Abstract
The concentration of the cell division proteins FtsZ, FtsA, and ZipA and their assembly into a division ring during the Escherichia coli B/r K cell cycle have been measured in synchronous cultures obtained by the membrane elution technique. Immunostaining of the three proteins revealed no organized structure in newly born cells. In a culture with a doubling time of 49 min, assembly of the Z ring started around minute 25 and was detected first as a two-dot structure that became a sharp band before cell constriction. FtsA and ZipA localized into a division ring following the same pattern and time course as FtsZ. The concentration (amount relative to total mass) of the three proteins remained constant during one complete cell cycle, showing that assembly of a division ring is not driven by changes in the concentration of these proteins. Maintenance of the Z ring during the process of septation is a dynamic energy-dependent event, as evidenced by its disappearance in cells treated with sodium azide.
During the cell cycle of exponentially growing Escherichia coli cells, several events occur in an ordered and periodic fashion; among them, the initiation of chromosome replication, nucleoid segregation, and cell division are the most conspicuous. One of these events is the localization of at least nine proteins in the cell center, forming a ring that constricts simultaneously as the septum grows (25, 29). Ring proteins assemble in a certain order, and recruitment of one protein to the ring depends on the localization of the preceding ones, with FtsZ being the first protein known to assemble into a Z ring. Assembly of the Z ring does not depend on any other known gene or protein. FtsA and ZipA follow and depend on FtsZ localization, although they are both independent of each other and are both necessary for the recruitment of the rest of the septal ring proteins (14, 29).
FtsZ is a structural homologue of eukaryotic tubulin (22), and like tubulin, it has GTPase and polymerization activities in vitro. The structure of the Z ring in vivo is not known, but it has been shown that it is highly dynamic, being continuously remodeled, and that this dynamic behavior is related to its GTPase activity (40). FtsA is a peripheral membrane protein that belongs to the actin/hsp70/sugar kinase family and binds ATP (37). ZipA is a membrane protein able to interact with FtsZ and to induce it to form filament bundles in vitro (13, 33).
Localization of the Z ring has been proposed to be governed by at least two mechanisms: nucleoid occlusion might prevent ring formation in the space occupied by the nucleoid (48), while the products of the minCDE genes inhibit ring formation at the cell poles (for a review, see reference 35). In addition, the timing of Z ring assembly seems to be linked to chromosome replication, either to the early stages, as in Bacillus subtilis (15), or to the termination phase, as in E. coli (6). However, it has been postulated also that FtsZ ring assembly is independent of chromosome replication and dependent on cell size (12).
While in eukaryotes, progression through the cell cycle is based on a balance between transcriptional regulation and controlled proteolysis (42), it is not known if a mechanism of this type is present in E. coli. Examination of protein levels during the E. coli cell cycle has so far revealed no evidence of a developmental program. In an early report, none of about 750 proteins detected did show any cell cycle-related variation (24); a more recent work showed variation in three out of 1,000 detected proteins (3), but the relation of these three proteins (the products of the genes dps, gapA, and pyrI) to the cell cycle is uncertain. More direct approaches have been applied to measure the levels of DnaK (18) and of the major penicillin-binding proteins of E. coli (47) during the cell cycle and have found them to be invariant. Nevertheless, as most of the cell division proteins are expressed at low levels, it seems unlikely that even a doubling in their levels would be detected by two-dimensional gel analysis (24).
The levels of ftsZ transcripts in E. coli oscillate during the cell cycle (10, 50), while transcription of ftsA and ftsZ genes is in its turn coupled to chromosome replication (41, 21). It has been found also that the levels of FtsZ are rate limiting for cell division (28, 46) and that they are inversely dependent on growth rate, so that the amount of FtsZ per cell is constant, independently of the cell size, as expected for a structural or regulatory component of the cell septum (2). This, together with the fact that the GTPase and polymerization activities of FtsZ are concentration dependent (39, 45), suggests that the intracellular levels of FtsZ might exert some control over the assembly of the Z ring. Oscillations in the levels of FtsZ have been described in other organisms, like Caulobacter crescentus (31), Prochlorococcus sp. (17), and Synechococcus elongatus (27). In this work, the levels of FtsZ, FtsA, and ZipA proteins and the assembly of these proteins in the septal ring have been analyzed in synchronous cultures of E. coli B/r K obtained by the membrane elution technique (16) and were found to remain constant along a cell cycle.
MATERIALS AND METHODS
Media and growth conditions.
