Abstract
The transcription of mecA, the gene required for oxacillin resistance in staphylococci, was quantified in a collection of 65 geographically and genetically diverse clinical and 8 defined laboratory Staphylococcus aureus isolates. mecA transcription was measured by real-time reverse transcription-PCR, confirmed by Northern blot analysis, and correlated with the presence and DNA sequence of the two mecA repressors, mecI and blaI. Isolates were first examined that contained mecI and/or blaI with wild-type sequence. BlaI provided significantly more repression of mecA transcription than did MecI, unrelated to blaI genetic location. Both together repressed mecA better than either one alone. In clinical isolates containing only wild-type mecI, mecA transcription repression was 10- to 25-fold less effective than that seen in previously studied constructs derived from strain N315. There was a difference in the mecI ribosomal binding site (RBS) between the clinical isolates (GGAA) and N315 (GGAG). The GGAA RBS was associated with 5.5- to 7.3-fold less mecA repression than GGAG in isogenic constructs. The values generated for wild-type repressors were compared to those in 26 isolates containing mecI mutations. mecA transcription appeared to be repressed only by BlaI in isolates with mecI nonsense and frameshift mutations. In contrast, mecI repression seemed to be partially or fully retained in many of the isolates with mecI and one isolate with blaI missense mutations, providing structure-function correlates with the site and type of mutation. We conclude that mecA repressor activity is highly variable in clinical S. aureus isolates due to mecI mutations, RBS polymorphisms, and unidentified genomic adaptations.
Two repressors, BlaI and/or MecI, regulate transcription of mecA, the gene required for oxacillin resistance in staphylococci (8, 15). mecA encodes a penicillin binding protein (PBP2a) that is poorly bound by β-lactam antibiotics and can perform essential functions of cell wall construction when β-lactams inactivate the cell's normal penicillin binding protein complement (3, 14). Two signal transducers, BlaR1 and MecR1, regulate repressor activity (20). MecR1 and BlaR1 are transmembrane β-lactam sensory transducers, having a surface-exposed β-lactam binding domain and a cytoplasmic zinc peptidase motif. Signal transduction is thought to occur following interaction of a β-lactam antibiotic with the binding domain, transmitting a signal through four membrane-spanning segments. This induces conformational change in the cytoplasmic metalloprotease domain causing autoproteolysis. Proteolytic cleavage of the inducer is followed by cleavage of the repressor, leading to derepression of target genes. The genes for signal transducers and repressors are contained in two-gene operons (blaR1-blaI and mecR1-mecI) that are divergently transcribed from their regulated genes (blaZ and mecA, respectively) (8, 13, 15). The repressors bind to specific palindromic sequences that overlap divergent promoters for mecA and mecR1 (8, 18). A helix-turn-helix DNA binding motif has been assigned to the amino terminus of the repressor protein based on structure predictions (11), and the carboxyl terminus has been assumed to be involved in repressor dimerization by analogy to other repressors and by the location of the repressor cleavage site (19, 20). However, the structure of neither MecI nor BlaI has been solved.
In a previous study (17), we showed that at least one repressor with a wild-type nucleotide sequence was present in 91 of 95 genetically diverse clinical Staphylococcus aureus isolates. We hypothesized that repression was evolutionarily conserved in order to prevent overproduction of a potentially toxic, regulated gene product. However, we did not assess the effectiveness of repression by directly measuring the transcription of regulated genes. A direct assessment of transcriptional regulation is of interest for several reasons. First, in an earlier study (16), members of our group identified an isolate containing mecI with wild-type repressor and promoter-operator sequences in which mecA transcription was eightfold greater than that in another isolate containing mecI with the same sequence. In the present study, we sought evidence for similar variations in mecA transcriptional repression among clinical isolates containing mecI genes with wild-type sequence. Second, we sought to confirm data generated with a single, genetically manipulated strain indicating that wild-type MecI and BlaI provided additive repression of mecA transcription when both were present in the same strain (15). Third, we wanted to assess the extent to which mutations in mecI or blaI affected repressor function. These data might provide valuable structure-function correlates to complement studies investigating the molecular basis of MecR1-BlaR1 and MecI-BlaI signal transduction.
However, all but one of the clinical isolates with mutant mecI also contained the corepressor gene, blaI, making it difficult to assess the contribution of individual repressor mutations to mecA regulation. In order to assess the function of mutant repressors, we first had to establish baseline values for transcription repression in isolates with blaI and mecI that had wild-type DNA sequence. We used real-time reverse transcription PCR (RT-PCR) as our method for transcriptional quantification. We identified groups of isolates that had all possible combinations of the two repressors with wild-type sequence: both together, both absent, and each present without the other. The values for mecA transcription among these isolates were used as standards with which to compare isolates with mecI mutations. As described in a previous publication (17), we have found, among the 26 clinical isolates with mecI mutations, a wide variety in the type and location of mutation, making it possible to attempt structure-function predictions.
MATERIALS AND METHODS
Source of isolates.
