Abstract
Pax-3 is a transcription factor that is expressed in the neural tube, neural crest, and dermomyotome. We previously showed that apoptosis is associated with neural tube defects (NTDs) in Pax-3-deficient Splotch (Sp/Sp) embryos. Here we show that p53 deficiency, caused by germ-line mutation or by pifithrin-α, an inhibitor of p53-dependent apoptosis, rescues not only apoptosis, but also NTDs, in Sp/Sp embryos. Pax-3 deficiency had no effect on p53 mRNA, but increased p53 protein levels. These results suggest that Pax-3 regulates neural tube closure by inhibiting p53-dependent apoptosis, rather than by inducing neural tube-specific gene expression.
Keywords: Pax-3, P53, neural tube, apoptosis
Pax-3 encodes a DNA-binding transcription factor that is expressed in neuroepithelium, presomitic mesoderm, and neural crest (Goulding et al. 1991; Chalepakis et al. 1994). Homozygous Sp/Sp embryos carry loss-of-function Pax-3 alleles and develop open neural tube defects (NTDs), specifically, exencephaly, spina bifida, or both, with 100% penetrance, and die midgestation (Auerbach 1954; Epstein et al. 1993). The embryonic lethality is caused by defective cardiac neural crest migration and consequent malformation of cardiac outflow tracts (Conway et al. 1997; Epstein et al. 2000). Heterozygous Sp/+ embryos are viable, but manifest white patches of fur on a dark coat background, caused by defective neural crest-derived melanocyte development. Because of the failure of Pax-3-expressing structures to properly form in Splotch embryos, it has been accepted that Pax-3 regulates expression of differentiation-specific genes during development of these structures. In support of this hypothesis, Pax-3 has been shown to be upstream of myogenic gene expression, for example, of the genes Myf-5 and MyoD, during skeletal muscle development (Maroto et al. 1997; Tajbakhsh et al. 1997) and to inhibit expression of myelin basic protein during Schwann cell development (Kioussi et al. 1995). However, regulation of these genes by Pax-3 may be indirect, as none of the genes that have been found to be over- or underexpressed as a function of Pax-3 have high-affinity Pax-3 binding sites, with the exception of one element on the MyoD promoter (Phelan and Loeken 1998).
We showed previously that in Sp/Sp embryos, NTDs are associated with neuroepithelial apoptosis (Phelan et al. 1997). This suggested that disruption of a Pax-3-dependent developmental program may cause the malformed structures to undergo apoptosis. An alternative explanation is that Pax-3 directly or indirectly inhibits apoptosis. Several other studies lend support to the latter interpretation. For example, apoptosis is prevalent in somites of Splotch embryos (Borycki et al. 1999), and inhibition of Pax-3 expression with antisense oligonucleotides, or expression of an engineered PAX-3 fused to a transcriptional repressor domain, causes apoptosis in cultured presomitic mesoderm, pediatric rhabdomyosarcoma (RMS), and melanoma (Barr et al. 1993; Galili et al. 1993; Shapiro et al. 1993; Bernasconi et al. 1996; Borycki et al. 1999; Scholl et al. 2001). A question that is raised by these observations is whether inhibition of apoptosis is an essential, or even the sole, function of Pax-3 during development or transformation.
The product of the p53 tumor suppressor gene mediates apoptosis in response to many genotoxic stresses (Appella and Anderson 2001). p53-dependent apoptosis is responsible for elimination of transformed cells and suppression of tumor growth in vivo; loss of p53 function is associated with poor clinical prognosis of many human malignancies (Fisher 1994). Very few studies have investigated whether apoptosis during embryogenesis is p53-mediated. Notably, deficiency of XRCC4 or DNA ligase IV, both of which participate in nonhomologous end-joining DNA double-strand break repair and V(D)J recombination, causes embryonic lethality and massive neuronal apoptosis, and these effects are rescued by p53 deficiency (Frank et al. 2000; Gao et al. 2000). This suggests that XRCC4 and DNA ligase IV participate in DNA repair during normal embryogenesis, and that in the absence of DNA repair, affected structures undergo p53-dependent apoptosis. However, involvement of p53 in apoptosis involving DNA strand breaks is consistent with its response to genotoxic stress. Whether apoptosis associated with malformations caused by other mechanisms, such as Pax-3 deficiency, is p53-mediated has not been determined.
