Abstract
Although the archaeal and eukaryotic nucleosome core histones evolved from a common ancestor, conserved lysine residues are present at DNA-binding locations in all four eukaryotic histones that are not present in the archaeal histones. Introduction of lysine residues at the corresponding locations into an archaeal histone, HMfB, generated a variant with increased affinity for DNA that formed more compact complexes with DNA. However, these complexes no longer facilitated the circularization of short DNA molecules and had lost the flexibility to wrap DNA alternatively in either a negative or positive supercoil.
Sequences are known for ∼30 members of the HMf family of archaeal histones. They range in length from 65 to 69 residues with 60 to >90% sequence identity (14, 16; http://www.biosci.ohio-state.edu/∼microbio/Archaealhistones). Thesesmall proteins are simply histone folds (1) comprised of three α-helices (α1, α2, and α3), which are separated by two β-strand loops (L1 and L2). They have structures very similar to those of the histone folds that form the central globular regions of the four eukaryotic nucleosome core histones, H2A, H2B, H3, and H4 (6, 7), and with both structure and primary sequence conservation (Fig. 1A), it seems clear that archaeal and eukaryotic histones have evolved from a common ancestor (1, 7, 14, 16). In both cases, dimer formation is required for histone fold stability, but archaeal histones form both homodimers and heterodimers with different partners, whereas the eukaryotic histones form only H2A-H2B and H3-H4 heterodimers. Similarly, homodimers of a single archaeal histone can assemble to form a histone core that can bind and wrap DNA in a positive or negative supercoil (9-11), whereas the eukaryotic histones cannot polymerize individually, and the eukaryotic nucleosome core always contains two H2A-H2B and two H3-H4 heterodimers with the DNA wrapped in a negative supercoil.
FIG. 1.
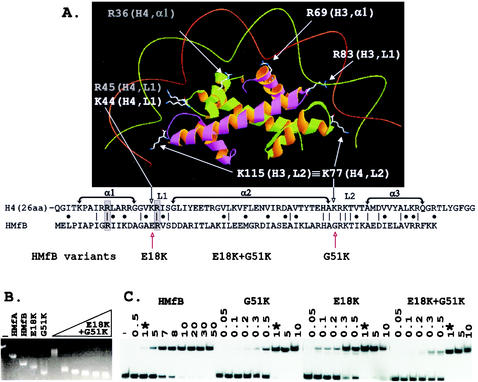
Histone fold-DNA complex, sequence alignment, and electrophoretic mobility shift assays of complexes formed by HMfB variants. (A) The histone folds of a histone H3-H4 heterodimer with conserved side chains positioned to bind DNA as in the eukaryotic nucleosome (7, 8). The side chains of lysines conserved at the same location in L2 in all four eukaryotic histones, illustrated in the figure by K115 in H3 and K77 in H4, extend the contacts of each histone fold dimer by one-half helical turn in each direction (8). As identified in the sequence alignment, the corresponding histone fold position is occupied by G51 in HMfB and by a glycine in almost all archaeal histones (16). The sequences that form α1, α2, α3, L1, and L2 of the histone folds of H4 and HMfB are indicated, with identical residues and conservative differences identified by vertical lines and dots, respectively. Conserved arginines located in α1 and L1 that also extend side chains to make DNA contacts (7, 8) are boxed and shaded. (B) Agarose gel electrophoresis of linear pBR322 (lane −) and of the complexes formed at 25°C by wild-type HMfA and HMfB (15, 16) and the HMfB, E18K, and G51K variants (18) at saturating histone-to-pBR322 DNA ratios is shown; adjacent lanes show the complexes formed by assembly of the HMfB E18K+G51K variant at histone-to-pBR322 DNA mass ratios of 1, 3, 4, 5, 6, and 7. The histone-DNA assembly conditions, agarose gel electrophoresis, and ethidium bromide staining protocols have been described in detail previously (15). (C) Autoradiogram of the polyacrylamide gel electrophoretic separations of the complexes formed at 25°C in reaction mixtures that contained 0.1 ng of 32P-labeled 110-bp clone 20 DNA (3), 1 ng of competitor sonicated herring sperm DNA, and the amounts (in nanograms) of HMfB or the HMfB variant listed above the gel. Lane −, protein-free DNA control; ∗, the lanes that contained the complexes formed under identical conditions by 1 ng of HMfB or 1 ng of a HMfB variant. The histone-DNA assembly conditions, polyacrylamide gel electrophoresis, and autoradiography procedures used have been described previously (3, 4).