The S. aureus clinical isolates examined in this study came from three sources, as detailed in a previous publication (17): 30 isolates, all of which were genetically distinct, came from the Public Health Research Institute (PHRI) collection, kindly provided by Barry Kreiswirth; 26 isolates, chosen for geographical and temporal diversity, came from the Virginia Commonwealth University collection; and 9 isolates, found to have wild-type mecI sequence and no blaI coregulation, were blood culture isolates from the SCOPE collection kindly provided by Sandy Tennant and Mike Edmond (5). In addition, eight isolates were defined, genetically manipulated strains used as comparators for RT-PCR analysis.
Isolates with wild-type mecI and blaI.
Isolates with both wild-type mecI and blaI together, neither repressor, or each without the other were chosen from among the clinical isolates and other well-characterized strains in our collection. The DNA sequence of the repressors was determined and described previously (17). The isolates with only wild-type mecI came from all three clinical collections (12 isolates); those with blaI only, containing the previously described IS1272-mediated deletion of mecI (2), were all from the PHRI collection, and each was of a unique spa-repeat type (12 isolates); those with both mecI and blaI were from the PHRI and VCU collections, and each was of a unique spa-repeat type (12 isolates); and those with neither repressor contained only the IS1272-mediated mecI deletion (three isolates) or laboratory-constructed mecI knockouts (three isolates). Well-characterized isolates (1, 12, 15, 16) were used as comparators in the appropriate groups: COL and 450 M (ΔmecI-ΔblaI), N315 (mecI blaI), 450 M/pI258 (ΔmecI-blaI) and N315P, BMS1, 450 M::630, and 450 M::522 (mecI+ blaI).
Isolates with mecI and blaI mutations.
The 26 isolates with mecI mutations and the single isolate with a mutation in blaI were described previously (17).
Media.
Mueller-Hinton broth and Mueller-Hinton agar (both from BBL Microbiology Systems, Cockeysville, Md.) and brain heart infusion broth and agar (Difco Laboratories, Detroit, Mich.), with and without selective additives (Sigma, St. Louis, Mo.; United States Biochemicals, Cleveland, Ohio), were used for subculture and maintenance of S. aureus strains.
Southern blot analysis.
The genetic location of blaI was determined by probing plasmid and chromosomal DNA of selected isolates with this gene. Plasmid and chromosomal DNA were extracted from bacterial isolates using Qiagen columns (Qiagen, Inc., Chatsworth, Calif.), electrophoresed through agarose, and transferred to a nylon membrane using capillary action. Probe DNA was labeled with digoxigenin and applied to membranes according to the manufacturer′s instructions (Roche Molecular Biochemicals, Indianapolis, Ind.). Signal was detected by exposing membranes to X-ray film. blaI was assumed to be plasmid encoded if the probe hybridized with a plasmid band of a size approximately equal to that of control plasmid pI258 and chromosomally encoded if the probe hybridized with only the chromosomal band. However, it is possible that in some isolates, plasmid DNA comigrated with chromosomal DNA and was mistakenly designated a chromosomal gene.
Northern blot analysis.
S. aureus was grown in 30 ml of brain heart infusion broth to an optical density at 600 nm of 0.6. Cultures were centrifuged, and the bacteria were resuspended in 1,000 μl of RLT buffer (RNeasy kit; Qiagen Inc.) and added to 2-ml FastPrep Blue tubes containing ceramic matrices (Bio 101, La Jolla, Calif.). The bacteria were lysed with a Fast Prep instrument (Bio101) at setting 6 for 40 s, placed on ice for 1 min, and centrifuged at 10,000 × g for 5 min at 4°C. The upper aqueous phase was aspirated, and total RNA was extracted using a Qiagen RNeasy kit. About 7 μg of RNA was separated by resolution through formaldehyde-containing 1% agarose. The intensities of the 23S and 16S rRNA were visualized using a 254-nm UV short-wave lamp, and quantities were adjusted so that the same amount of RNA was loaded for each bacterium. RNA was transferred from agarose to positively charged nylon membranes (Stratagene, La Jolla, Calif.) by capillary action as previously described (15). Labeling and hybridization were done by use of the digoxigenin labeling and detection kits according to the manufacturer′s instructions (Roche Molecular Biochemicals) and exposed to X-ray film.
Real-time RT-PCR.