Results and Discussion
Inactivation of p53 rescues Pax-3-deficient embryos from neural tube defects
To investigate the involvement of p53 in apoptosis and NTD caused by Pax-3 deficiency, Sp/+ and p53+/− mice were crossed, and then double heterozygous progeny were mated to introduce a variable p53 genotype onto Sp/Sp embryos. When examined on embryonic day 10.5 (E10.5), as expected, all of the Sp/Sp embryos that were wild type at the p53 locus had developed open NTDs (Table 1). Remarkably, loss of both p53 alleles prevented NTDs in all Sp/Sp p53−/− embryos. Even p53 heterozygosity was sufficient to prevent NTD in 42% of Sp/Sp embryos. All Sp/Sp embryos whose NTDs were prevented by loss of one or both p53 alleles were indistinguishable from wild-type embryos, whereas Sp/Sp p53+/− embryos that were malformed were indistinguishable from Sp/Sp embryos on a wild-type p53 background. There was no significant effect of heterozygous or homozygous p53 mutation in Sp/+ embryos or in embryos that were wild type at the Splotch locus.
Table 1.
p53 deficiency suppresses NTD in Sp/Sp embryos
| Genotype
|
Normal
|
NTD
|
|---|---|---|
| Sp/Sp p53+/+ | 0 | 3 |
| Sp/Sp p53+/− | 20 | 28 |
| Sp/Sp 53−/− | 5 | 0 |
| +/Sp p53+/+ | 9 | 0 |
| +Sp p53+/− | 54 | 5 |
| +Sp p53−/− | 5 | 1 |
| +/+ p53+/+ | 3 | 0 |
| +/+ p53+/− | 22 | 1 |
| +/+ p53−/− | 3 | 1 |
Embryos from Sp/+ p53+/− matings were obtained on day 10.5 and scored for NTD (exencephaly and/or spina bifida). Yolk sac DNA was used for PCR at the Pax-3, p53, Zfy, and Nfl loci. There was no difference in the gender distribution in defective p53−/− or p53+/− embryos. There was a significant effect of Splotch genotype in p53+/+ embryos (P < 0.0006); in Sp/Sp embryos, the effect of p53 mutation to suppress NTD was statistically significant (P < 0.02); in Sp/− embryos and in embryos that were wild-type at the Splotch locus, there was no significant effect of p53 mutation.
To further test the involvement of p53 in NTD caused by Pax-3 deficiency, the effects of a p53 inhibitor, pifithrin-α, were tested. Pifithrin-α inhibits p53-dependent transcription and apoptosis (Komarov et al. 1999). The precise mechanisms are not known, but given that nuclear accumulation of p53 is reduced, this suggests that pifithrin-α stimulates nuclear export, inhibits nuclear import, or decreases p53 stability. Pregnant Sp/+ females that had been mated with Sp/+ males were administered pifithrin-α during formation of the neural tube (E8.5 and E9.5). As shown in Table 2, pifithrin-α prevented NTD in 55% of Sp/Sp embryos, whereas all Sp/Sp embryos whose mothers had been injected with saline developed NTD. As with Sp/Sp embryos whose NTDs were prevented by mutant p53 alleles, pifithrin-α-treated embryos without NTD were indistinguishable from embryos that were wild type at the Splotch locus, whereas those with NTD were indistinguishable from Sp/Sp embryos. The all-or-none effect of p53 deficiency on neural tube development suggests that there is a minimum threshold of p53 necessary to activate a program leading to an NTD, and that below this threshold, normal neural tube development ensues.
Table 2.
p53 inhibitor suppresses NTD in Sp/Sp embryos
| Genotype
|
PFT
|
Normal
|
NTD
|
|---|---|---|---|
| Sp/Sp | − | 0 | 4 |
| Sp/Sp | + | 6 | 5 |
| Sp/+ | − | 3 | 0 |
| Sp/+ | + | 3 | 0 |
| +/+ | − | 3 | 0 |
| +/+ | + | 2 | 0 |
Embryos were obtained from Sp/+ matings and scored for NTD as described for Table 1. Pregnant mice were injected with pifithrin-α (PFT) or saline on days 8.5 and 9.5. Yolk sac DNA was used for genotype determination at the Pax-3 locus. The effect of PFT on Sp/Sp embryos was statistically significant (P < 0.05).