In the crystal structure (7), the histone folds of each histone dimer in the eukaryotic nucleosome contact DNA over ∼2.5 sequential helical turns, and residues in α1, L1, and L2 appear to be directly responsible for DNA binding (Fig. 1A). However, experimental validation and quantitation of the importance of individual residue interactions with DNA are confounded by the complexity of the nucleosome. Histone variants can be generated, but they must be assembled with a heterodimer partner and assayed for DNA binding and complex formation within the context of a histone core that also contains a second copy of this variant heterodimer plus two other histone heterodimers with wild-type DNA binding properties. To pursue this issue, we investigated the DNA binding and complex assembly properties of archaeal histone HMfB variants that had residue changes introduced at sites conserved in both the archaeal and eukaryotic histones (18). Consistent with the crystal structure (7), nonconservative substitutions of residues that were predicted to participate directly in DNA binding generated HMfB variants with reduced abilities to bind and compact DNA (18). Overall, the archaeal histones are most similar to the histone fold of eukaryotic histone H4 (Fig. 1A). However, none of the archaeal histones have a lysine residue at the DNA-binding site in L1 occupied by K44 in H4, and only the five paralogs in Methanoccocus jannaschii have a lysine at the DNA-binding site in L2 occupied by K77 in H4, a position that is also filled by a lysine in all three other eukaryotic histones, namely, by K74 in H2A, K82 in H2B, and K115 in H3 (Fig. 1A). Given the common ancestry, it seemed surprising that homologs of DNA-binding lysine residues conserved in all eukaryotic histones were not also present in the archaeal histones. Specifically, a negatively charged residue (E18) occupies the corresponding position in L1 in HMfB, and a glycine (G51) fills the corresponding position in L2 (Fig. 1A). HMfB variants, E18K and G51K, were therefore constructed, and intriguingly, these had increased affinity for DNA and formed complexes with DNA that were more compact than those formed by wild-type HMfB (18). With this result, it seemed important to determine if having lysines at these sites compromised the unique properties of archaeal histones, namely, their abilities to form complexes that circularize DNA molecules as short as 80 bp (2, 15) and to form complexes that can wrap DNA alternatively and interchangeably in either a negative or positive supercoil (9-11).
Preparation of recombinant HMfB and HMfB variants.
After isopropyl β-d-thiogalactopyranoside (IPTG)-induced synthesis in Escherichia coli JM105, preparations of recombinant HMfB and the HMfB variants E18K and G51K were purified and quantitated as previously described (15). A Quik-Change site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) was used to change codon 18 of the hmfB gene in the expression plasmid pKS323 that encodes the HMfB G51K variant from GAA to AAA (18). The presence of the desired mutation was confirmed by sequencing, the resulting plasmid was transformed into E. coli JM105, and synthesis of the encoded HMfB E18K+G51K variant was induced by addition of 400 μM IPTG to a culture of this transformant growing in Luria-Bertani medium that contained 100 μg of ampicillin/ml. The HMfB E18K+G51K variant was purified by the procedure used for HMfB (15), except that the heparin-Sepharose column was preequilibrated with 45 mM potassium citrate and 50 mM Tris-HCl (pH 8), and bound proteins were eluted with a linear gradient of 45 to 200 mM potassium citrate in 50 mM Tris-HCl (pH 8). Circular dichroism spectra of the purified HMfB and HMfB E18K, G51K, and E18K+G51K variants were measured with an AVIV 62A-DS spectropolarimeter (Aviv, Lakewood, N.J.) (15, 18). They were essentially identical (not shown), consistent with these proteins’ having very similar and equally folded structures in solution.
Electrophoretic mobility gel shift assays.
