Abstract
Senescence limits the proliferative capacity of primary cells in culture. We describe here a genetic screen to identify genes that allow bypass of this checkpoint. Using retroviral cDNA expression libraries, we identify BCL6 as a potent inhibitor of senescence. BCL6 is frequently activated in non-Hodgkin's lymphoma, but its mechanism of action has remained unclear. BCL6 efficiently immortalizes primary mouse embryonic fibroblasts and cooperates with RAS in oncogenic transformation. BCL6 overrides the senescence response downstream of p53 through a process that requires induction of cyclin D1 expression, as cyclin D1 knockout fibroblasts are specifically resistant to BCL6 immortalization. We show that BCL6 expression also dramatically extends the replicative lifespan of primary human B cells in culture and induces cyclin D1 expression, indicating that BCL6 has a similar activity in lymphoid cells. Our results suggest that BCL6 contributes to oncogenesis by rendering cells unresponsive to antiproliferative signals from the p19ARF–p53 pathway.
Keywords: Senescence, BCL6, LAZ3, cyclin D1, p19ARF, p53, lymphoma
The proliferative capacity of most primary cells in culture is limited by the induction of senescence. This state of irreversible growth-arrest is characterized by expression of a number of senescence-associated markers, such as senescence-associated β-galactosidase, plasminogen-activator inhibitor 1 (PAI-1), p19ARF, p53, p21cip1, and p16INK4A (Sherr and DePinho 2000). In most rodent cells, induction of the tumor suppressor genes p19ARF and p53 is critical to the induction of senescence, as mutation of either gene allows escape from replicative senescence and causes immortalization (Harvey et al. 1993; Kamijo et al. 1997). Consistent with a critical role for the p19ARF–p53 pathway in the induction of senescence, it was shown recently that the transcriptional repressors BMI-1 and TBX-2 inhibit senescence through down-regulation of p19ARF expression (Jacobs et al. 1999, 2000). In contrast, genetic ablation of p16INK4A alone or p21cip1 in mouse embryonic fibroblasts (MEFs) does not render cells resistant to the induction of senescence (Pantoja and Serrano 1999; Krimpenfort et al. 2001), even though enforced expression of p16INK4A or p21cip1 (McConnell et al. 1998; Dai and Enders 2000) induces a senescent-like state in many cell types.
These data suggest that the p19ARF–p53 pathway is a critical regulator of the senescence response in murine fibroblasts, whereas the p16INK4A–pRb pathway is not. Genetic inactivation of the p16INK4A or Rb tumor suppressor genes alone is not sufficient to immortalize MEFs (Krimpenfort et al. 2001; Peeper et al. 2001). However, simultaneous ablation of both Rb and the related p107, or inactivation of all three Rb gene family members (Rb, p107, and p130) does render MEFs immortal, even though such cells have high levels of p19ARF and p53 (Dannenberg et al. 2000; Peeper et al. 2001). These data indicate that the Rb family proteins not only act upstream of the p19ARF–p53 pathway, through regulation of p19ARF by E2F (Bates et al. 1998), but also downstream, by rendering cells insensitive to p53 signaling.
BCL6 encodes a transcriptional repressor, which is frequently activated by chromosomal translocation in non-Hodgkin's lymphoma (Ye et al. 1993; Chang et al. 1996; Staudt 2000). The chromosomal translocations involving BCL6 invariably affect the promoter only and leave the ORF of BCL6 intact. BCL6 is required for normal B and T-cell development (Ye et al. 1997), but its broad expression suggests that it also has a role outside of the lymphoid compartment (Bajalica-Lagercrantz et al. 1998). Several studies have attempted to elucidate the molecular pathways that are targeted by BCL6. DNA microarray analyses have identified several targets of BCL6, including cyclin D2, but the relevance of these targets for the activity of BCL6 has not been elucidated (Shaffer et al. 2000). Therefore, the molecular pathways that are regulated by BCL6 during oncogenesis are still unclear.
Here we use an unbiased functional approach to identify genes that override the senescence response. Unexpectedly, we find that BCL6 efficiently inhibits senescence by conferring resistance to antiproliferative signals from the p19ARF–p53 pathway.