Oligonucleotide primers and probes for mecA and 16S rRNA were designed with Primer Express 1.0 software form ABI Prism (Perkin-Elmer Applied Biosystems, Foster City, Calif.) and purchased from Megabase Inc (Evanston, Ill.). The probes consisted of an oligonucleotide labeled at the 5′ end with the reporter dye 6-carboxyfluorescein and with the quencher dye N,N′,N′-tetramethyl-6 carboxytetramethylrhodamine at the 3′ end. RT-PCR was done with the TaqMan One-Step RT-PCR Master Mix Reagents kit as described by the manufacturer (PE Applied Biosystems, Foster City, Calif.). The RT-PCR mixture (25 μl contained 6.25 U of Multiscribe reverse transcriptase, 10.0 U of RNase inhibitor, 500 nM (each) gene-specific primer, 100 nM (each) probe, and 25 ng of total RNA template. Amplification and detection of specific products were performed with the ABI Prism 7700 sequence detection system (PE Applied Biosystems) with the following cycle profile: 1 cycle at 48°C for 30 min, 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s and 60°C for 1 min. The critical threshold cycle (Ct) is defined as the cycle at which the fluorescence becomes detectable above background levels and is inversely proportional to the logarithm of the initial number of template molecules. A standard curve was plotted for each primer-probe set with Ct values obtained from amplification of known quantities of RNA isolated from strain S. aureus 450 M. The standard curves were used to transform Ct values to the relative number of RNA molecules. The amount of contaminating chromosomal DNA in each sample was determined with the control reactions that did not contain reverse transcriptase. The quantity of cDNA for each experimental gene was normalized to the quantity of 16S cDNA in each sample. Each RNA sample was run in triplicate. The forward and reverse primers for mecA RT-PCR were GTTAGATTGGGATCATAGCGTCATT and TGCCTAATCTCATATGTGTTCCTGTAT, respectively, and for 16S RT-PCR they were TCCGGAATTATTGGGCGTAA and CCACTTTCCTCTTCTGCACTCA, respectively. The probes, labeled with carboxyfluorescein at the 5′ end and N,N′,N′-tetramethyl-6 carboxytetramethylrhodamine at the 3′ end, were TTCCAGGAATGCAGAAAGACCAAAGCATGA for mecA and AAGCCCACGGCTCAACCG for 16S RNA.
Assay for β-galactosidase activity.
β-galactosidase activity was assessed in cell extracts, produced by homogenization of bacteria with glass beads for 3 min in a mini-bead-beater (BioSpec Products, Bartlesville, Okla.) using o-nitrophenyl-β-d-galactopyranoside as a substrate, as previously described (18).
Statistical analysis.
Differences between the RT-PCR values in one group versus another were assessed by analysis of variance using SPSS for Windows (SPSS Inc., Chicago, Ill.).
RESULTS
Transcription of mecA in isolates with wild-type repressor sequence.
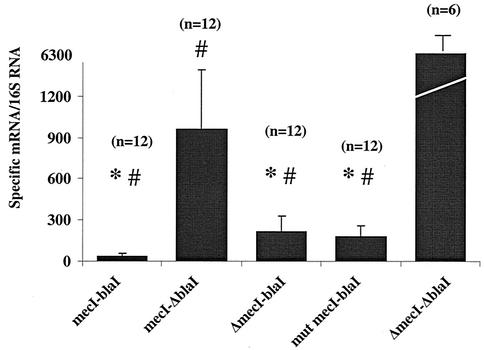
Each RT-PCR value for the wild-type isolates and for the mutants described below is the result of the analysis of a single RNA sample for most isolates. However, the reproducibility of the RT-PCR results was supported by analysis of a second, independently prepared RNA sample on 35 isolates (10 wild types, 17 mutants, and 8 comparators). Repeat values for missense, frameshift and nonsense mutants are shown in Table 1. In addition, as described below, 36 of the 73 RNA samples analyzed by RT-PCR were also examined by Northern blotting, and the relative magnitude and difference between samples was confirmed in each case. The RT-PCR values in the groups of isolates with wild-type mecI and blaI sequence are shown in Fig. 1. The mecA-mecR1 intergenic regions, containing the promoter-operator sequences, were wild type and identical for all isolates studied. The mean value of each group was compared to that for each other group. Transcription repression mediated by BlaI alone and that mediated by BlaI plus MecI were significantly greater than that mediated by MecI alone (P < 0.05). All isolates containing both repressors together also provided greater repression than BlaI alone, but that difference did not reach statistical significance due to the wide variation in values. The absence of both repressors led to a significantly greater transcription of mecA than was seen when either repressor was present (P < 0.001). The effect of genetic location on blaI-mediated repression was also investigated. Among the isolates with blaI and mecI, blaI was in the chromosome in five isolates and on a plasmid in seven, including the comparator type strain, N315. The mecA transcription values (mean ± standard deviation) were the following: chromosome, 50.7 ± 15.2; plasmid, 41.2 ± 9.2). In isolates with blaI alone, the repressor was in the chromosome in seven isolates and on a plasmid in six isolates, including the comparator laboratory strain, 450 M/pI258. Values were the following: chromosome, 217 ± 103; plasmid, 149 ± 102. There was no significant difference in blaI-mediated repression related to genetic location.
TABLE 1.