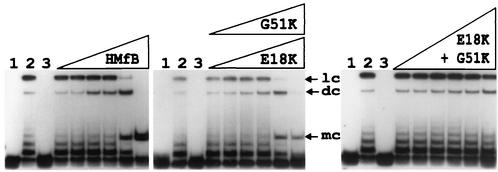
Archaeal histone assembly on DNA molecules longer than ∼2 kbp results in complexes that migrate faster during agarose gel electrophoresis than the protein-free DNA molecule (15). The extent of this compaction is reduced by residue substitutions that reduce the affinity of HMfB for DNA (18). However, the complexes formed by the HMfB E18K and G51K variants had increased electrophoretic mobilities, consistent with forming complexes more compact than those formed by wild-type HMfB (18), and the HMfB E18K+G51K variant formed complexes with linear pBR322 DNA that migrated even faster than those formed by the HMfB E18K and G51K variants (Fig. 1B). Archaeal histone binding to shorter DNA molecules results in complexes with decreased electrophoretic mobilities relative to that of the histone-free DNA, and this can be detected and quantitated by a more conventional polyacrylamide gel retardation assay (15). As shown in Fig. 1C, the E18K, G51K, and E18K+G51K variants had very similar affinities for a 110-bp DNA molecule, and affinities that were ∼10-fold higher than the affinity of the wild-type HMfB (apparent Kd, 2.7 ± 0.5 nM) for this short DNA molecule (3, 4).
DNA circularization.
To determine if complexes formed by the HMfB variants still retained the ability to facilitate end-to-end ligation of DNA molecules, complexes were assembled on an 88-bp 32P-labeled linear DNA molecule with 4-nucleotide complementary single-strand extensions that was used previously for such assays (2, 15). These complexes were then incubated with T4 DNA ligase, and after protein removal, the DNA molecules generated were separated by electrophoresis and visualized by autoradiography. As documented for HMfB (2), complexes formed by the HMfB E18K and G51K variants also facilitated the end-to-end ligation of this DNA molecule, resulting in 92-bp covalently closed circular DNA molecules, although fewer such circular DNAs were generated by the variants than by wild-type HMfB. In contrast, ligase was unable to circularize this 88-bp DNA molecule when held in a complex by the HMfB E18K+G51K variant (Fig. 2).
FIG. 2.
Autoradiogram of the electrophoretic separation of the DNA products generated by incubation of HMfB and the HMfB variants with an 88-bp DNA and DNA ligase. The 88-bp DNA molecule was generated by SpeI and XbaI digestion of pLITMUS28Δ10; it was 32P labeled and used in circularization assays as previously described (2, 15). Lane 1, 0.5-ng aliquot of the 32P-labeled DNA; lane 2, products of incubation of this DNA with T4 DNA ligase for 12 h at 16°C in the absence of histones; lane 3, products of incubation of an aliquot of this DNA with 10 ng of HMfB or an HMfB variant followed by deproteinization without exposure to DNA ligase. The adjacent lanes contained the DNA products resulting from reaction mixtures that contained 0.5 ng of DNA and 0.5, 1, 3, 5, 10, or 25 ng of the indicated histone; the products were incubated for 12 h at 16°C with T4 DNA ligase and then subjected to deproteinization. Almost identical results were obtained with the E18K (shown) and G51K variants. Linear concatemer (lc), dimer circle (dc), and monomer circle (mc) ligation products are identified (3). Note that monomer circles were not generated when aliquots of the DNA were incubated with DNA ligase in the absence of an archaeal histone (lanes 2).
DNA topology assays.
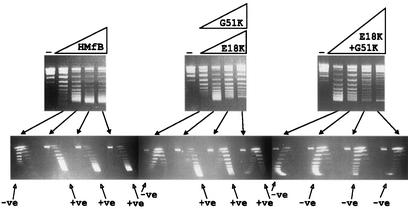
To determine if adding the lysine residues also decreased the topological flexibility of the complexes formed, complexes were assembled on relaxed circular pUC18 DNA molecules in 10 mM Tris-HCl buffer (pH 8) containing 50 mM NaCl, 2 mM K3PO4, and 1 mM EDTA. Under these solution conditions, at low histone-to-DNA ratios, wild-type HMfB forms complexes in which the DNA is wrapped in a negative supercoil, but this changes spontaneously to positive supercoiling with increasing histone addition (9-11). The HMfB E18K and G51K variants also formed complexes in which the DNA topology changed from negative to positive supercoiling with increasing histone addition. However, the DNA in the complexes formed by the HMfB E18K+G51K variant remained negatively supercoiled at all histone-to-DNA ratios (Fig. 3).