Results and Discussion
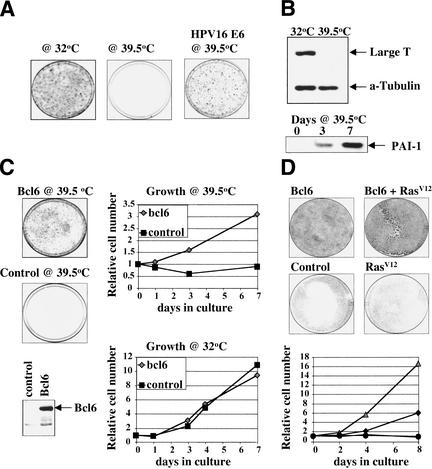
To identify genes that allow bypass of replicative senescence, we generated conditionally immortalized MEFs using a temperature-sensitive mutant of SV40 large T antigen (Lee et al. 1995). The tsT antigen-expressing MEFs (tsT-MEFs) are immortalized at the permissive temperature (32°C), but enter synchronously into a senescence-like state after shift to the nonpermissive temperature (39.5°C), due to rapid disappearance of T antigen (Fig. 1A,B). That these cells undergo a growth arrest reminiscent of replicative senescence is supported by the fact that several senescence markers, including p21cip1, PAI-1 and senescence-associated β-galactosidase, are induced upon shift to the nonpermissive temperature (Figs. 1B and 3A, below; data not shown). Because T antigen inactivates both pRb and p53 through direct binding, we asked whether inactivation of p53 alone is sufficient to inhibit the induction of senescence at the nonpermissive temperature. Figure 1A shows that expression of HPV16 E6, which binds and degrades p53 (Scheffner et al. 1990), allowed continued growth at the nonpermissive temperature, indicating that tsT-MEFs entered into a p53-dependent senescence program after temperature shift.
Figure 1.
tsT-MEFs enter into a p53-dependent senescence program after shift to the nonpermissive temperature. (A) tsT-MEFs were cultured at 32°C, shifted to the nonpermissive temperature (39.5°C), and stained 10 d later. (Right) tsT-MEFs were infected with the E6 retrovirus at 32°C, shifted to nonpermissive temperature and stained 10 d later. (B) Abundance of SV40 T antigen and α-tubulin in tsT-MEFs at both temperatures was determined by Western blotting. (Bottom) Expression of senescence marker PAI-1 in tsT-MEFs at various time points after shift to the nonpermissive temperature. (C) The full-length BCL6 cDNA was cloned in pBabe-hygro retroviral vector and used to infect tsT-MEFs at 32°C. Expression of BCL6 was detected by Western blotting. After 2 d, infected cells were shifted to the nonpermissive temperature, and stained after 10 d. Growth curves are shown of tsT-MEFs infected with BCL6-expressing or control retrovirus at 32°C and 39.5°C. (D) Primary FVB MEF infected with BCL6 alone or in combination with a RAS oncogene escape replicative senescence. In contrast, control (pBabe-hygro infected) and RASV12-infected MEFs fail to proliferate. Growth curves of passage 5 MEFs infected with indicated retroviruses are shown. (♦) Bcl6; (▪) control; (▴) Bcl6/Ras V12; (●) Ras V12.
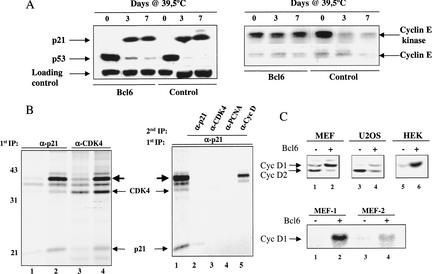
Figure 3.