Characterization of mecI mutations and quantitation of specific mecA mRNA
| Mutation class | Strain | Mutation in mecI gene
|
Amt of specific mecA mRNA/16S mRNA | |
|---|---|---|---|---|
| Nucleotide position (change) | Amino acid changea | |||
| Nonsense | SA41 | 202 (C→T) | Gln 68→stop codon | 330 |
| SA35 | 202 (C→T) | Gln 68→stop codon | 285 | |
| SA7 | 202 (C→T) | Gln 68→stop codon | 179, 129b | |
| SA33 | 202 (C→T) | Gln 68→stop codon | 176 | |
| SA18 | 202 (C→T) | Gln 68→stop codon | 145, 83 | |
| SA12 | 202 (C→T) | Gln 68→stop codon | 142, 139 | |
| SA20 | 202 (C→T) | Gln 68→stop codon | 117 | |
| SA9 | 202 (C→T) | Gln 68→stop codon | 116 | |
| SA16 | 202 (C→T) | Gln 68→stop codon | 102 | |
| SA19 | 202 (C→T) | Gln 68→stop codon | 141 | |
| SA2 | 202 (C→T) | Gln 68→stop codon | 90 | |
| SA11 | 202 (C→T) | Gln 68→stop codon | 43 | |
| SA57 | 343 (G→T) | Glu 115→stop codon | 226, 301 | |
| Frameshift | SA10 | 193 (extra A) | Lys 65→12 aa to stop | 306 |
| SA13 | 64 (extra A) | Lys 22→7 aa to stop | 1,172 | |
| SA42 | 250 (extra A) | Lys 84→5 aa to stop | 834, 801 | |
| SA56 | 91 (extra T) | Ile 30→8 aa to stop | 330, 219 | |
| Missense | SA5 | 32 (C→T) | Ala 11→Val | 21, 28 |
| SA15 | 35, 32 | |||
| SA24 | 261, 256 | |||
| SA14 | 347 (T→C) | Leu 116→Ser | 202, 193 | |
| SA36 | 1,907, 940 | |||
| SA46 | 22 (A→G) | Iso 8→Val | 21, 35 | |
| SA78 | 125 (C→T) | Pro 42→Leu | 64, 75 | |
| SA30 | 116 (A→G) | Asp 39→Gly | 40, 52 | |
| SA54 | 152 (G→T) | Arg 51→Ile | 1,101, 1,227 | |
| SA47 | 370 (T→A) | +18 aa to stop | 66, 79 | |
| SA27 | 142 (C→T) | Leu 48→Phe | 66, 49 | |
aa, amino acids.
For some isolates two independent RNA samples, prepared on different days, were analyzed to assess reproducibility of RT-PCR values for mecA-specific mRNA.
FIG. 1.
Quantitation of mecA mRNA by Taqman Real Time RT-PCR in S. aureus isolates. Relative values of mecA mRNA over 16S mRNA are shown on the vertical axis, while groups of isolates with different repressor genotypes are shown on the horizontal axis. Each filled bar designates the mean of that group; n represents the number of strains in each group; and standard deviations are indicated by error brackets. The repressor genotype of each group is indicated below the bar. Δ indicates that the repressor is absent or deleted; mut mecI is the group of isolates with nonsense mutations. ∗, transcription repression of mecA significantly greater than that mediated by mecI alone (mecI-ΔblaI; P < 0.05); #, transcription repression of mecA significantly greater than that seen in the absence of repressor (ΔmecI-ΔblaI; P < 0.001).
The mean ± standard deviation (960 ± 431) mecA transcription values for isolates containing mecI alone were far greater than that seen for the laboratory comparator strains, N315P and 450 M::630 (49 and 12, respectively). For 7 of 12 isolates, the mecA transcription values ranged from 1,077 to 1,592. The comparator strains are derived from N315, the strain providing the first published sequence of mecR1 and mecI (10) and the first published S. aureus genomic sequence (12). N315P is N315 cured of its penicillinase-producing plasmid. 450 M::630 contains the N315 mecR1 mecI mecA-mecR1 promoter-operator and a portion of mecA introduced into the chromosomal lipase gene (geh) of strain 450 M (mecR1-mecI deletion), providing mecA repression in trans. The lacZ gene was fused to the first 50 bp of mecA in the 450 M::630 construct to provide a reporter as an additional measure of mecA transcription, all as previously described (18). A possible explanation for the transcription difference between N315-derived and clinical isolates was found when the ribosomal binding site (RBS) for mecI from N315 (GGAG) was compared to that for clinical isolates (GGAA) (see Fig. 4). All 23 isolates containing wild-type mecI that were examined by RT-PCR had the GGAA RBS. Among 73 isolates examined in this study, only 11 isolates contained a GGAG RBS, all of which were in isolates containing mecI mutations (five nonsense, three frameshift, and three missense).
FIG. 4.
Nucleotide sequence of the ribosomal binding site (RBS) for the mecI repressor genes of S. aureus N315 (sequence 1, top) and S. aureus BMS1 (sequence 2, bottom). These sequences are representative of identical sequence variants seen among clinical isolates. The RBS sequence is boxed. The ATG start codon of the mecI gene is in bold and italics, while the overlapping TAA translational stop codon of the preceding mecR1 gene is in bold and underlined.