FIG. 3.
One- and two-dimensional agarose gel electrophoretic separations of the topoisomers generated by HMfB or HMfB variants assembled on relaxed, circular pUC18 DNA molecules. The procedures used to assemble complexes, remove plectonemic supercoils, deproteinize, and separate pUC18 topoisomers by one- and two-dimensional agarose gel electrophoresis have been described previously (9, 10). The control lanes (−) contained relaxed pUC18 DNA; adjacent lanes contained the pUC18 topoisomers generated by HMfB or HMfB variant assembly on aliquots of this DNA at histone-to-DNA mass ratios of 0.4, 0.6, 0.8, and 1. Almost identical results were obtained with the E18K variant (shown) and G51 variant. Topoisomers were separated in the first dimension (upper gels) on the basis of linking number, with increasing linking number resulting in increasing mobility, and in the second dimension (lower gels) on the basis of negative (−ve) or positive (+ve) supercoiling. Nicked and relaxed circular DNAs migrated together and formed the band near the top of each gel.
Discussion.
The results obtained demonstrate that E18K, G51K, and E18K+G51K substitutions increase the affinity of an archaeal histone for DNA (Fig. 1B and C) and are consistent with the nucleosome crystal structure predictions that the side chains of lysines at these histone fold locations participate directly in DNA binding (Fig. 1A) (7, 8). To form a stable complex with DNA, a minimum of two (HMfB)2 homodimers must polymerize (2, 4, 9, 14), and DNA binding by such an archaeal histone tetramer protects ∼60 bp from micrococcal nuclease digestion (4, 13). If the DNA molecule is ≥80 bp, the ends of the bound DNA in such a complex can be ligated readily to form a circular DNA (3), and the flexibility of these complexes also allows the DNA to be bound in either a negative or positive supercoil and to switch between these configurations depending on the assembly conditions (9-11). Tetramers formed by the HMfB E18K and G51K variants must have four additional lysines appropriately positioned for DNA binding, and tetramers formed by the HMfB E18K+G51K variant will have eight such additional lysines. With four additional lysine-DNA interactions possible, the E18K and G51K variants formed complexes in which the ends of an 88-bp molecule were less often appropriately positioned for ligase-catalyzed circularization (Fig. 2), but these variants still did form complexes that could switch from negative to positive DNA supercoiling (Fig. 3). However, with eight additional DNA-binding interactions possible per histone tetramer, the complexes formed by the HMfB E18K+G51K variant had apparently no flexibility. The ends of the 88-bp molecule were never held in appropriate proximity and/or orientation for joining by ligase, and DNA binding resulted only in negative supercoiling.
Lysines are nevertheless conserved at these histone-fold locations in the nucleosome core histones of all eukaryotes, arguing that their acquisition must have been beneficial at a very early stage in eukaryotic evolution before eukaryotic divergence. The ability of archaeal histones to dimerize with alternative partners (16) and to assemble cores with different histone compositions and DNA affinities (4, 9) was apparently sacrificed with genome expansion in favor of forming a more rigid and regularly organized chromatin. The N- and C-terminal sequences (histone tails) that extend from the histone folds of the eukaryotic histones, regions that are not present in the archaeal histones, must then have evolved to facilitate the destabilization of eukaryotic chromatin for replication and transcription. These tails carry the targets for the posttranslation histone acetylation, phosphorylation, methylation, and ubiquitinylation events that regulate gene expression and replication in eukaryotes (12, 19). Intriguingly, however, although archaeal histones do not have histone tails, archaeal genomes do encode proteins with sequences related to those of eukaryotic histone acetylases and deacetylases (14). This includes the genomes of Crenarchaea that do not have histones, and it has been shown that Sso10b/Alba, a nucleoid protein with no structural relationship to histones (20), is acetylated in vivo in Sulfolobus solfataricus and that this acetylation reduces the affinity of Sso10b/Alba for DNA (5). It seems possible, therefore, that lysine acetylation evolved very early in the archaeal lineage as a generic mechanism to modulate DNA binding by nucleoid proteins, possibly before the use of the histone fold for DNA packaging and almost certainly before the evolution of histone tails. However, consistent with the lack of histone tails, all attempts to acetylate HMfB in vitro using yeast histone acetylase have been unsuccessful, and we have also been unable to acetylate calf thymus histones in vitro using His-tagged recombinant MTH0817, a Methanothermobacter thermautotrophicus protein (17) that has a sequence related to the sequences of the eukaryotic ELP3 family of histone acetylases (Y. Xie, M. R. Parthun, and J. N. Reeve, unpublished results).