BCL6 rescues senescence downstream of p53. (A) Western blot analysis of p21cip1 and p53 protein levels in tsT-MEFs expressing either BCL6 or control virus at indicated time points (days) after shift to the nonpermissive temperature. Cyclin E protein level was determined by Western blotting; Cyclin E-associated kinase activity was determined on cyclin E immunoprecipitates using histone H1 as a substrate. (B) (Left) Cells expressing either BCL6 or control virus were labeled with [35S]methionine and lysates were precipitated with either p21cip1 or CDK4 antibody and separated on SDS–polyacrylamide gels. (Lanes 1,3) FVB-MEFs (passage 5); (lanes 2,4) FVB MEFs expressing BCL6 (also at passage 5). (Right) [35S]methionine cell lysates of BCL6 cells were immunoprecipitated first with p21cip1 antibody in nonionic detergent, protein complexes were released by boiling in 1% SDS and reimmunoprecipitated with indicated antibodies. Individual proteins are indicated with arrows. (C) (Top) Western blot analysis for the expression of cyclin D1 and cyclin D2 in MEFs, human U2OS cells (osteosarcoma), and primary human embryonic kidney (HEK). (Lanes 1,3,5) Control; (lanes 2,4,6) BCL6-expressing derivatives. (Bottom) Northern blot analysis for the expression of cyclin D1 in Balb/c and FVB MEFs; (lanes 1,3) control; (lanes 2,4) BCL6-expressing MEFs.
To identify novel genes that allow bypass of this p53-dependent senescence program, we infected tsT-MEFs at 32°C with three high-complexity retroviral cDNA expression libraries. After infection, cells were replated at low density at the nonpermissive temperature. Continuously growing colonies appeared at low frequency only after infection with a cDNA expression library generated from peripheral blood lymphocytes of a patient suffering from polycythaemia vera, a myeloproliferative disorder. Integrated proviruses in rescued colonies were mobilized by Moloney virus superinfection as described (Jacobs et al. 2000) and cDNA inserts were recovered by PCR after second-round selection, and identified by DNA sequence analysis. Four independent colonies were found to carry a full-length BCL6 cDNA. The BCL6 gene is frequently activated by promoter translocation in non-Hodgkin's lymphoma and encodes a sequence-specific transcriptional repressor (Ye et al. 1993; Chang et al. 1996).
Figure 1C shows that BCL6 expressed in tsT-MEFs does not confer a growth advantage at 32°C, but selectively allows continued proliferation at 39.5°C (Fig. 1C). Importantly, when expressed in primary MEFs of FVB genetic background (at 37°C), BCL6 was very efficient in inhibiting both spontaneous senescence and premature senescence induced by a RASV12 oncogene (Serrano et al. 1997; Fig. 1D). Coexpression of BCL6 and RASV12 caused complete oncogenic transformation, as these cells also proliferated in soft agar and formed tumors in nude mice (data not shown). We have cultured both BCL6-immortalized MEFs and BCL6 + RAS-transformed MEFs for several months, indicating that BCL6 does not act merely to postpone senescence.
Induction of p19ARF expression, leading to activation of p53, is a critical step in the senescence response (Kamijo et al. 1997). To study how BCL6 inhibits senescence, we monitored the expression of the key components of the senescence pathway in BCL6-immortalized MEFs in comparison with late passage (pre-senescent) primary MEFs. Figure 2 shows that late passage primary MEFs express p19ARF protein, which causes a partial activation of p53, as evidenced by the high-level expression of the p53 target p21cip1 in these cells (Fig. 2, cf. lanes 1 and 5). Significantly, all six BCL6-immortalized MEF cell lines expressed as high levels of p19ARF as the late passage primary MEFs, indicating that BCL6 does not inhibit the induction of p19ARF expression to induce immortalization. Furthermore, BCL6-immortalized MEFs also expressed high levels of the p53 target gene p21cip1, indicating that BCL6 also does not prevent the activation of p53 by p19ARF. Together, these data suggest that activation of the p19ARF–p53 pathway, which occurs normally during the senescence response, is not suppressed by BCL6.
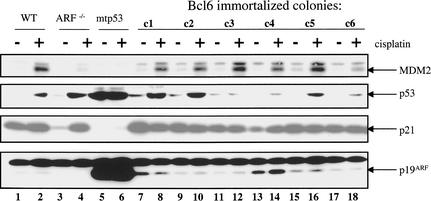
Figure 2.