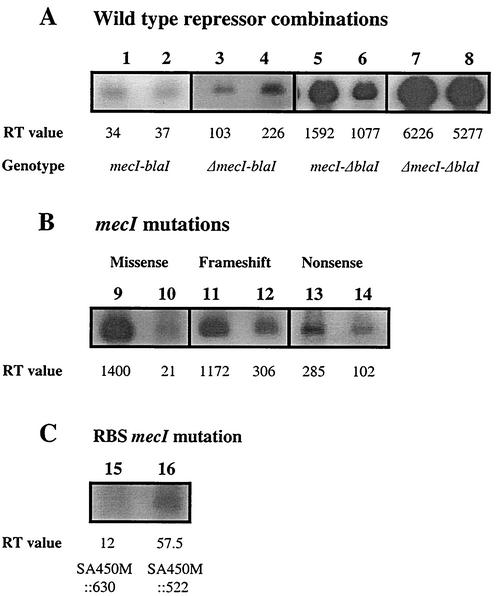
In order to assess the contribution of mecI RBS sequence polymorphisms to differences in mecI-mediated mecA repression, we compared mecA transcription in isogenic constructs, 450 M::630 and 450 M::522. 450 M::522 came from isolate BMS1 (15) and differed from 450 M::630 only in that the mecI GGAA RBS was substituted for the N315 GGAG sequence. Repression of mecA transcription was 8.2-fold greater for 450 M::630 (mean, 7.0) than for 450 M::522 (mean, 57.5). These RT-PCR values were confirmed by Northern blotting (see Fig. 3) and by β-galactosidase activity (471 U of activity for 450 M::630 and 3,142 U for 450 M::522, a 7.3-fold difference). However, the RBS differences did not entirely explain the difference in mecA transcription between some isolates with only wild-type mecI and others. The mean transcription value among seven of these isolates (1,274; range, 1,097 to 1,592) was 26- and 131-fold greater than that for either N315P or 450 M::630, respectively.
FIG. 3.
Representative Northern blot of mecA transcription in S. aureus isolates. (A) Isolates with wild-type repressor combinations: both repressors together (mecI-bla1), SA3 (lane 1) and SA8 (lane 2); each repressor without the other (ΔmecI-blaI), SA79 (lane 3) and SA37 (lane 4), (mecI-ΔblaI), SA92 (lane 5) and SA85 (lane 6); both repressors absent (ΔmecI-ΔblaI), SA450M (lane 7) and SAN315PΔI (lane 8). (B) mecI mutations: missense mutations, SA36 (lane 9) and SA46 (lane 10); frameshift mutations, SA13 (lane 11) and SA10 (lane 12); nonsense mutations, SA35 (lane 13) and SA16 (lane 14). (C) mecI with ribosome binding site (RBS) sequence GGAG [SA 450M::630 (lane 15)] and GGAA [SA450M::522 (lane 16)]. All images were acquired from their original gels by using the FluorChem imaging system (Alpha Innotech) and FluorChem version 2.0 software. Labeling was added using Corel Photo-Paint software, version 8.
Characterization of mecI mutations.
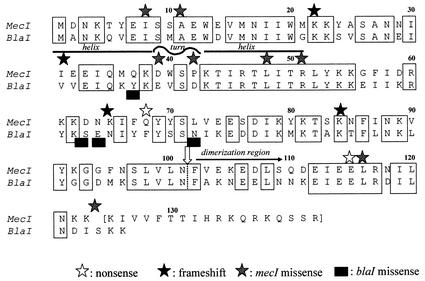
The 28 mutant mecI genes could be divided into three classes (Table 1 and Fig. 2). Class one (nonsense) mutations occurred in 13 isolates; 12 of the mutations were identical. In each of the 12 isolates, a substitution at nucleotide 202 (C→T) introduced a translational stop replacing a glutamine residue. In the 13th isolate, a translational stop was introduced at nucleotide 343, eight amino acids from the carboxyl terminus of the protein.
FIG. 2.
Optimal alignment of the MecI and BlaI repressor protein amino acid sequences showing sites and types of mutations, indicated by stars. Identical residues are shown between boxes. Additional information about nucleotide and amino acid changes is found in Table 1. The sites of cleavage of mecI and blaI are indicated by a vertical white arrow; the putative dimerization domain is indicated by a black horizontal arrow; and the putative helix-turn-helix DNA binding motif is indicated by horizontal lines over the appropriate residues.
Class two (frameshift) mutations, occurring in four isolates, introduced an A at a different site in three genes and a T in a fourth gene. The addition of adenines occurred in a group of from four to six adenine repeats; the addition of thymine created four dinucleotide TA repeats. The frameshift in these four isolates introduced a translational stop from four to eight amino acids after the mutation.
Class three (missense) mutations were found in 11 isolates. Two and three isolates had identical mutations (Leu116Ser and Ala11Val, respectively), while the final six isolates each had a unique mutation. In one of these isolates, SA47 (stop124Lys), the translational termination of the open reading frame was replaced by a lysine, extending the protein sequence by 18 amino acids. Five of the eight unique mutations in mecI were in amino acids that were conserved between mecI and blaI. There was a single isolate that contained mutant blaI; mecI was deleted in this isolate. There were four missense mutations in this isolate, each at a residue that was not conserved between blaI and mecI. At one position, the blaI mutation changed the amino acid to the one found at the comparable position in mecI (E64N) (Fig. 2).
Transcription of mecA in isolates with mecI and blaI mutations.