Inactivation of p53 rescues Pax-3-deficient embryos from apoptosis
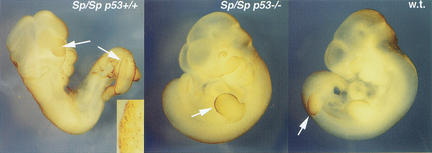
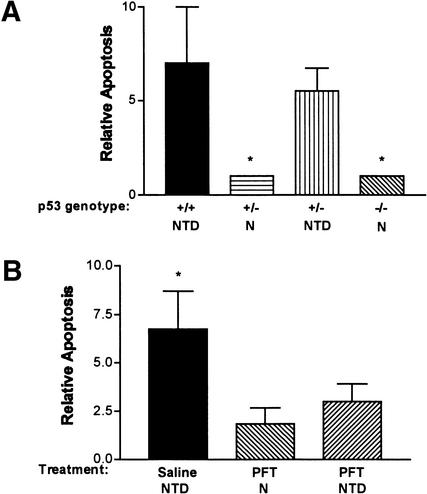
To determine whether p53 loss of function prevented apoptosis as well as NTD, Sp/Sp embryos were assayed for apoptosis by a whole-mount TUNEL procedure. As shown in Figure 1, numerous apoptotic cells were observed at defective sites of Sp/Sp p53+/+ embryos. However, Sp/Sp p53−/− and wild-type embryos were indistinguishable, and neuroepithelial apoptosis was not observed. Apoptotic cells were detectable along the apical ectodermal ridge of the limb buds in the Sp/Sp p53−/− embryos as well as the wild-type embryos, indicating that the apoptosis leading to digit formation is not p53-dependent. To quantitatively compare severity of apoptosis in embryos with variable p53 function, neuroepithelial apoptosis in Sp/Sp embryos was scored blindly on a scale of 1–10. As shown in Figure 2A, the apoptotic index of embryos with NTD was greater than in normal embryos, and high apoptosis scores were only observed in malformed embryos with one or two wild-type p53 alleles. Similar results were obtained upon TUNEL analysis of embryos in which p53 had been inhibited with pifithrin-α (Fig. 2B).
Figure 1.
Embryonic day 10.5 (E10.5) embryos following TUNEL assay to visualize apoptotic cells. (Left) Sp/Sp p53+/+ embryo (TUNEL score = 8). Exencephaly and spina bifida, with large numbers of apoptotic cells, indicated by arrows. (Inset) TUNEL-positive cells from the region of the spina bifida. (Center) Normal Sp/Sp p53−/− embryo (TUNEL score = 2). Apoptotic cells can be visualized along the apical ectodermal ridge of the limb bud (indicated by arrow), as expected for this stage of development, but not along fused neural tube. (Right) Wild-type (+/+ p53+/+) embryo (TUNEL score = 2). Apoptotic cells are also visible on limb bud (indicated by arrow), but not along neural tube. (Brown staining along dorsal surface of head of wild-type (w.t.) embryo is artefactual owing to tissue compression and translucency.)
Figure 2.
Effect of p53 inhibition on apoptosis in Sp/Sp embryos. (A) Relative apoptosis scores (on a scale of 1–10) in Sp/Sp embryos as a function of p53 genotype and normal or abnormal morphology. (N) Normal neural tube; (NTD) exencephaly and/or spina bifida. (*) P < 0.05 versus p53+/+ or p53+/− with NTD. (B) Relative apoptosis scores in Sp/Sp embryos whose mothers had been administered saline or pifithrin-α (PFT). Mean apoptosis scores for normal embryos and embryos with exencephaly and/or spina bifida are shown separately. (*) P < 0.05 versus PFT normal embryos.
These results show that loss of p53 function, by genetic or chemical means, prevented both apoptosis and NTD caused by Pax-3 deficiency. Given that neural tube development was normal in p53-deficient embryos despite the absence of Pax-3, this profoundly alters the concept by which Pax-3 controls neural tube development. These observations indicate that the apoptosis and NTD in Sp/Sp embryos do not result from the failure of a Pax-3-dependent neural tube-specific program. Rather, neural tube morphogenesis occurs by a mechanism that is Pax-3-independent, and Pax-3 simply keeps the cells alive until the program is complete by directly or indirectly inhibiting apoptosis by a p53-dependent mechanism.