The p19ARF–p53 pathway is intact in BCL6-immortalized MEFs. Western blot analysis of p53-pathway proteins in wild-type MEFs (P = 7) and BCL6-immortalized MEFs (P = 15), both before and 15 h after exposure to 40 μM cisplatin. Immortal MEFs of p19ARF−/− mice (ARF−/−) and spontaneously immortalized MEFs harboring a mutant p53 protein (mtp53) are shown for comparison.
Establishment of MEFs as continuously growing cell lines is almost invariably accompanied by loss of the p53 or p19ARF tumor suppressors. Several lines of evidence indicate that neither p19ARF nor p53 is mutated in late passage BCL6-immortalized MEFs. First, as p19ARF mutation in MEFs invariably leads to loss of protein expression (Kamijo et al. 1997; Zindy et al. 1998), the presence of clearly detectable levels of p19ARF in BCL6-immortalized MEFs argues that the p19ARF locus is still intact in these cells (Fig. 2). Second, the finding that expression of p19ARF leads to expression of the p53 target gene p21cip1 in BCL6-immortalized MEFs indicates that both p19ARF and p53 are functional. Third, when late passage BCL6-immortalized MEFs were exposed to the DNA-damaging agent cisplatin, all six lines of BCL6-immortalized MEFs induced p53 and its downstream target MDM2 (Barak et al. 1993), again indicating that p53 is not mutated in these cells (Fig. 2). Together, these data indicate that late passage BCL6-immortalized MEFs are not under selective pressure to mutate the p19ARF–p53 pathway.
To study the molecular mechanism by which BCL6 allows escape from senescence further, we expressed BCL6 in tsT-MEFs and monitored the p53 response following shift to the nonpermissive temperature. Figure 3A shows that both control and BCL6-expressing tsT-MEFs experience a dramatic reduction in p53 protein level upon shift to 39.5°C, resulting from the liberation of p53 from a T antigen complex. This, in turn, causes p53 to become active and activate its target p21cip1 (el Deiry et al. 1994; Fig. 3A). Importantly, p21cip1, MDM2, and Bax are induced to the same extent in BCL6-expressing and control tsT-MEFs (Fig. 3A; data not shown), again indicating that BCL6 acts downstream of the p53 pathway to override senescence. Unexpectedly, cyclin E-associated kinase activity was not inhibited as a result of p21cip1 induction in BCL6-expressing cells after temperature shift, whereas, as expected, cyclin E kinase activity was inhibited in control cells (Fig. 3A). As cyclin E and CDK2 protein levels were not elevated by BCL6 (Fig. 3A; data not shown), one possible model would be that p21cip1 is sequestered in BCL6 cells, rendering the protein unavailable to inhibit cyclin E/CDK2 kinase activity.
To examine whether BCL6 induces a sequestering protein that interacts with p21cip1, we labeled both BCL6 MEFs and control MEFs with [35S]methionine and performed a low-stringency immunoprecipitation (to preserve protein interactions) with p21cip1 antibody. Figure 3B shows that a protein of ∼35 kD (indicated by bold arrow) is associated with p21cip1 in BCL6 MEFs, but to a much lesser extent in control MEFs. A protein of the same electrophoretic mobility is also more abundant in a CDK4 immunoprecipitate of BCL6 MEFs (Fig. 3B), raising the possibility that this p21cip1-associated protein is a D-type cyclin. To address this, we carried out a sequential immunoprecipitation experiment. Figure 3B shows that the 35-kD p21cip1-associated protein reacted with an antibody against D-type cyclins. Subsequent analyses indicated that the 35-kD protein is most likely cyclin D1, as the cyclin D1 protein level is up-regulated by BCL6 in three different cell types (Fig. 3C), whereas in agreement with an earlier study, cyclin D2 was down-regulated by BCL6 (Shaffer et al. 2000). This up-regulation of cyclin D1 is at the level of transcription, as cyclin D1 mRNA was also increased by BCL6 in MEFs (Fig. 3C).