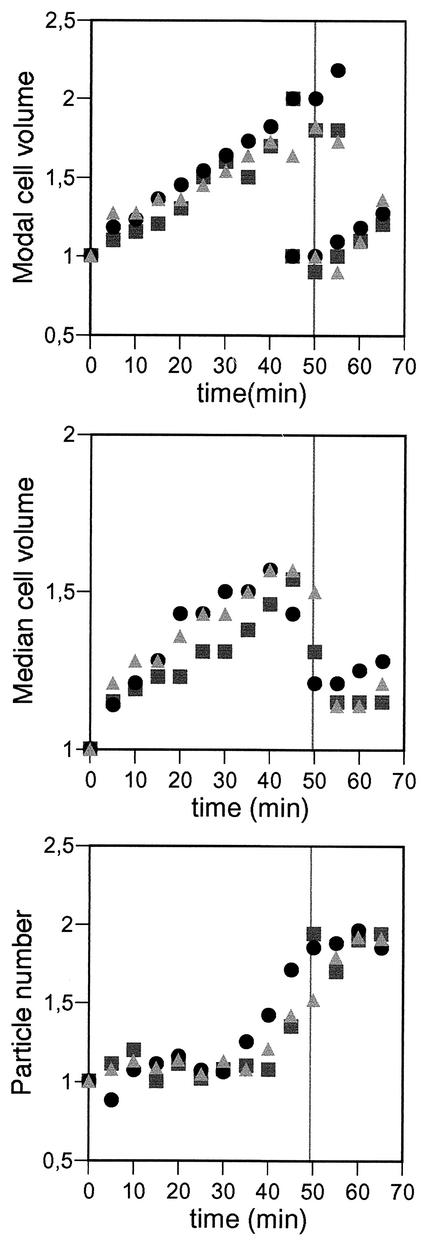
E. coli B/rK (C. Helmstetter) was used for the synchronization experiments, and E. coli MC1061 (36) was used for the energy depletion experiment. Cells were grown at 37°C in M9 minimal medium (36) supplemented with 0.2% glucose or in Luria-Bertani (LB) medium (36). Newborn E. coli B/r K cells were obtained by the membrane elution technique (16) from cultures growing exponentially in M9 minimal medium supplemented with 0.2% glucose at 37°C. Samples were collected for 1.5 min every 5 min during a period spanning more than one doubling time. All the samples were then incubated at 37°C with shaking for the rest of the sampling period; they were then fixed in 0.75% formaldehyde for particle counting and immunofluorescence analysis or centrifuged and resuspended in lysis buffer for Western blotting. This procedure allows reconstruction of a full cell division cycle from the samples while maintaining the sampling size at values sufficient for the subsequent analysis. Synchrony of all the cultures was considered to be satisfactory, as it complied with the following criteria: as shown in Fig. 1, the median and modal cell volumes and cell numbers measured relative to those obtained for the first sampled portion in three independent experiments showed that the cell volume distributions were unimodal during the first 40 min. At minute 45, some cells had already divided, because the cell number started to increase and the cell volume distribution became bimodal, containing a major fraction of large cells and a minor one of small newborn cells. At minute 50, both fractions were of similar sizes, and consequently, the median of the cell volume distribution decreased. At minute 55, most of the cells had divided, the cell number had doubled, and the median of the cell volume distribution had decreased almost to the initial level, although the distribution was still bimodal, with a minor peak of large, presumably undivided, cells. This is in agreement with a doubling time of 49 min when measured in a nonsynchronous culture under the same conditions. In addition, the degree of synchrony was calculated as described by Scherbaum (38) as F = [(n − n0)/n0] · [1 − (t/τ)], where n0 and n are the initial and final particle numbers, t is the time span during which synchronous division takes place, and τ is the generation time. For the three experiments shown in Fig. 1, the values calculated were 0.75, 0.76, and 0.82.
FIG. 1.
Degree of synchrony of E. coli B/r K cultures obtained by the membrane elution method. The quality of the synchronization was monitored by measuring cell numbers and cell volumes with a Coulter counter and a Coulter channel analyzer. The modal and median cell volume and the number of cells are shown relative to the initial values. Typical cell numbers for the first point were 1 · 106 to 2 · 106 cells/ml in a volume of 10 ml. The gray vertical lines mark the doubling time for nonsynchronous cultures of this strain under the same experimental conditions. Each symbol represents results for an individual experiment.
Cell parameter measurements, photography, and immunofluorescence microscopy.
Cells were fixed in 0.75% formaldehyde. Cell number and cell volume were measured with a ZM Coulter counter with a 30-μm-diameter orifice connected to a C1000 Channelyzer (both from Coulter Electronics). For each sample, the mode or modes and the median of the cell volume distributions were recorded and are referred to as modal and median cell volumes. For immunofluorescence microscopy, cells were prepared as described by Addinall and Lutkenhaus (1). The final lysozyme concentration used was 8 μg/ml, and the permeabilization time was 1 min. The primary antibody used for FtsZ immunolocalization was MAb4, a monoclonal antiserum raised against FtsZ (43). The anti-FtsZ polyclonal antiserum MVJ9 and the anti-EF-Tu polyclonal antiserum MVJ4 have been described previously (30). The anti-FtsA polyclonal antiserum MVM1 and the anti-ZipA polyclonal antiserum MVC1 were obtained by immunization of rabbits with FtsA and ZipA purified as described by Yim et al. (49) and RayChaudhuri and Park (32), respectively. Cy3-conjugated anti-mouse or anti-rabbit serum (Amersham Pharmacia Biotech) was used as the secondary antibody. Cells were observed by fluorescence microscopy with a Zeiss Axiolab HBO 50 microscope with a 100× immersion oil lens. Images were captured with a Sensys charge-coupled device camera (Photometrics) fitted with an HQ:CY3 filter (excitation, 545/30 nm; emission, 610/75 nm; beam splitter, 565LP). The software used for image capture was IPLab Spectrum, and Adobe Photoshop version 5.5 software was used for processing. Images were analyzed with Object-Image version 1.62 (N. Vischer, University of Amsterdam). One hundred cells were analyzed for each time point.