Pax-3 down-regulates p53 protein, but not mRNA
PAX-5, as well as its paralogs PAX-2 and PAX-8, inhibits p53 gene expression, an effect that is mediated by PAX-5 binding to the p53 promoter (Stuart et al. 1995). Thus, in human astrocytomas, p53 deficiency caused by PAX-5-dependent transcriptional inhibition may contribute to tumor development. Similarly, the PAX-5 paralog PAX-8 may inhibit p53 gene expression in differentiated thyroid carcinomas (Puglisi et al. 2000). However, unlike PAX-5 and its paralogs, PAX-2 and PAX-8, there are no identifiable binding sites for Pax-3 within a 530-bp upstream region of the murine p53 gene (Bienz-Tadmor et al. 1985). On the other hand, regulation of p53-dependent apoptosis is primarily posttranslational, involving protein modifications such as phosphorylation or acetylation that regulate its stability (Appella and Anderson 2001). Therefore, an effect of Pax-3 on p53 protein levels could also be possible.
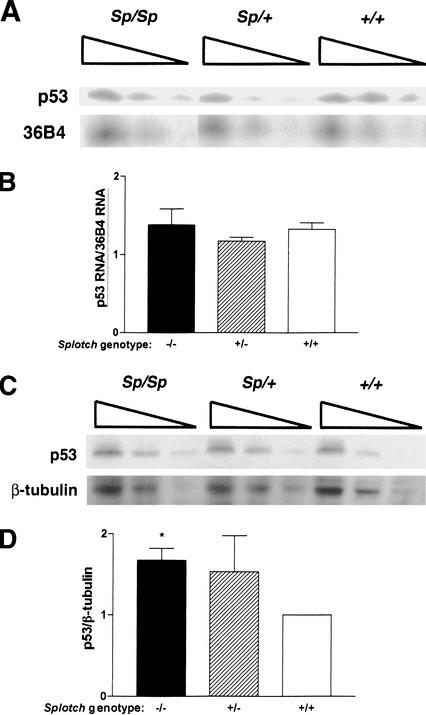
To test whether Pax-3 might regulate either p53 mRNA or protein levels, E10.5 embryos from Sp/+ matings were assayed for p53 mRNA by semiquantitative RT–PCR, or for p53 protein by Western blot analysis. There was no difference in p53 mRNA in embryos of different Splotch genotypes (Fig. 3A,B). However, p53 protein levels were increased almost twofold in Sp/Sp and Sp/+ embryos compared with wild-type embryos (comparing the amount of p53 protein from each genotype at intermediate dilution and the absence of p53 protein in wild-type embryos at greatest dilution), although only the differences between wild-type and Sp/Sp levels were statistically significant (Fig. 3C,D). It should be noted that, on E10.5, only the neuroepithelium, neural crest, and dermomyotome express Pax-3. Therefore, a twofold difference in p53 protein levels in the whole embryo may underestimate the magnitude by which Pax-3 affects p53 levels in individual Pax-3-expressing cells. Further investigation will be necessary to fully understand how Pax-3 regulates p53, as well as p53-dependent transcription and apoptosis. In addition, it will be important to determine whether inhibition of p53-dependent apoptosis is required for development of other Pax-3-dependent tissues such as neural crest and somitic mesoderm derivatives.
Figure 3.
Effect of Splotch genotype on p53 mRNA and protein levels. (A) Semiquantitative RT–PCR analysis of reverse-transcribed RNA from Sp/Sp, Sp/−, and wild-type embryos. PCR was performed using p53 or 36B4-specific primers and fourfold serial dilutions of RT reaction products. (B) Quantitation of p53 cDNA normalized to 36B4 cDNA from three embryos of each genotype. There was no significant effect of Splotch genotype on p53 expression (P = 0.55). (C) Western blot analysis of p53 protein in Sp/Sp, Sp/−, and wild-type embryos. Twofold serial dilutions of protein extracts were electrophoresed, and immunoblotting was performed using antibodies directed against p53 or β-tubulin. (D) Quantitation of p53 normalized to β-tubulin from three embryos of each genotype. (*) P < 0.02 versus wild type.