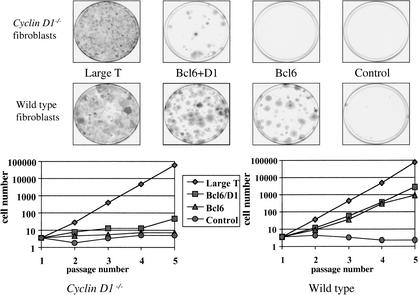
To address the relevance of the observed up-regulation of cyclin D1 by BCL6, we asked whether BCL6 could immortalize MEFs derived from cyclin D1 knockout mice (Sicinski et al. 1995). Figure 4 shows that BCL6-immortalized primary MEFs from wild-type mice, but not from littermate cyclin D1−/− mice. In contrast, T antigen immortalized both with equal efficiency. Importantly, coexpression of cyclin D1 allowed BCL6 to immortalize cyclin D1−/− MEFs. Together, these data provide genetic evidence that cyclin D1 is a critical downstream target of BCL6 in immortalization.
Figure 4.
BCL6 is unable to immortalize cyclin D1 knockout MEFs. Colony formation assay on primary MEFs at passage 5 infected with the BCL6 or BCL6 and cyclin D1 encoding retroviruses, and stained 10 d after infection. SV40 T antigen virus was used as a positive control (Large T). (Top four dishes) cyclin D1 knockout MEFs; (bottom four dishes) genotype-matched wild-type MEFs. Growth curves of cyclin D1−/− and matched wild-type control MEFs infected with the indicated retroviruses is shown at bottom. Passage numbers indicated reflect the number of passages after retroviral infection at P = 5.
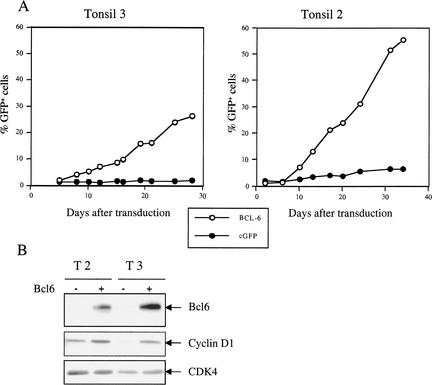
To ask whether BCL6 can also extend the proliferative capacity of primary B cells in culture, we infected EBV-negative human primary tonsillar B cells with a retroviral vector expressing both BCL6 and GFP or with control GFP vector. Infected cells were subsequently cultured in the presence of CD40 ligand, IL-2, and IL-4, and the percentage of GFP-positive cells was monitored over time. Figure 5 shows that both BCL6 and control virus infected primary human B cells at an efficiency of ∼2%, as judged by the percentage GFP-positive cells. In contrast to control GFP virus-infected cells, in two independent experiments, the fraction of BCL6-expressing cells increased steadily over time, indicating that BCL6-expressing B cells had a selective growth advantage (Fig. 5). Under the culture conditions used, primary human tonsillar B cells typically proliferate for up to 40 d. In contrast, we have been able to culture BCL6-infected B cells for over 4 mo without any signs of reduced proliferation (data not shown). These cells were CD19+; CD3−; CD56−, indicating that they were of B-cell origin. Furthermore, they expressed both immunoglobulin κ and λ light chains, indicative of the polyclonal nature of the B-cell population (data not shown). This indicates that expression of BCL6 also significantly extends the replicative lifespan of primary human B cells in culture. Importantly, BCL6-expressing B cells expressed higher levels of cyclin D1 protein as their mock-infected counterparts, suggesting that cyclin D1 up-regulation may also contribute to the increased proliferative capacity of BCL6-expressing human B cells (Fig. 5).
Figure 5.
BCL6 confers a growth advantage to primary human B cells. (A) Two independent batches of primary human tonsillar B cells were infected with retroviral vectors expressing either GFP (●) or BCL6 and GFP (○). The percentage of GFP-positive cells was followed over time in culture. (B) Western blot analysis of BCL6, cyclin D1, and control CDK4 expression in primary human B cells in culture and BCL6-infected derivatives.