Immunoblotting.
Cells were lysed by resuspension in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer and heated for 5 min at 95°C. Proteins were separated by SDS-PAGE in 10% acrylamide gels and transferred to Immobilon-P membranes (pore size, 0.45 μm; Millipore Co.) with a Milliblot graphite electroblotter system (Millipore Co.). Incubation of the membranes with antisera followed standard protocols (36). The rabbit antibodies were detected with protein A coupled to peroxidase (Bio-Rad) and developed with the BM chemiluminescence blotting substrate (POD; Roche Molecular Biochemicals). Autoradiographs were analyzed with NIH Image version 1.61 software (W. Rasband, National Institutes of Health). Protein levels were always expressed as the ratio of their band volumes to that of EF-Tu and made relative to the value obtained for the first point.
For the quantification of FtsZ, FtsA, and ZipA, exponential nonsynchronous cultures of E. coli B/rK were collected at an optical density at 600 nm of 0.2 to 0.3. A portion was fixed with formaldehyde for particle counting, and another portion was centrifuged, resuspended in lysis buffer, and heated to 100°C for 5 min. Quantification was done by SDS-PAGE and immunoblotting. The gels included extracts from independent cultures and a set of standards with known amounts of purified FtsZ, FtsA, or ZipA. FtsZ was expressed and purified as described by Rivas et al. (34). FtsA was expressed and purified according to the procedure of Yim et al. (49). ZipA was expressed and purified as described by Ray Chaudhuri (33). Proteins were quantified by the method of Bradford with a commercial assay (Bio-Rad).
RESULTS
Timing of FtsZ ring assembly during the cell cycle.
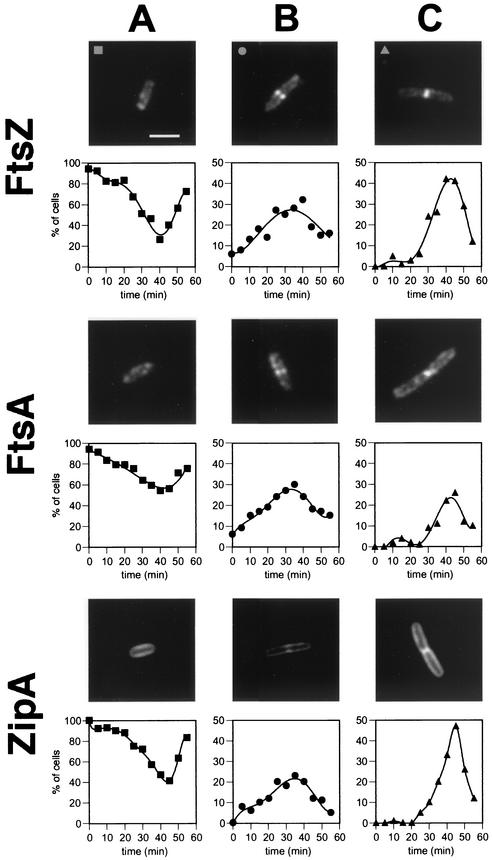
To determine the time of assembly of FtsZ, FtsA, and ZipA into a midcell ring during the cell cycle of E. coli B/rK, samples of synchronous cultures (Fig. 1) obtained as described in Materials and Methods were analyzed. For FtsZ, immunofluorescence microscopy revealed the presence of three types of cells: cells without visible rings (Fig. 2A), cells with two dots at opposite sites in the cell center (Fig. 2B), and cells with a sharp band in their centers (Fig. 2C). The two-dot structures have been interpreted as the two-dimensional projections of a three-dimensional ring with a relatively low amount of fluorescent material and have been called open rings, while the sharp bands, called closed rings, are interpreted as rings that contain more fluorescent material (6). It has been postulated also that both open and closed structures occur sequentially in the cell as accumulation of fluorescent material progresses along the cycle. Some of the cells without rings showed one single weak fluorescent dot at midcell that might be the precursor of the open ring structure, but these single dots appeared diffuse and proved difficult to quantify reliably, and therefore these cells were counted as cells without rings.
FIG.2.
