receptors specifically recruit the HDAC3-containing SMRT and N-CoR complexes to their target genes. Both the Sin3 and Mi-2/NURD complexes contribute to this repression, not because they are actively recruited by unliganded NRs, but because they contribute to the global dynamic deacetylation of chromatin. Both the Sin3 and Mi-2/NURD complexes are well suited for a role in global chromatin deacetylation, because both complexes contain RbAp46 and RbAp48 subunits, which have capacity to interact directly with histones (Parthun et al. 1996), and because both complexes are abundant and are primarily localized in the nucleus. This model unifies well the current biochemical data that both Sin3 and SMRT/N-CoR are present in distinct HDAC-containing complexes and the functional data that the Sin3 and Mi-2/NURD complexes are involved in repression by unliganded NRs. Furthermore, this model may be generally applicable to other repressors.
Figure 5.
A working model for the involvement of three class I HDAC-containing complexes in repression by unliganded NRs. In this model, only HDAC3-containing SMRT and N-CoR complexes were actively recruited through a direct interaction between unliganded TR and corepressors SMRT or N-CoR, which in turn lead to a local chromatin deacetylation and repression. On the other hand, both the HDAC1/2-containing Sin3 and Mi-2/NURD complexes are not recruited by unliganded receptors. Instead, they contribute to repression by unliganded receptors through their ability to deacetylate chromatin in a global fashion as a result of their intrinsic association with chromatin.
Materials and methods
Plasmid constructs
The 4xUAS-TRβA reporter was generated by inserting four copies of 17mer Gal4 DNA-binding site into the NdeI site in the TRβA promoter (−445 to transcriptional start site; Wong et al. 1995). The TK-CAT reporter was generated by subcloning the EcoRI–EcoR fragment containing the TK promoter from the pBLCAT2 into pBluescript II. The MMTV-CAT reporter was constructed by replacing the TK promoter in the TK-CAT reporter with a 1.5 kb XhoI–BamH1 fragment containing the MMTV LTR. The preparation of ssDNA of reporters was as described (Wong et al. 1995). To make Gal4–TR and Gal4–Mad1 expression constructs, the DNA fragment encoding Gal4–DBD (amino acids 1–147) was first cloned into pSP64 poly(A) (Promega). The DNA fragments corresponding to the ligand-binding domain of Xenopus TRβA (amino acids 86–369) and amino acids 1–47 of Mad1 were then inserted in-frame with the Gal–DBD to generate Gal4–TR and Gal4–Mad1. The reporter pTRβA and TR and RXR expression plasmids have been described previously (Wong et al. 1995).
Microinjection of Xenopus oocytes and subsequent analyses of protein expression, chromatin structure, and transcription
Preparation and microinjection of mRNAs and reporter DNA into stage VI Xenopus oocytes were as described previously (Wong et al. 1995). In general, mRNA was injected at a concentration of 100 ng/μL (18.4 nL/oocyte) 2–3 h before the injection of reporter ssDNA (50 ng/μL, 18.4 nL/oocyte). The injected oocytes were incubated at 18°C overnight and processed for transcriptional analysis by primer extension and assay for chromatin structure by partial micrococcal nuclease digestion as described previously (Wong et al. 1995). The internal control for transcription is the primer extension product of the Xenopus storage histone H4 mRNA. Western analyses for Gal–DBD fusion proteins were carried out by use of a Gal–DBD-specific antibody from Santa Cruz Biotechnology (sc-510).