We show here that BCL6 acts as an immortalizing oncogene by rendering fibroblasts resistant to the anti-proliferative signals emanating from the p19ARF–p53 pathway during the senescence response. We find that BCL6 circumvents these antiproliferative signals, at least in part, through up-regulation of cyclin D1 expression. This is critical for BCL6-immortalizing activity, as cyclin D1 knockout MEFs specifically resist BCL6-mediated immortalization. Our work adds BCL6 to a short list of oncogenes that strictly require cyclin D1 as a downstream target to mediate their oncogenic effects. Other genes that depend on cyclin D1 for oncogenic activity include ras and neu, whereas oncogenes like c-myc and Wnt-1 do not require cyclin D1 as a downstream target (Robles et al. 1998; Yu et al. 2001). However, our data by no means rule out that BCL6 has targets other than cyclin D1 in suppression of senescence.
The defect in germinal center formation of BCL6 knockout mice and the finding that BCL6 regulates genes involved in lymphocyte physiology have suggested that BCL6 regulates lymphocyte differentiation and the immune response (Ye et al. 1997; Shaffer et al. 2000). Our study suggests that during lymphomagenesis, BCL6 does not act primarily to regulate differentiation, but targets the p19ARF–p53 pathway. The importance of the p19ARF–p53 pathway in lymphomagenesis is illustrated by the finding that both p19ARF and p53 knockout mice have a high incidence of spontaneous lymphomas (Donehower et al. 1992; Kamijo et al. 1997). Suppression of this pathway may therefore contribute significantly to lymphomagenesis. Consistent with this, p19ARF-deficient murine preB cells proliferate indefinitely in vitro (Randle et al. 2001). Furthermore, several studies have found an absence of p53 mutations in BCL6-expressing human lymphomas (Ariatti et al. 2000). This suggests that these lymphomas have a reduced requirement to acquire mutations in p53, which is consistent with our finding that BCL6 acts downstream of p53. It is also important to point out that expression of BCL6 is not restricted to the lymphoid compartment only, which points at a broader role for BCL6 (Bajalica-Lagercrantz et al. 1998). We have observed expression of BCL6 in breast cancer (van't Veer et al. 2002) and SAGE databases provide further evidence that BCL6 is expressed in tumors of nonlymphoid origin. Thus, BCL6 may also act as an oncogene in solid tumors.
The p19ARF–p53 pathway is activated in a variety of cell types in response to a number of cellular stresses, including senescence. It has been suggested that the senescence response results primarily from the stress that cells experience when cultured in vitro (referred to as culture shock; Sherr and DePinho 2000). The isolation of a bona fide human oncogene in our genetic screen validates the cell system that we have developed and suggests that the stress that lymphoid cells experience during lymphomagenesis may be very similar to the stress invoked in vitro by the senescence response.
Materials and methods
Generation of MEFs expressing temperature-sensitive large T antigen
Primary Balb/c MEFs were infected at passage 2 at 32°C with pMESVTS retrovirus (Lee et al. 1995). G418-resistant colonies were assayed for the expression of large T antigen by Western blot analysis. Positive colonies yielding the lowest number of spontaneous background colonies at the nonpermissive temperature (39.5°C) were chosen for the functional genetic screens.
Retroviral rescue screens
To identify genes that allow bypass of senescence at the nonpermissive temperature, we infected tsT-MEFs at 32°C with retroviral cDNA libraries (Berns et al. 2000). One day post-infection, the cells were split 1 in 12, allowed to attach at 32°C, and then transferred to 39.5°C. Several individual colonies were visible as soon as 8 d after temperature shift. The colonies were picked and expanded. Recovery of integrated proviruses for second-round selection has been described (Jacobs et al. 2000). After second-round selection, retrovirally encoded cDNAs were recovered by PCR with retrovirus-specific primers and identified by sequence analysis.
Cell culture, growth curves, and retroviral infection
All cells were maintained in DMEM supplemented with 10% FCS. For immortalization assays and growth curves, primary FVB MEFs were infected with either empty pBabe-Hygro vector, or vectors encoding BCL6 or H-RASV12. Four days after infection, 2.5 × 104-infected cells were plated into 12-well plates in duplicate. At various time points, cells were processed for colorimetric analysis of cell proliferation as described (Serrano et al. 1997). Values were normalized to the optical density at day 0. Phoenix packaging cells were used to generate ecotropic retroviruses as described (Serrano et al. 1997). Low passage (p1–p3) MEFs were used to infect with viral supernatant supplemented with 4 μg/mL polybrene. Cyclin D1 knockout MEFs (Sicinski et al. 1995) and matched wild-type littermate controls of C57/bl6 genetic background were used at passage 5 for immortalization experiments.