Immunofluorescence microscopy showing the three different cell types after staining with the corresponding antisera and the fraction of cells showing each of the three patterns of localization of FtsZ, FtsA, and ZipA during one complete cell cycle. Cells taken at different times from the synchronous cultures were fixed and analyzed by immunofluorescence microscopy using specific antibodies. One hundred cells were analyzed for each time point. (A) Newborn cells showed no rings. (B) The rings were seen first as two fluorescent dots at both sides of the cell, i.e., the open ring. (C) Shortly before division, the ring is seen as a sharp and continuous image, i.e., the closed ring. Note the different scales in the y axes. The bar in panel A represents 2 μm and applies to all panels.
The pattern of immunolocalization of FtsA and ZipA was nearly identical to that of FtsZ, showing also open and closed ring images (not shown). It is known that the localization of FtsZ is independent of both FtsA and ZipA, while the localization of these two proteins is dependent on FtsZ. If the localization of these three proteins is sequential, the assembly must occur in less than 5 min, as we do not find any significant delay in the visualization of the three different rings, which appear at similar frequencies along the course of the experiments.
The frequencies of the three cell types (cells with no rings, open rings, and closed rings) followed different time courses along the cycle (Fig. 2). Cells with no rings were predominant among the population of newly born cells (95% for FtsZ and FtsA and 100% for ZipA). The population of cells with open rings showed a broad temporal distribution, increasing gradually until minute 40 for FtsZ and minute 35 for FtsA and ZipA and then decreasing, therefore spanning the whole cell cycle, though always at a rather low frequency. The distribution of cells with a closed ring was more sharply defined, being very low before minute 30 and then increasing rapidly to reach a maximum at minute 40 (FtsZ) and at minute 45 (FtsA and ZipA) (Fig. 2). A temporal sequence can be defined, starting with the newborn cells without rings that become cells with open rings as they grow and the division proteins assemble and finally develop into cells with closed rings before constriction and division. This sequential assembly would be consistent with the interpretation of the open and closed rings appearing as a result of continuous protein accumulation in the rings.
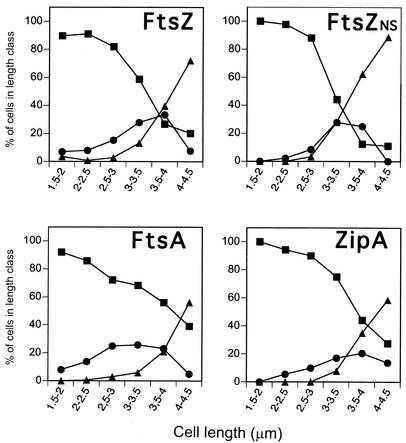
For the three proteins, the population of ringless cells was high even at late times (Fig. 2). This could be due to a poor efficiency of the immunostaining reaction caused by a low affinity of the polyclonal antisera used or simply could be a consequence of imperfect synchrony, so that at the late times during the cycle, those cells that divide disappear immediately from the populations of cells with rings and are added as two newly born cells to the cells without rings. To evaluate the two possibilities, the frequency of the three cell types was plotted as a function of cell length for these cell populations (Fig. 3). For comparison, the same analysis was done for the FtsZ rings in a nonsynchronous exponential phase culture of E. coli B/rK grown under the same conditions. For the three proteins, the shorter (younger) cells were predominantly cells without rings; open rings appeared in medium size cells, and the longer (older) cells were predominantly cells with closed rings. Moreover, the plots of FtsZ ring cell types obtained from synchronous and nonsynchronous cultures were nearly identical, showing that the large fraction of cells without rings is not an artifact of the synchronization procedure but reflects the distribution of cells in a culture. The fraction of cells with FtsZ rings (open and closed) in the nonsynchronous culture was 35%, and this fraction increased to 65% when the cells were grown in LB medium. This is in agreement with the data of den Blaauwen et al. (6), showing that there is a direct linear relation between the beginning of FtsZ ring assembly and the cell duplication time, so that assembly starts at a younger relative cell age for shorter generation times; as a consequence, the faster the culture grows, the larger will be the fraction of cells with rings. Therefore, from the data shown in Fig. 3, it can be concluded that for FtsZ and ZipA, the relatively large fraction of cells without rings found at late ages is contributed by the newborn cells resulting from division in which, as expected, the rings are not detectable. For FtsA, there is nevertheless an excess of long cells without rings, suggesting that in this case, the efficiency of labeling is lower than for the other two proteins. It is interesting that this inefficient labeling decreases the numbers of cells with rings detected but does not seem to alter the time courses of the open and closed ring frequency distributions.
FIG. 3.
Cell types as a function of cell length. Cell length was measured for all the cells of the experiments shown in Fig. 2 and for a steady-state exponentially growing, nonsynchronous culture (FtsZNS). Then the frequencies of the three cell types identified by immunofluorescence were plotted as a function of cell length. There was a small fraction of cells (less than 2% of the population) longer than 5 μm; these were mostly cells without rings that were considered anomalous and were not included. Squares, cells without rings; circles, cells with open rings; triangles, cells with closed rings.
Levels of cell division proteins during the cell cycle.