Implications for Pax-3 in tumorigenesis
The inhibition of p53-dependent apoptosis by Pax-3 has implications for tumorigenesis, as well as development. Expression of a PAX-3/FKHR or PAX-7/FKHR fusion protein appears to be a key step in the development of pediatric rhabdomyosarcoma (Bernasconi et al. 1996; Fredericks et al. 2000). In addition, transcriptionally active PAX-3 is expressed in human melanomas, but not in surrounding normal tissue (Barr et al. 1999; Galibert et al. 1999; Vachtenheim and Novotna 1999; Scholl et al. 2001). In the mouse, p53 inactivation has been shown to cooperate with activated RAS to cause melanoma (Bardeesy et al. 2001; Yang et al. 2001). Although loss of function of the p53 gene is associated with many malignancies, the evidence presented here indicates that an alternative way to cause functional p53 deficiency is by reactivating or ectopically expressing Pax-3. Therefore, in human melanoma or rhabdomyosarcoma, PAX-3 inhibition of p53-dependent apoptosis may be critical to tumor establishment.
It should be noted that several Pax proteins (Pax-1, Pax-2, Pax-3, Pax-6, and Pax-8), which differ in their paired domain sequences and the presence of a complete or partial homeodomain, have the capacity to transform fibroblasts and induce tumors (Maulbecker and Gruss 1993). Furthermore, as noted above, PAX-5 inhibits p53 transcription, a process that may lead to astrocytoma (Stuart et al. 1995). Hence, induction of p53 loss of function by different mechanisms may be common to all Pax proteins during development, and may be an integral process leading to tumorigenesis when Pax proteins are inappropriately expressed.
Materials and methods
Mice
Heterozygous Splotch (Sp/+) mice on a C57Bl/6J background, and heterozygous p53 knockout (p53+/−) mice on a FVB background were obtained from Jackson Laboratories. Embryos were recovered on E10.5 to assay p53 mRNA or protein, or apoptosis and NTD. Embryos to be used for TUNEL assay were fixed in 4% paraformaldehyde, and embryos for RT–PCR or Western blot analysis were stored at −80°C. Pifithrin-α (Calbiochem) was administered by intraperitoneal injection of 2.2 mg/kg dissolved in PBS on E8.5 and E9.5.
TUNEL assay
Apoptosis was assayed by a whole-mount TUNEL procedure as described (Phelan et al. 1997). Apoptosis specifically localized to the neural tube was scored by an individual who was blinded to embryo genotype. A scale of 1–10 was used, using negative (embryo reacted without terminal transferase enzyme) and positive (embryo nicked with DNase prior to TUNEL procedure) controls as standards for scores of 1 and 10, respectively.
Genotype analysis
DNA was extracted from yolk sacs or tails using DNAzol (Molecular Research Center) to determine the genotype of individual embryos or pups. The genotype of the Splotch allele was determined as described (Machado et al. 2001); the genotype of the p53 allele was as described (Jacks et al. 1994) with modifications as described at http://aretha.jax.org/pub-cgi/protocols/protocols.sh?objtype=protocol&protocol_id=125; gender was determined by using primers to the Zfy and Nf1 genes (Sah et al. 1995); however, there was no interaction of gender and p53 genotype on exencephaly in embryos on a FVB background.
RT–PCR analysis
Semiquantitative reverse transcription PCR (RT–PCR) analysis was performed using RNA from individual embryos whose genotype had been determined using yolk sac DNA as described above; 500 ng of RNA was reverse transcribed as described (Phelan et al. 1997). PCR was performed using primers specific to p53 (Toda et al. 1998) or to 36B4 (Hill et al. 1998), which was used as a control, and fourfold serial dilutions of RT reaction products (6.25–0.39 ng for p53, 24–1.5 pg for 36B4) and cycling conditions as described (Hill et al. 1998; Toda et al. 1998). p53 cDNA was expressed relative to 36B4 cDNA by scanning densitometry of bands that were present in a linear range following autoradiography of PCR products.
Western blot analysis
Immunoblot analysis of p53 protein was performed using twofold serial dilutions of protein (250–62.5 μg) from individual embryos and p53 antibodies (Ab-1 and Ab-3 from Calbiochem, both diluted 1:500) and goat-anti-mouse secondary antibody (Pierce, diluted 1:5000). p53 was normalized to β-tubulin, which was detected with a primary anti-tubulin antibody (Santa Cruz Biotechnology, diluted 1:1000) and goat-anti-rabbit secondary antibody (Pierce, diluted 1:2500). Secondary antibodies were detected by chemiluminescence (Pierce).