Western blotting, kinase assays, and immunoprecipitations
Cell extracts were prepared in RIPA lysis buffer, assayed for protein concentration, and 20 μg of extract was resolved on 10%–12% SDS–polyacrylamide gels. Proteins were transferred to PVDF membranes. Primary antibodies used for Western blotting were C-19 for p21cip1, H-295 for cyclin D1, M-20 for cyclin D2, M-20 for cyclin E, and H-135 for PAI-1, all from Santa Cruz and M7211 (Dako) for BCL6. The p122 and p419 antibodies were used for the detection of p53 and T antigen, respectively. Enhanced chemiluminescence (Amersham) was used for detection of proteins.
Radioactive labeling of cells and immunoprecipitation has been described previously (Shvarts et al. 1996). After labeling, lysates (in NETN buffer) were used for immunoprecipitation with either C-19 p21cip1 antibody or a mix of H-22 and C-22 CDK4 antibodies (Santa Cruz). For sequential immunoprecipitations, antibody complexes were disassociated by boiling in 1% SDS for 5 min. Released proteins were diluted in RIPA lysis buffer and re-precipitated with C-19 for p21, Pc-10 for PCNA, M-20 for D-type cyclins, or a mix of H-22 and C-22 for CDK4.
Immune-complex kinase assays were performed as described (Dulic et al. 1992) by use of cyclin E M-20 antibody (Santa Cruz) and histone H1 as a substrate.
For Northern blotting, total RNA was extracted with RNAzol (Tel Test), separated on 1% agarose gel, transferred to nitrocellulose, and hybridized with murine cyclin D1 cDNA probe.
Primary human B cell isolation and culture
Human tonsils were obtained from tonsillectomies. T cells were depleted by use of anti-CD4 and anti-CD8 microbeads. Next, cells were incubated with anti-CD19 FITC conjugated and anti-CD3 phycoerythrin (PE) conjugated, followed by sorting of the CD19+ CD3− population. The resulting B cells were 95%–98% pure upon reanalysis. B cells were cultured in Iscove's medium together with 10% FCS at 37°C as described (Banchereau et al. 1991). Briefly, CD40L-expressing L cells, 80 Gray irradiated, were seeded 5.104 cells per well in 24-well plates. The 5.105-sorted B cells were added together with IL-2 (20 U/mL) and IL-4 (50 ng/mL). After 1 wk, the cells were used for retroviral transduction. After transduction, B cells were cultured again with irradiated CD40L-expressing L cells, IL2 and IL4.
Retroviral transduction of human B cells
Retroviral transductions of B cells were performed as described (Heemskerk et al. 1999). Briefly, 5.105 B cells in 0.25 mL Iscove's medium plus 10% FCS were mixed with 0.25 mL of thawed retroviral supernatant (pLZRS-BCL6-IRES-GFP) and were plated in fibronectin-coated nontissue culture-treated 24-well plates, and incubated for 6 h at 37°C. Next, 0.25 mL of supernatant was removed, and 0.25 mL of fresh retroviral supernatant was added and incubated at 37°C overnight.
Acknowledgments
We thank Norman Drinkwater for the kind gift of the tsT antigen vector, Peter Sicinski for cyclin D1 knockout fibroblasts, Xiaoling Liu for the murine cyclin D1 probe, Reuven Agami and Daniel Peeper for critical reading of the manuscript, and Marielle Hijmans for carrying out Western blots. This work was supported by grants from the Dutch Cancer Society and the Human Frontiers Science Program and the National Institutes of Health. G.Q.D. is the Birnbaum Scholar of the Leukemia and Lymphoma Society of America and a recipient of a Burroughs Wellcome Career Award in the Biomedical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bernards@nki.nl; FAX 31-20-512-1954.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.929302.