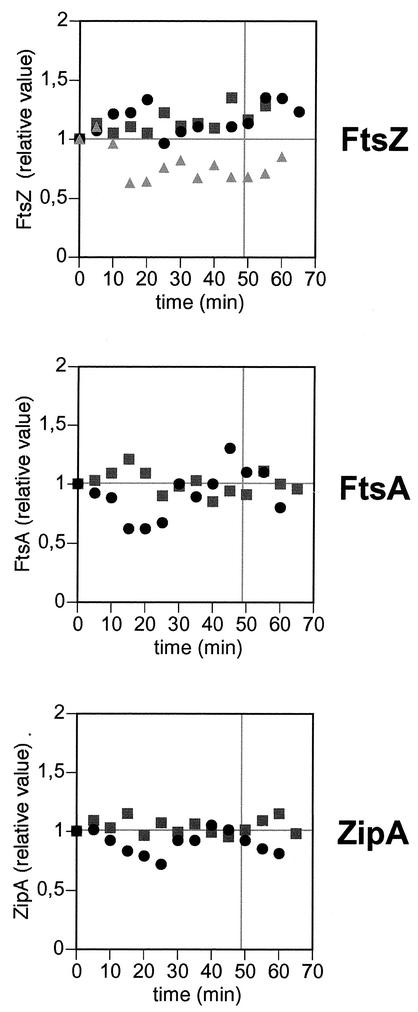
The levels of FtsZ, FtsA, and ZipA along one complete cell cycle were analyzed by Western blotting (36) in the synchronous cultures. The low cell densities obtained in the synchronous cultures (106 cells/ml, 10 ml per time point) made it difficult to obtain a reliable direct measurement of the number of molecules per cell, because even small sample losses may significantly affect the result. Therefore the elongation factor Tu (EF-Tu) was used as an internal marker to correct for sample losses and loading errors, and all the measurements were made relative to the level of the abundant EF-Tu. As EF-Tu levels are directly related to the cell mass (9), the ratios of FtsZ, FtsA, and ZipA to EF-Tu can be taken as relative measurements of the protein concentrations in the cells (units of protein per unit of cell mass). Comparison of the different experiments showed that there were no consistent trends or oscillations in the concentrations of these proteins that could be related with progression through the cell cycle (Fig. 4). For FtsZ, the three experiments shown had coefficients of variation of 8.8, 11.1, and 18.8%, while the coefficient for a series of repeated loadings of a reference sample was 11.7%. Therefore, the dispersion of the data within every experiment could be ascribed to experimental error, and it can be concluded that there are no periodicities larger than this in the concentrations of the three cell division proteins analyzed.
FIG. 4.
Relative levels of FtsZ, FtsA, and ZipA during one complete cell cycle. Samples from the synchronous cultures were taken at different times and analyzed by Western blotting with specific antibodies. Anti-EF-Tu was included in all the blots. The levels of FtsZ, FtsA, and ZipA were then measured relative to the EF-Tu band, and the ratios were made relative to the ratio at time zero; therefore, the levels of the three cell division proteins are expressed as relative concentrations. The symbols reflect results of three independent experiments for FtsZ (top) and results of two independent experiments for FtsA (middle) and ZipA (bottom). The gray vertical lines mark the doubling times of this strain in asynchronous cultures under these experimental conditions.
The concentrations of the three proteins were measured in nonsynchronous cultures of E. coli B/rK in steady-state growth under the same set of conditions. The cell dimensions were measured in contrast-phase microscopy images with the help of Object-Image version 1.62 software (N. Vischer, University of Amsterdam). At least 100 cells were measured in each case. The average cell length and diameter were 2.88 and 0.84 μm, and the cell volume was calculated by assuming cells to be cylinders with hemispherical caps. The measurements were done in three independent exponentially growing steady-state cultures. An average of 3,200 molecules of FtsZ per cell were present in this strain under these conditions. This is consistent with the value of around 5,000 molecules per cell estimated by Pla et al. (30) in E. coli K-12 strains MC1061 and W3110 and much lower than the value of 15,000 reported by Lu et al. (23) in E. coli BL21 (note that the figure of 20,000 FtsZ molecules per cell, usually found in secondary sources in the literature, refers to unpublished data cited in reference 5). The average number of FtsA molecules per cell derived from our calculation was 740, nearly four times higher than previous calculations (44), and gives an FtsZ/FtsA ratio of 5 to 1, much lower than previous estimates but similar to the one measured in B. subtilis (8). Finally, the number of ZipA molecules per cell was 1,500, similar to the estimate of 1,000 calculated by Hale and de Boer (13).
FtsZ ring assembly is energy dependent.
Stricker et al. (40) have shown that the Z ring is a highly dynamic structure subjected to continuous remodeling and, moreover, that this remodeling depends on the GTPase activity of FtsZ. This is consistent with previous in vitro studies of FtsZ assembly (26) and suggests a role for the energetic metabolism in ring assembly and maintenance.