All of the isolates with mecI mutations but one contained blaI with wild-type sequence, as shown in a previous study (17). Thus, the mecA transcription results for the isolates with mecI mutations (mut mecI-blaI) were compared to those for two groups of isolates (ΔmecI-blaI) and (mecI-blaI).
The nonsense mutation at amino acid 68, found in 12 isolates, was predicted to inactivate MecI, leaving only BlaI as the sole mecA transcriptional regulator. The data were consistent with this prediction. The values for mecA transcription in this group (mut mecI blaI, 181.6 ± 76; range, 102 to 330) were not statistically different from values for those in the group having BlaI but no MecI (ΔmecI blaI, 216 ± 117; P > 0.05). They were also not statistically different from values for the entire group having both MecI and BlaI (mecI-blaI). However, while the transcription values of all 11 isolates containing both repressors (mecI blaI) were less than 70 (mean, 34; range, 3 to 65), 11 of the 12 isolates with BlaI only (ΔmecI blaI) (mean, 216; range, 103 to 431) and 10 of the 12 isolates with mecI nonsense (mut mecI blaI) mutations (mean, 182; range, 102 to 330) had values greater than 100. The difference between the subset of values in the 11 isolates with both repressors (mecI blaI) and those of the other two groups (ΔmecI blaI; mut mecI blaI) with mecA transcription values greater than 100 (11 and 12 isolates, respectively) was significantly different (P < 0.05). This suggests that values greater than 100 resemble those for the group with BlaI but not MecI and those less than 70 resemble those for the group with two functional repressors. Therefore, we made the assumption that isolates with mecA transcription values of <70 or >100 were likely to have a functional or inactive mecI repressor, respectively.
The mecA transcription values of the four isolates with frameshift mutations were all greater than 200 (306, 330, 801, and 1,172), making it likely that the mutation completely inactivated mecI, as predicted (Table 1).
There were nine different missense mutations in 11 isolates (Table 1). Mutations in 7 of the 11 isolates had mecA transcription values that were less than 70 (mean, 45; range, 21 to 66), while mutations in four isolates yielded values ranging from 201 to 1,907. One of the isolates with high mecA transcription values (261 and 256) had the same missense mutation (A11V) as two isolates with low transcription values (22 and 35). There were no differences in blaI or mec promoter-operator sequences or mecI RBS sequences among these three isolates.
Northern blot analysis of mecA transcription.
The RT-PCR results for some isolates were confirmed by Northern blot analysis. The same RNA used for RT-PCR analysis was examined by Northern blotting for the following isolates: six with the same missense mutation, all four with frameshift mutations, six with nonsense mutations, and two each with wild-type repressor combinations (both together, each without the other, and neither). The relative intensity of the mecA transcript by Northern blot analysis was consistent with the relative values generated using RT-PCR in each instance. Representative Northern blot results are shown in Fig. 3.
Sequence analysis of mecR1 and induction of mecA transcription.
In four isolates containing mecI without blaI and mecA transcription values >1,000, the DNA sequence of mecR1 was determined. In all four isolates the sequence was wild- type, identical to that of N315. In addition, all of the isolates with mecI mutations were induced overnight by growing them in 0.3 μg of oxacillin/ml, and mecA transcription was determined by RT-PCR. Although there was a wide range of transcription values following induction, mecA transcription values increased at least twofold for each isolate. This suggests that BlaR1 was functional in these strains, because mutation would have led to constitutive BlaI-mediated repression.
DISCUSSION
The present study illustrates the importance of studying clinical bacterial isolates as support for observations made using defined laboratory constructs. This is particularly true when assessing gene regulation and regulatory networks that affect antibiotic resistance. The pressure of exposure to antibiotics in hospitals may alter gene expression in ways that cannot be predicted using defined laboratory strains. Oxacillin-resistant staphylococci must be able to express high-level resistance to β-lactam antibiotics in order to survive in hospital environments where β-lactam use is high. Many staphylococci express resistance in a low-level or heterotypic manner and switch to homotypic or high-level-resistance expression following β-lactam exposure (7). This switch requires an increase in mecA transcription mediated by induction through MecR1 and/or BlaR1. Induction involves cleavage of the cognate repressor (MecI for MecR1 and BlaI for BlaR1) and is specific for each inducer's homologous repressor (15, 20). However, relief of MecI-mediated repression by induction through MecR1 is slow, taking hours, and incomplete, never achieving a level of mecA transcription seen when all repressors are missing or inactivated. In contrast, relief of BlaI-mediated repression by induction through BlaR1 is rapid, taking minutes, and complete (15). Thus, when MecI represses mecA transcription, the staphylococcal cell is at a clinical disadvantage when exposed to β-lactam antibiotics, unable to respond rapidly. As we have shown in this study, in some clinical isolates MecI is inactivated or modified by deletion or mutation, leaving mecA regulated only by BlaI and BlaR1. However, 37% of the clinical isolates that we examined in an earlier study (17) contained mecI genes and mec promoter-operator target sequences that were identical to those shown in laboratory constructs to provide full mecA repression.