References
- Ariatti C, Vivenza D, Capello D, Migliazza A, Parvis G, Fassone L, Buonaiuto D, Savinelli F, Rossi D, Saglio G, et al. Common-variable immunodeficiency-related lymphomas associate with mutations and rearrangements of BCL-6: Pathogenetic and histogenetic implications. Hum Pathol. 2000;31:871–873. doi: 10.1053/hupa.2000.7626. [DOI] [PubMed] [Google Scholar]
- Bajalica-Lagercrantz S, Piehl F, Farnebo F, Larsson C, Lagercrantz J. Expression of the BCL6 gene in the pre- and postnatal mouse. Biochem Biophys Res Commun. 1998;247:357–360. doi: 10.1006/bbrc.1998.8551. [DOI] [PubMed] [Google Scholar]
- Banchereau J, de Paoli P, Valle A, Garcia E, Rousset F. Long-term human B cell lines dependent on interleukin-4 and antibody to CD40. Science. 1991;251:70–72. doi: 10.1126/science.1702555. [DOI] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Koh E, Daley GQ, Bernards R. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene. 2000;19:3330–3334. doi: 10.1038/sj.onc.1203639. [DOI] [PubMed] [Google Scholar]
- Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai CY, Enders GH. p16 INK4a can initiate an autonomous senescence program. Oncogene. 2000;19:1613–1622. doi: 10.1038/sj.onc.1203438. [DOI] [PubMed] [Google Scholar]
- Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G1 control causing immortalization and increased cell turnover under growth-restricting conditions. Genes & Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Dulic V, Lees E, Reed SI. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- el Deiry W, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill DE, Wang Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, Giovanella BC, Tainsky A, Bradley MA, Donehower LA. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- Heemskerk MH, Hooijberg E, Ruizendaal JJ, van der Weide MM, Kueter E, Bakker AQ, Schumacher TN, Spits H. Enrichment of an antigen-specific T cell response by retrovirally transduced human dendritic cells. Cell Immunol. 1999;195:10–17. doi: 10.1006/cimm.1999.1520. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van De Vijver MJ, Koh EY, Daley GQ, et al. Senescence bypass screen identifies TBX2, which represses cdkn2a (p19ARF) and is amplified in a subset of human breast cancers. Nat Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi W, Loonstra A, Berns A. Loss of Cdk2n2A(p16INK4a) confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- Lee GH, Ogawa K, Drinkwater NR. Conditional transformation of mouse liver epithelial cells. An in vitro model for analysis of genetic events in hepatocarcinogenesis. Am J Pathol. 1995;147:1811–1822. [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Starborg M, Brookes S, Peters G. Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr Biol. 1998;8:351–354. doi: 10.1016/s0960-9822(98)70137-x. [DOI] [PubMed] [Google Scholar]
- Pantoja C, Serrano M. Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene. 1999;18:4974–4982. doi: 10.1038/sj.onc.1202880. [DOI] [PubMed] [Google Scholar]
- Peeper DS, Dannenberg JH, Douma S, te Riele H, Bernards R. Escape from premature senescence is not sufficient for oncogenic transformation by Ras. Nat Cell Biol. 2001;3:198–203. doi: 10.1038/35055110. [DOI] [PubMed] [Google Scholar]
- Randle DH, Zindy F, Sherr CJ, Roussel MF. Differential effects of p19Arf and p16Ink4a loss on senescence of murine bone marrow-derived preB cells and macrophages. Proc Natl Acad Sci. 2001;98:9654–9659. doi: 10.1073/pnas.171217498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles AI, Rodriguez PM, Glick AB, Trempus C, Hansen L, Sicinski P, Tennant RW, Weinberg RA, Yuspa SH, Conti CJ. Reduced skin tumor development in cyclin D1-deficient mice highlights the oncogenic ras pathway in vivo. Genes & Dev. 1998;12:2469–2474. doi: 10.1101/gad.12.16.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, DePinho RA. Cellular senescence: Mitotic clock or culture shock? Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, et al. MDMX: A novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Parker SB, Li T, Fazeli A, Gardner H, Haslam SZ, Bronson RT, Elledge SJ, Weinberg RA. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- Staudt LM. The molecular and cellular origins of Hodgkin's disease. J Exp Med. 2000;191:207–212. doi: 10.1084/jem.191.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]