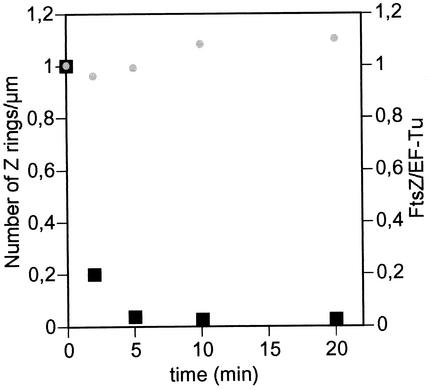
To study the energy dependence of ring assembly, the levels of FtsZ and the number of cells with Z rings were measured in an exponential culture of E. coli K-12 MC1061 in which respiration had been inhibited by addition of 15 mM sodium azide. Azide addition produces a fast depletion of the energetic pool of the cell. The levels of FtsZ remained constant for at least 20 min, while the FtsZ rings (open and closed ones) disappeared in slightly more than 2 min (Fig. 5). This confirms that the Z ring is not a static complex that can last once assembled but a dynamic one that requires a continuous energy input to persist. We conclude that it is not enough for the cell to divide to trigger FtsZ ring assembly once in a cell cycle; assembly must be actively sustained along septation, probably until completion of the septum.
FIG. 5.
Energy dependence of FtsZ ring assembly. A steady-state exponential culture of E. coli MC1061 was grown in LB at 37°C to an optical density at 600 nm of 0.2. At time zero, sodium azide was added to a final concentration of 15 mM, and samples were taken for further analysis at the times indicated. Black squares, number of FtsZ rings per unit length relative to the initial value; gray circles, FtsZ/EF-Tu ratio relative to the initial value.
DISCUSSION
Using synchronously growing cultures of E. coli B/r K obtained by the membrane elution technique, we have analyzed the levels of the FtsZ, FtsA, and ZipA in these cells as well as the course of assembly of the three proteins into a ring at midcell during the cell cycle. We have found that the concentration of the three proteins is constant throughout the cycle.
If the processes of assembly and disassembly of the Z ring were driven by a passive, protein concentration-dependent mechanism of polymerization-depolymerization, then the concentration of FtsZ in the cell should be higher at some critical time during the cycle. The critical concentration of FtsZ for in vitro polymerization stays in the order of 1 to 2 μM (45, 39), well below the concentration of around 3.5 μM that we measured in vivo in a nonsynchronous culture grown under the same conditions as the synchronous one. We found that the relative concentration of FtsZ stays constant during one full cell cycle, i.e., the observed minor variations falling within experimental error cannot be interpreted as periodic oscillations able to trigger ring assembly. Comparison of the results in Fig. 2 and 4 shows that a population formed entirely by newborn cells without rings (times 0 and 60) contains the same relative amount of FtsZ as a population in which around 80% of the cells have a central ring, either open or closed (minute 40). If the concentration of these proteins is held constant, then the amount of protein per cell will increase as the cell volume increases and will oscillate following the cell cycle. This supports the notion that in E. coli, division requires a fixed amount of FtsZ and that it takes place once this amount is reached, independently of the concentration of the protein (2, 30). As discussed by Koch (19), the mechanisms that trigger cell division cannot operate as concentration-dependent enzyme systems; instead, they must be sensitive to the amount of some cell components, the amount of FtsZ in this case. The molecular mechanism that would constitute the Z ring assembly checkpoint remains elusive.
The lack of periodicity in the concentration of cell division proteins during the cell cycle, and in particular that of FtsZ, was unexpected and raises questions concerning the role of the oscillation of the ftsZ gene transcription during the cell cycle (10). It has been argued that this oscillation might result simply from transcription silencing during replication of this region of the chromosome (50), but this would not explain the phenotype observed when ftsZ is dissociated from its natural promoters and placed under the control of an inducible promoter (10, 28). It has been proposed also that the oscillation might result from a mechanism linking cell division and chromosome replication (21, 41). The fact that an oscillating pattern of mRNA is translated into a steady pattern of protein concentration indicates that there are posttranscriptional regulatory mechanisms controlling the levels of FtsZ. Indeed, it has been found that translation of ftsZ mRNA can be inhibited by two different antisense RNAs, namely, dicF, which is encoded by a prophage gene (4), and stfZ, which comprises part of ftsA and part of ftsZ (7). Moreover, the possibility that not all the FtsZ immunoreactive protein is equally active has to be considered. If there were some posttranslational processing either to activate or to inactivate FtsZ, then it might be possible to mask an oscillation in the relatively minor levels of the active form within the total FtsZ population that would consequently remain largely invariant. In fact, heterogeneity in FtsZ preparations has been reported (23, 32), although the origin of this heterogeneity and its physiological relevance are not known.
Another mechanism that might work to control FtsZ activity independently of its concentration might be the balance between a series of positive and negative regulators of polymerization. Two of these regulators have been identified recently in B. subtilis, ZapA (11) and EzrA (20), which are, respectively, FtsZ polymer-stabilizing and -destabilizing proteins. A zapA homologue is also found in E. coli and many other bacteria, while ezrA has a much more restricted distribution and is not found in E. coli, in which other proteins, like MinC, might work as negative regulators.