Acknowledgments
This research was supported by NIH grant GM53185.
REFERENCES
- 1.Arents, G., and E. N. Moudrianakis. 1995. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA 92:11170-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, K. A., C. Chow, and J. N. Reeve. 1999. Histone stoichiometry and DNA circularization in archaeal nucleosomes. Nucleic Acids Res. 27:532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, K. A., S. L. Pereira., J. Widom, and J. N. Reeve. 2000. Archaeal histone selection of nucleosome positioning sequences and the procaryotic origin of histone-dependent genome evolution. J. Mol. Biol. 303:25-34. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, K. A., F. Marc, K. Sandman, and J. N. Reeve. 2002. Both DNA and histone fold sequences contribute to archaeal nucleosome stability. J. Biol. Chem. 277:9293-9301. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148-151. [DOI] [PubMed] [Google Scholar]
- 6.Decanniere, K., A. M. Babu, K. Sandman, J. N. Reeve, and U. Heinemann. 2000. Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus. J. Mol. Biol. 303:35-47. [DOI] [PubMed] [Google Scholar]
- 7.Luger, K., A. W. Mäder, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 8.Luger, K., and T. J. Richmond. 1998. DNA binding within the nucleosome core. Curr. Opin. Struct. Biol. 8:33-40. [DOI] [PubMed] [Google Scholar]
- 9.Marc, F., K. Sandman, R. Lurz, and J. N. Reeve. 2002. Archaeal histone tetramerization determines DNA affinity and the direction of DNA supercoiling. J. Biol. Chem. 277:30879-30886. [DOI] [PubMed] [Google Scholar]
- 10.Musgrave, D. R., K. M. Sandman, and J. N. Reeve. 1991. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc. Natl. Acad. Sci. USA 88:10397-10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musgrave, D. R., P. Forterre, and A. Slesarev. 2000. Negative constrained DNA supercoiling in archaeal nucleosomes. Mol. Microbiol. 35:341-349. [DOI] [PubMed] [Google Scholar]
- 12.Narliker, G. J., H.-Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 13.Pereira, S. L., and J. N. Reeve. 1999. Archaeal nucleosome positioning sequence from Methanothermus fervidus. J. Mol. Biol. 289:675-681. [DOI] [PubMed] [Google Scholar]
- 14.Reeve, J. N. 2003. Archaeal chromatin and transcription. Mol. Microbiol. 48:587-598. [DOI] [PubMed]
- 15.Sandman, K., K. Bailey, S. L. Pereira, D. Soares, W.-T. Li, and J. N. Reeve. 2001. Archaeal histones and nucleosomes. Methods Enzymol. 334:116-129. [DOI] [PubMed] [Google Scholar]
- 16.Sandman, K., and J. N. Reeve. 2001. Chromosome packaging by archaeal histones. Adv. Appl. Microbiol. 50:75-99. [DOI] [PubMed] [Google Scholar]
- 17.Smith, D. R., L. A. Doucette-Stamm, C. DeLoughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. Church, C. J. Daniels, J. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. The complete genome sequence of Methanobacterium thermoautotrophicum strain ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soares, D. J., K. Sandman, and J. N. Reeve. 2000. Mutational analysis of archaeal histone-DNA interactions. J. Mol. Biol. 297:39-47. [DOI] [PubMed] [Google Scholar]
- 19.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 20.Wardleworth, B. N., R. J. Russell, S. D. Bell, G. L. Taylor, and M. F. White. 2002. Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 21:4654-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]