ChIP assay
The ChIP assays for recruitment of corepressor complexes and histone acetylation were essentially as described (Shang et al. 2000) with the following modifications. After overnight incubation, the groups of injected oocytes were treated with 1% formaldehyde for 10 min and homogenized in buffer (50 μL/oocyte) (20 mM Tris at pH 7.6, 60 mM KCl, 3 mM CaCl2, and 1 mM DTT) by pipetting. Micrococcal nuclease (1 U/50 μL extract) was added to the homogenates to digest chromatin into 400–500-bp length fragments. The digestion was stopped by addition of 10 mM EGTA and centrifuged. The extracts were used for ChIP assay (each reaction with 100 μL extract and 1–2 μL of individual antibody) as described (Shang et al. 2000). The antibodies against acetylated H3 and H4 and HDAC3 were purchased from Upstate Biotechnology. The antibodies against Xenopus Sin3A and HDAC1 were kind gifts from Drs. Yun-Bo Shi and Peter Jones (NICHD/NIH, Bethesda, MD). The antibodies against SMRT, N-CoR, and CHD4 have been described previously (Xue et al. 1998; Li et al. 2000). The final PCR reactions were carried out with inclusion of 1 μCi of [32P]dCTP in each PCR reaction, and the PCR product was visualized by autoradiography after fractionation by use of a 6% native polyacrylamide gel. The quantification of ChIP results were carried out by use of a PhosphorImager and by setting the value in control samples as 1.
Acknowledgments
We thank Drs. Yun-Bo Shi and Peter Jones for Sin3 and HDAC1 antibodies, Don Ayers for Mad1 cDNA, and Dr. Sophia Tsai for critical reading and comments. This work was supported by NIH grants to J.W. and NICHD (5K22HD01238–02) to P.W. This work is dedicated to the memory of Alan Wolffe.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jwong@bcm.tmc.edu; FAX (713) 790-1275.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.962502.
References
- Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Almouzni G, Wolffe AP. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes & Dev. 1993;7:2033–2047. doi: 10.1101/gad.7.10.2033. [DOI] [PubMed] [Google Scholar]
- Eilers AL, Billin AN, Liu J, Ayer DE. A 13-amino acid amphipathic alpha-helix is required for the functional interaction between the transcriptional repressor Mad1 and mSin3A. J Biol Chem. 1999;274:32750–32756. doi: 10.1074/jbc.274.46.32750. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes & Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes & Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The smrt and n-cor corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassig CA, Tong JK, Fleischer TC, Owa T, Grable PG, Ayer DE, Schreiber SL. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metabol. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- Huang EY, Zhang J, Miska EA, Guenther MG, Kouzarides T, Lazar MA. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes & Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes & Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Ordentlich P, Downes M, Evans RM. Corepressors and nuclear hormone receptor function. Curr Top Microbiol Immunol. 2001;254:101–116. doi: 10.1007/978-3-662-10595-5_5. [DOI] [PubMed] [Google Scholar]
- Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: Links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Wolffe AP. A necessary good: Nuclear hormone receptors and their chromatin templates. Mol Endocrinol. 2001;15:1–16. doi: 10.1210/mend.15.1.0589. [DOI] [PubMed] [Google Scholar]
- Vermaak D, Wade PA, Jones PL, Shi YB, Wolffe AP. Functional analysis of the SIN3-histone deacetylase RPD3-RbAp48-histone H4 connection in the Xenopus oocyte. Mol Cell Biol. 1999;19:5847–5460. doi: 10.1128/mcb.19.9.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, Wong J, Pruss D. Activators and repressors: Making use of chromatin to regulate transcription. Genes Cells. 1997;2:291–302. doi: 10.1046/j.1365-2443.1997.1260323.x. [DOI] [PubMed] [Google Scholar]
- Wong J, Shi YB, Wolffe AP. A role for nucleosome assembly in both silencing and activation of the Xenopus TRβA gene by the thyroid hormone receptor. Genes & Dev. 1995;9:2696–2711. doi: 10.1101/gad.9.21.2696. [DOI] [PubMed] [Google Scholar]
- Wong J, Patterton D, Imhof A, Guschin D, Shi YB, Wolffe AP. Distinct requirements for chromatin assembly in transcriptional repression by thyroid hormone receptor and histone deacetylase. EMBO J. 1998;17:520–534. doi: 10.1093/emboj/17.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]