Other models postulate that ring assembly is initiated at potential division sites (nucleation sites), and development of these sites is cell cycle dependent. Among them, the nucleoid occlusion model proposes that termination of DNA replication could be the trigger for cell division (48). According to these models, Z ring assembly is triggered by some previous process and is independent of the FtsZ concentration.
We find that the maintenance of the Z ring depends on energy. This fact, together with the highly dynamic behavior of FtsZ in the Z ring (40), suggests that the mechanisms that control the state of the FtsZ protein in the cell are likely to act along a significant part of the cell cycle, either at preventing premature assembly of the Z ring in young cells or to trigger and remodel it as they progress into division.
Acknowledgments
We are grateful to D. RayChaudhuri for the gift of pET15-ZIP, and to N. Nanninga for MAb4. The excellent technical work of Mercedes Casanova and Pilar Palacios is acknowledged.
Grants to M.V.'s laboratory included BIO97-1246 Plan Nacional de I+D from Ministerio de Educación y Cultura and Cell Factory BIO4-CT96-0122 Project from the European Commission. S.R. and J.M. were pre- and postdoctoral fellows of the Comunidad Autónoma de Madrid.
REFERENCES
- 1.Addinall, S. G., and J. Lutkenhaus. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178:7167-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldea, M., T. Garrido, J. Pla, and M. Vicente. 1990. Division genes in Escherichia coli are expressed coordinately to cell septum requirements by gearbox promoters. EMBO J. 9:3787-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechtloff, D., B. Grünenfelder, T. Åkerlund, and K. Nordström. 1999. Analysis of protein synthesis rates after initiation of chromosome replication in Escherichia coli. J. Bacteriol. 181:6292-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouché, F., and J. P. Bouché. 1989. Genetic evidence that DicF, a second division inhibitor encoded by the Escherichia coli dicB operon, is probably RNA. Mol. Microbiol. 3:991-994. [DOI] [PubMed] [Google Scholar]
- 5.Dai, K., and J. Lutkenhaus. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 174:6145-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Blaauwen, T., N. Buddelmeijer, M. E. G. Aarsman, C. M. Hameete, and N. Nanninga. 1999. Timing of FtsZ assembly in Escherichia coli. J. Bacteriol. 181:5167-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewar, S. J., and W. D. Donachie. 1993. Antisense transcription of the ftsZ-ftsA gene junction inhibits cell division in Escherichia coli. J. Bacteriol. 175:7097-7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feutch, A., I. Lucet, M. D. Yudkin, and J. Errington. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40:115-125. [DOI] [PubMed] [Google Scholar]
- 9.Furano, A. 1975. Content of elongation factor TU in Escherichia coli. Proc. Natl. Acad. Sci. USA 72:4780-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrido, T., M. Sánchez, P. Palacios, M. Aldea, and M. Vicente. 1993. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 12:3957-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gullbrand, B., and K. Nordström. 2000. FtsZ ring formation without subsequent cell division after replication runout in Escherichia coli. Mol. Microbiol. 36:1349-1359. [DOI] [PubMed] [Google Scholar]
- 13.Hale, C. A., and P. A. de Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175-185. [DOI] [PubMed] [Google Scholar]
- 14.Hale, C. A., and P. A. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harry, E. J., J. Rodwell, and G. Wake. 1999. Co-ordinating DNA replication and cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol. Microbiol. 33:33-40. [DOI] [PubMed] [Google Scholar]
- 16.Helmstetter, C. E. 1969. Methods for studying the microbial division cycle. Methods Microbiol. 1:327-363. [Google Scholar]
- 17.Holtzendorff, J., F. Partensky, S. Jacquet, F. Bruyant, D. Marie, L. Garzarek, I. Mary, D. Vaulot, and W. R. Hess. 2001. Diel expression of cell cycle-related genes in synchronized cultures of Prochlorococcus sp. strain PCC 9511. J. Bacteriol. 183:915-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hupp, T. R., J. D. Keasling, S. Cooper, and J. M. Kaguni. 1994. Synthesis of DnaK protein during the division cycle of Escherichia coli. Res. Microbiol. 145:99-109. [DOI] [PubMed] [Google Scholar]
- 19.Koch, A. 2002. Control of the bacterial cell cycle by cytoplasmic growth. Crit. Rev. Microbiol. 28:61-77. [DOI] [PubMed] [Google Scholar]
- 20.Levin, P. A., I. G. Kurtser, and A. D. Grossman. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, G., K. Begg, A. Geddes, and W. D. Donachie. 2001. Transcription of essential cell division genes is linked to chromosome replication in Escherichia coli. Mol. Microbiol. 40:909-916. [DOI] [PubMed] [Google Scholar]