Statistical analysis
Data were analyzed by 1-Way Analysis of Variance and Neuman Keuls post-hoc test, or χ-square analysis, using Prism 3 software (GraphPad Software).
Acknowledgments
This work was supported by grants from the National Institutes of Health, the Juvenile Diabetes Foundation, and the March of Dimes Birth Defects Foundation to M.R.L. We are grateful to Peter Howley for critical comments on the manuscript, to Guo Jun Zhang for advice on p53 immunoblotting procedures, and to Rakhi Patel and Rebecca Clark for technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mary.loeken@joslin.harvard.edu; FAX (617) 732-2541.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.969302.
References
- Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- Auerbach R. Analysis of the developmental effects of a lethal mutation in the house mouse. J Exp Zool. 1954;127:305–329. [Google Scholar]
- Bardeesy N, Bastian BC, Hezel A, Pinkel D, DePinho RA, Chin L. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol Cell Biol. 2001;21:2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FG, Galili N, Holick J, Biegel JA, Rovera G, Emanuel BS. Rearrangement of the PAX3 paired box gene in the paediatric solid tumor alveolar rhabdomyosarcoma. Nat Genet. 1993;3:113–117. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- Barr FG, Fitzgerald JC, Ginsberg JP, Vanella ML, Davis RJ, Bennicelli JL. Predominant expression of alternative PAX3 and PAX7 forms in myogenic and neural tumor cell lines. Cancer Res. 1999;59:5443–5448. [PubMed] [Google Scholar]
- Bernasconi M, Remppis A, Fredericks WJ, Rauscher FJ, Schafer BW. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc Natl Acad Sci. 1996;93:13164–13169. doi: 10.1073/pnas.93.23.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz-Tadmor B, Zakut-Houri R, Libresco S, Givol D, Oren M. The 5′ region of the p53 gene: Evolutionary conservation and evidence for a negative regulatory element. EMBO J. 1985;4:3209–3213. doi: 10.1002/j.1460-2075.1985.tb04067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycki A-G, Li J, Jin F, Emerson CP, Jr, Epstein JA. Pax3 functions in cell survival and in pax7 regulation. Development. 1999;126:1665–1674. doi: 10.1242/dev.126.8.1665. [DOI] [PubMed] [Google Scholar]
- Chalepakis G, Jones FS, Edelman GM, Gruss P. Pax-3 contains domains for transcription activation and transcription inhibition. Proc Natl Acad Sci. 1994;91:12745–12749. doi: 10.1073/pnas.91.26.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway SJ, Henderson DJ, Copp AJ. Pax3 is required for cardiac neural crest migration in the mouse: Evidence from the splotch (Sp2H) mutant. Development. 1997;124:505–514. doi: 10.1242/dev.124.2.505. [DOI] [PubMed] [Google Scholar]
- Epstein DJ, Vogan KJ, Trasler DG, Gros P. A mutation within intron 3 of the Pax-3 gene produces aberrantly spliced mRNA transcripts in the splotch (Sp) mouse mutant. Proc Natl Acad Sci. 1993;90:532–536. doi: 10.1073/pnas.90.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JA, Li J, Lang D, Chen F, Brown CB, Jin F, Lu MM, Thomas M, Liu E, Wessels A, et al. Migration of cardiac neural crest cells in Splotch embryos. Development. 2000;127:1869–1878. doi: 10.1242/dev.127.9.1869. [DOI] [PubMed] [Google Scholar]
- Fisher DE. Apoptosis in cancer therapy: Crossing the threshold. Cell. 1994;78:539–542. doi: 10.1016/0092-8674(94)90518-5. [DOI] [PubMed] [Google Scholar]
- Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- Fredericks WJ, Ayyanathan K, Herlyn M, Friedman JR, Rauscher FJ., III An engineered PAX3-KRAB transcriptional repressor inhibits the malignant phenotype of alveolar rhabdomyosarcoma cells harboring the endogenous PAX3-FKHR oncogene. Mol Cell Biol. 2000;20:5019–5031. doi: 10.1128/mcb.20.14.5019-5031.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert MD, Yavuzer U, Dexter TJ, Goding CR. Pax3 and regulation of the melanocyte-specific tyrosinase-related protein-1 promoter. J Biol Chem. 1999;274:26894–26900. doi: 10.1074/jbc.274.38.26894. [DOI] [PubMed] [Google Scholar]