In the present study, when we examined baseline, uninduced repression of mecA in clinical isolates that contained only mecI with wild-type DNA sequence, mecA transcription was from 6- to 131-fold greater than that seen with a well-studied comparator strain (N315P) and a laboratory construct (450 M::630). A single nucleotide difference in the mecI RBS (GGAA versus GGAG) (Fig. 4) between clinical isolates and laboratory strains was shown to be responsible for some of the difference in mecA transcription, increasing it from 7.3- to 8.2-fold in isogenic constructs. However, the magnitude of increase in mecA transcription due to mecI RBS mutations does not completely explain the 26- to >100-fold increases in transcription seen in some clinical isolates compared to laboratory constructs. It is unlikely that changes in MecR1 account for these differences, since the mecR1 sequence was wild type in four of these isolates. It is likely, therefore, that uninduced transcription of mecA can be greatly increased by mechanisms that do not involve mutations in mecI, its RBS, or its promoter-operator target. Additional genes outside of the regulatory operon may be involved in repression and induction, as proposed initially by Cohen and Sweeney (4) and as recently documented for beta-lactamase induction in Bacillus sp. (6). Identification of accessory genes and gene products involved in repression and induction of mecA transcription will be required for a full understanding of this regulatory network.
One observation made with laboratory constructs was confirmed in this study. In a previous study, members of our group had observed that there appeared to be an additive or synergistic interaction between BlaI and MecI, and the two repressors were shown to form heterodimers in the yeast two-hybrid assay (15). In the present study, in clinical isolates containing both wild-type mecI and blaI, mecA transcription was at a lower level than in isolates containing either one alone. This occurred despite the apparent reduced effect of wild-type mecI alone. The additive activity of the two repressors may have been due to direct interactions between the proteins at the mec promoter-operator. We demonstrated in a previous study (15) that increasing either MecI oligomerization or mecI gene dosage increased mecA transcription repression. The addition of BlaI to MecI could effectively increase the amount of available repressor protein and the number of protein-protein interactions.
A major goal of the present study was to assess the effect that the various mecI mutations identified in clinical isolates had on mecA transcription as an aid to structure-function analysis of the repressor molecule. This goal was made difficult by the presence of corepressors, MecI and BlaI, in most of the isolates in which mecI was mutant and by the possible contribution, noted above, of additional unknown genes involved in repressor activity. However, evaluation of isolates with wild-type repressor sequence as comparators for isolates with mutations enabled us to make some observations about the effect of mutations on mecI function. First, nonsense and frameshift mutations were predicted to truncate the protein at positions that would have removed the carboxyl terminus. The cleavage site that inactivates the repressor following induction, presumably by preventing dimerization, is 22 amino acids from the carboxyl terminus in BlaI (19) and in the same relative position in MecI (G. Archer and M. Bosilevac, unpublished observations) (Fig. 2). Thus, all of the nonsense and frameshift mutations were predicted to inactivate repressor activity. The mecA transcription values for these isolates were similar to those for isolates in which BlaI alone regulated transcription, suggesting that MecI function was absent.
Second, we found four blaI missense mutations in a single isolate containing this repressor alone. The value for mecA transcription for this isolate (431) was slightly higher than the highest value in isolates containing blaI alone with wild-type sequence (346) but more than 10-fold below the value of isolates with no repressor (mean, 6,302). This suggests that these mutations did not markedly affect repressor function. The mutations (Y37N, S63Y, E64N, and N72I) were all at amino acids that were nonconserved between MecI and BlaI, two proteins that have 68% amino acid identity overall (Fig. 2). One mutation changed the amino acid from the one found in BlaI to its counterpart in MecI (E64N). It is reasonable to assume that the areas of nonidentity between the two proteins define parts of structure that are either nonessential for repressor function or that can be highly polymorphic and still not abolish activity.
Third, we examined mecA transcription in isolates that had eight different mecI missense mutations. Six of the mutations changed amino acids that were conserved between MecI and BlaI. The mutations at six nucleotides, changing five amino acids and one stop codon (A11V, I7V, D39G, P42L, L48,F and stop124K) in seven isolates, had mecA transcription values that were <70, not significantly different from values seen in a subset of isolates with two wild-type repressors. This suggests that these mutations left much of MecI repressor activity intact. Three mutations replaced amino acids conserved between BlaI and MecI, and although two (A11V and I7V) were in the amino terminus of the protein in an area without identified function, the other (L48F) was in a region that is predicted to be the helix-turn-helix conformation that mediates DNA binding. Conservation of function in this case may be more related to the nature of the amino acid substitution than to its location. The other three mutations either were at amino acids that were different between MecI and BlaI (D39G and P42L) (Fig. 2) or extended the protein by 19 amino acids by mutating the translational stop. One of these mutations has been studied previously (D39G) (18). Examination of purified protein and isogenic constructs containing this mutation showed that the mutation reduced repressor activity sixfold. However, transcription was still repressed sevenfold over that seen in the absence of repressor. The clinical isolate in which this mecI mutation was found also contained blaI. The RT-PCR transcription values provide evidence that MecI-BlaI additive repression can take place even when one of the proteins has a mutation that reduces its activity.