- 22.Löwe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 23.Lu, C., J. Stricker, and H. P. Erickson. 1998. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima-quantitation, GTP hydrolysis, and assembly. Cell Motil. Cytoskeleton 40:71-86. [DOI] [PubMed] [Google Scholar]
- 24.Lutkenhaus, J., B. A. Moore, M. Masters, and W. D. Donachie. 1979. Individual proteins are synthesized continuously throughout the Escherichia coli cell cycle. J. Bacteriol. 138:352-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 26.Mingorance, J., S. Rueda, P. Gomez-Puertas, A. Valencia, and M. Vicente. 2001. Escherichia coli FtsZ polymers contain mostly GTP and have a high nucleotide turnover. Mol. Microbiol. 41:83-91. [DOI] [PubMed] [Google Scholar]
- 27.Mori, T., and C. H. Johnson 2001. Independence of circadian timing from cell division in cyanobacteria. J. Bacteriol. 183:2439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palacios, P., M. Vicente, and M. Sánchez. 1996. Dependency of Escherichia coli cell-division size, and independency of nucleoid segregation on the mode and level of ftsZ expression. Mol. Microbiol. 20:1093-1098. [DOI] [PubMed] [Google Scholar]
- 29.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pla, J., M. Sánchez, P. Palacios, M. Vicente, and M. Aldea. 1991. Preferential cytoplasmic location of FtsZ, a protein essential for Escherichia coli septation. Mol. Microbiol. 5:1681-1686. [DOI] [PubMed] [Google Scholar]
- 31.Quardokus, E. M., N. Din, and Y. V. Brun. 1996. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc. Natl. Acad. Sci. USA 93:6314-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.RayChaudhuri, D., and J. T. Park. 1992. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251-254. [DOI] [PubMed] [Google Scholar]
- 33.RayChaudhuri, D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18:2372-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivas, G., A. López, J. Mingorance, M. J. Ferrándiz, S. Zorrilla, A. P. Minton, M. Vicente, and J. M. Andreu. 2000. Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer. J. Biol. Chem. 275:11740-11749. [DOI] [PubMed] [Google Scholar]
- 35.Rothfield, L. I., Y. L. Shih, and G. King. 2001. Polar explorers: membrane proteins that determine division site placement. Cell 106:13-16. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sánchez, M., A. Valencia, M. J. Ferrándiz, C. Sander, and M. Vicente. 1994. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 13:4919-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scherbaum, O. H. 1959. A comparison of the degree of synchronous multiplication in various microbial systems. J. Protozool. Suppl. 6:17. [Google Scholar]
- 39.Sossong, T. M., Jr., M. R. Brigham-Burkey, P. Hensley, and K. H. Pearce, Jr. 1999. Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry 38:14843-14850. [DOI] [PubMed] [Google Scholar]
- 40.Stricker, J., P. Maddox, E. D. Salmon, and H. P. Erickson. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. USA 99:3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tormo, A., A. Dopazo, A. G. de la Campa, M. Aldea, and M. Vicente. 1985. Coupling between DNA replication and cell division mediated by the FtsA protein in Escherichia coli: a pathway independent of the SOS response, the “TER” pathway. J. Bacteriol. 164:950-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Udvardy, A. 1996. The role of controlled proteolysis in cell-cycle regulation. Eur. J. Biochem. 240:307-313. [DOI] [PubMed] [Google Scholar]
- 43.Voskuil, J. L., C. A. Westerbeek, C. Wu, A. H. Kolk, and N. Nanninga. 1994. Epitope mapping of Escherichia coli cell division protein FtsZ with monoclonal antibodies. J. Bacteriol. 176:1886-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, H., and R. C. Gayda. 1992. Quantitative determination of FtsA at different growth rates in Escherichia coli using monoclonal antibodies. Mol. Microbiol. 6:2517-2524. [DOI] [PubMed] [Google Scholar]
- 45.Wang, X., and J. Lutkenhaus. 1993. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol. Microbiol. 9:435-442. [DOI] [PubMed] [Google Scholar]
- 46.Ward, J. E., Jr., and J. Lutkenhaus. 1985. Overproduction of FtsZ induces minicell formation in E. coli. Cell 42:941-949. [DOI] [PubMed] [Google Scholar]
- 47.Wientjes, F. B., T. J. M. Olijhoek, U. Schwarz, and N. Nanninga. 1983. Labeling pattern of major penicillin-binding proteins of Escherichia coli during the division cycle. J. Bacteriol. 153:1287-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woldringh, C. L., E. Mulder, P. G. Huls, and N. Vischer. 1991. Toporegulation of bacterial division according to the nucleoid occlusion model. Res. Microbiol. 142:309-320. [DOI] [PubMed] [Google Scholar]
- 49.Yim, L., G. Vandenbussche, J. Mingorance, S. Rueda, M. Casanova, J.-M. Ruysschaert, and M. Vicente. 2000. Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J. Bacteriol. 182:6366-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou, P., and C. Helmstetter. 1994. Relationship between ftsZ gene expression and chromosome replication in Escherichia coli. J. Bacteriol. 176:6100-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]