- Galili N, Davis R, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, III, Emanuel BS, Rovera G, Barr FG. Fusion of a fork head domain gene to PAX3 in the solid tumor alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- Goulding MD, Chalepakis G, Deutsch U, Erselius JR, Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AL, Phelan SA, Loeken MR. Reduced expression of Pax-3 is associated with overexpression of cdc46 in the mouse embryo. Dev Genes Evol. 1998;208:128–134. doi: 10.1007/s004270050163. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, William BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Kioussi C, Gross MK, Gruss P. Pax3: A paired domain gene as a regulator in PNS myelination. Neuron. 1995;15:553–562. doi: 10.1016/0896-6273(95)90144-2. [DOI] [PubMed] [Google Scholar]
- Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- Machado AF, Zimmerman EF, Hovland DN, Jr, Weiss R, Collins MD. Diabetic embryopathy in C57BL/6J mice. Altered fetal sex ratio and impact of the splotch allele. Diabetes. 2001;50:1193–1199. doi: 10.2337/diabetes.50.5.1193. [DOI] [PubMed] [Google Scholar]
- Maroto M, Reshef R, Munsterberg AE, Koester S, Goulding M, Lassar AB. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell. 1997;89:139–148. doi: 10.1016/s0092-8674(00)80190-7. [DOI] [PubMed] [Google Scholar]
- Maulbecker CC, Gruss P. The oncogenic potential of Pax genes. EMBO J. 1993;12:2361–2367. doi: 10.1002/j.1460-2075.1993.tb05890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan S, Loeken M. Identification of a new binding motif for the paired domain of Pax-3 and unusual characteristics of spacing and of bipartite recognition elements on binding and transcription activation. J Biol Chem. 1998;273:19153–19159. doi: 10.1074/jbc.273.30.19153. [DOI] [PubMed] [Google Scholar]
- Phelan SA, Ito M, Loeken MR. Neural tube defects in embryos of diabetic mice: Role of the Pax-3 gene and apoptosis. Diabetes. 1997;46:1189–1197. doi: 10.2337/diab.46.7.1189. [DOI] [PubMed] [Google Scholar]
- Puglisi F, Cesselli D, Damante G, Pellizzari L, Beltrami CA, Di Loreto C. Expression of Pax-8, p53 and bcl-2 in human benign and malignant thyroid diseases. Anticancer Res. 2000;20:311–316. [PubMed] [Google Scholar]
- Sah VP, Attardi LD, Mulligan JG, Williams BO, Bronson RT, Jacks T. A subset of p53-deficient embryos exhibit exencephaly. Nat Genet. 1995;10:175–180. doi: 10.1038/ng0695-175. [DOI] [PubMed] [Google Scholar]
- Scholl FA, Kamarashev J, Murmann OV, Geertsen R, Dummer R, Schafer BW. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 2001;61:823–826. [PubMed] [Google Scholar]
- Shapiro DN, Sublett JE, Li B, Downing JR, Naeve CW. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res. 1993;53:5108–5112. [PubMed] [Google Scholar]
- Stuart ET, Haffner R, Oren M, Gruss P. Loss of p53 function through PAX-mediated transcriptional repression. EMBO J. 1995;14:5638–5645. doi: 10.1002/j.1460-2075.1995.tb00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- Toda I, Wickham LA, Sullivan DA. Gender and androgen treatment influence the expression of proto-oncogenes and apoptotic factors in lacrimal and salivary tissues of MRL/lpr mice. Clin Immunol Immunopathol. 1998;86:59–71. doi: 10.1006/clin.1997.4466. [DOI] [PubMed] [Google Scholar]
- Vachtenheim J, Novotna H. Expression of genes for microphthalmia isoforms, Pax3 and MSG1, in human melanomas. Cell Mol Biol. 1999;45:1075–1082. [PubMed] [Google Scholar]
- Yang FC, Merlino G, Chin L. Genetic dissection of melanoma pathways in the mouse. Semin Cancer Biol. 2001;11:261–268. doi: 10.1006/scbi.2000.0376. [DOI] [PubMed] [Google Scholar]