Three final isolates had two mecI mutations that markedly altered transcriptional repression. Both mutations substituted amino acids that were conserved between MecI and BlaI, one (R52I) in the DNA binding domain and the other (L116S) in the carboxyl terminus, a region thought to be critical for dimerization (9). It is interesting that a nonsense mutation at the preceding amino acid (E115stop) in another clinical isolate altered mecA transcription to the same degree as the L116S missense mutation in one of two isolates with the same mutation (transcription values of 226 versus 201 and 194, respectively). These data support the functional importance of the carboxyl terminus of the protein and suggest that the L116S substitution has a major effect on repressor activity.
The structure-function clues gleaned from the analysis of repressor mutations in clinical isolates will have to await data from the crystal structure for confirmation of suggestions outlined above. However, the data should help the analysis of crystals and provide sites at which the molecules could be modified for more detailed structure-function analysis.
Acknowledgments
This study was supported in part by Public Health Service grant R37 AI35705 from the National Institutes of Allergy and Infectious Diseases.
REFERENCES
- 1.Archer, G. L., D. M. Niemeyer, J. A. Thanassi, and M. J. Pucci. 1994. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob. Agents Chemother. 38:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, G. L., J. A. Thanassi, D. M. Niemeyer, and M. J. Pucci. 1996. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 40:924-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers, H. F., B. J. Hartman, and A. Tomasz. 1985. Increased amounts of a novel penicillin-binding protein in a strain of methicillin-resistant Staphylococcus aureus exposed to nafcillin. J. Clin. Investig. 76:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, S., and H. M. Sweeney. 1968. Constitutive penicillinase formation in Staphylococcus aureus owing to a mutation unlinked to the penicillinase plasmid. J. Bacteriol. 95:1368-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 6.Filee, P., K. Benlafya, M. Delmarcelle, G. Moutzourelis, J. M. Frere, A. Brans, and B. Joris. 2002. The fate of the BlaI repressor during the induction of the Bacillus licheniformis BlaP beta-lactamase. Mol. Microbiol. 44:685-694. [DOI] [PubMed] [Google Scholar]
- 7.Finan, J. E., A. E. Rosato, T. M. Dickinson, D. Ko, and G. L. Archer. 2002. Conversion of oxacillin-resistant staphylococci from heterotypic to homotypic resistance expression. Antimicrob. Agents Chemother. 46:24-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory, P. D., R. A. Lewis, S. P. Curnock, and K. G. Dyke. 1997. Studies of the repressor (BlaI) of beta-lactamase synthesis in Staphylococcus aureus. Mol. Microbiol. 24:1025-1037. [DOI] [PubMed] [Google Scholar]
- 9.Hardt, K., B. Joris, S. Lepage, R. Brasseur, J. O. Lampen, J. M. Frere, A. L. Fink, and J. M. Ghuysen. 1997. The penicillin sensory transducer, BlaR, involved in the inducibility of beta-lactamase synthesis in Bacillus licheniformis is embedded in the plasma membrane via a four-alpha-helix bundle. Mol. Microbiol. 23:935-944. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu, K., K. Asada, E. Suzuki, K. Okonogi, and T. Yokota. 1992. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 298:133-136. [DOI] [PubMed] [Google Scholar]
- 11.Joris, B., K. Hardt, and J. M. Ghuysen. 1994. Induction of B-lactamase and low affinity penicillin binding protein 2′ synthesis in gram positive bacteria, p. 505-515. In J. M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Science, New York, N.Y.
- 12.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 13.Lewis, R. A., and K. G. Dyke. 2000. MecI represses synthesis from the beta-lactamase operon of Staphylococcus aureus. J. Antimicrob. Chemother. 45:139-144. [DOI] [PubMed] [Google Scholar]
- 14.Matsuhashi, M., M. D. Song, F. Ishino, M. Wachi, M. Doi, M. Inoue, K. Ubukata, N. Yamashita, and M. Konno. 1986. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J. Bacteriol. 167:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKinney, T. K., V. K. Sharma, W. A. Craig, and G. L. Archer. 2001. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and beta-lactamase regulators. J. Bacteriol. 183:6862-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niemeyer, D. M., M. J. Pucci, J. A. Thanassi, V. K. Sharma, and G. L. Archer. 1996. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J. Bacteriol. 178:5464-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosato, A. E., B. N. Kreiswirth, W. A. Craig, W. Eisner, M. W. Climo, and G. L. Archer. 2003. mecA/blaZ corepressors in clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 47:1460-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma, V. K., C. J. Hackbarth, T. M. Dickinson, and G. L. Archer. 1998. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. J. Bacteriol. 180:2160-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittman, V., H. C. Lin, and H. C. Wong. 1993. Functional domains of the penicillinase repressor of Bacillus licheniformis. J. Bacteriol. 175:7383-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, H. Z., C. J. Hackbarth, K. M. Chansky, and H. F. Chambers. 2001. A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science 291:1962-1965. [DOI] [PubMed] [Google Scholar]