Abstract
The predicted polypeptide product of open reading frame sso2387 from the archaeon Sulfolobus solfataricus, SsoPK2, displayed several of the sequence features conserved among the members of the “eukaryotic” protein kinase superfamily. sso2387 was cloned, and its polypeptide product was expressed in Escherichia coli. The recombinant protein, rSsoPK2, was recovered in insoluble aggregates that could be dispersed by using high concentrations (5 M) of urea. The solubilized polypeptide displayed the ability to phosphorylate itself as well as several exogenous proteins, including mixed histones, casein, bovine serum albumin, and reduced carboxyamidomethylated and maleylated lysozyme, on serine residues. The source of this activity resided in that portion of the protein displaying homology to the catalytic domain of eukaryotic protein kinases. By use of mass spectrometry, the sites of autophosphorylation were found to be located in two areas, one immediately N terminal to the region corresponding to subdomain I of eukaryotic protein kinases, and the second N terminal to the presumed activation loop located between subdomains VII and VIII. Autophosphorylation of rSsoPK2 could be uncoupled from the phosphorylation of exogenous proteins by manipulation of the temperature or mutagenic alteration of the enzyme. Autophosphorylation was detected only at temperatures ≥60°C, whereas phosphorylation of exogenous proteins was detectable at 37°C. Similarly, replacement of one of the potential sites of autophosphorylation, Ser548, with alanine blocked autophosphorylation but not phosphorylation of an exogenous protein, casein.
The reversible alteration of the functional properties of strategically selected proteins by phosphorylation-dephosphorylation represents one of nature's most widely employed means for controlling cellular processes (reviewed in reference 30). The scientific literature provides numerous examples of the prominent role played by this versatile regulatory mechanism in members of the Eucarya (reviewed in references 17, 18, and 24) and the Bacteria (reviewed in references 2, 7, and 22). By contrast, we know very little concerning the chemical nature, enzymatic catalysts, or physiological roles of the protein phosphorylation-dephosphorylation events that take place in members of the so-called third domain of life, the Archaea. Such knowledge is important not only for understanding how these biologically diverse organisms adapt to the extreme environments in which they typically reside but also for tracing the origins and evolution of a fundamentally important regulatory mechanism.
The available evidence suggests that protein phosphorylation is a fairly general phenomenon among the Archaea. Phosphorylated proteins have been detected in several halophilic, methanogenic, and thermophilic archaeons (11, 26, 48, 62, 63, 65, 66, 68, 69), and in several cases the observed patterns of protein phosphorylation exhibited the type of environmentally sensitive changes suggestive of regulatory control (48, 62, 68, 69). A CheA-like two-component cascade responsible for modulating chemo- and phototaxis has been characterized for the halophilic archaeon Halobacterium halobium (52, 53), while protein-serine/threonine phosphatases belonging to the PPP family of enzymes first discovered in the Eucarya have been described for the archaeons Sulfolobus solfataricus (32, 37), Methanosarcina thermophila TM-1 (49, 67), and Pyrodictium abyssi TAG11 (41). More recently, a dual-specificity protein-tyrosine phosphatase was described for Thermococcus kodakaraensis KOD1 (26).
Many basic questions concerning the phosphorylation-dephosphorylation of archaeal proteins remain unanswered. For example, how do members of the Archaea catalyze the phosphorylation of proteins on the hydroxyl amino acids serine, threonine, and tyrosine? The so-called “eukaryotic” protein kinase family represents the dominant source of protein-serine/threonine/tyrosine kinase activity in the Eucarya and is present in many members of the Bacteria as well (38, 59). While the genomes of most archaeons contain open reading frames (ORFs) whose potential protein products exhibit some of the characteristic features of the eukaryotic protein kinase superfamily (30, 38, 50, 59, 64), the level of overall sequence identity was extremely low. More importantly, it has yet to be determined whether any of these deduced protein kinases possess the catalytic capabilities inferred from their primary sequence. In this study we report that the recombinantly produced protein product of one such ORF, sso2387, from the extreme acidothermophilic archaeon S. solfataricus exhibited the ability to phosphorylate itself as well as exogenous proteins.
MATERIALS AND METHODS
Materials.
Purchased materials included [γ-32P]ATP and [γ-32P]GTP from NEN Research Products (Boston, Mass.), protein assay reagent and prestained standards for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Bio-Rad, Richmond, Calif.), chelating Sepharose (Pharmacia Biotech, Piscataway, N.J.), sequencing grade modified trypsin (Promega, Madison, Wis.), genomic DNA from S. solfataricus P2 (American Type Culture Collection, Rockville, Md.), and anti-Xpress antibody and TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.). Tamoxifen and other inhibitors, poly-l-lysine, poly-l-arginine, spermine, spermidine, poly(Glu:Tyr), and poly(Glu4:Tyr) were from Sigma-Aldrich (St. Louis, Mo.). Oligonucleotide primers were synthesized by Life Technologies Inc. (Gaithersburg, Md.). General laboratory reagents and culture media were from Fisher (Pittsburgh, Pa.) or Sigma-Aldrich.
Routine procedures.
Protein concentrations were determined as described by Bradford (8) by using premixed reagent and a standardized solution of bovine serum albumin (BSA). SDS-PAGE was performed as described by Laemmli (35). Polyacrylamide gels were stained with Coomassie brilliant blue as described by Fairbanks et al. (16). Electronic autoradiography was performed by using a Packard (Meriden, Conn.) Instantimager. Oligonucleotide primers were designed with the aid of the WWW Primer Picker 3 program from the Whitehead Institute (Cambridge, Mass.). Reduced, carboxyamidomethylated and maleylated lysozyme (RCM-lysozyme) was prepared as described by Tonks et al. (73).
Cloning of ORF sso2387.
ORF sso2387 was cloned by using the materials provided in the TOPO TA cloning kit (Invitrogen) according to the manufacturer's protocols. Briefly, sso2387 was amplified by PCR using genomic DNA (0.55 μg) from S. solfataricus as a template and 6 pmol each of a forward and a reverse oligonucleotide primer. The sequences of the forward and reverse primers were, respectively, 5′ATGGGGGAGTGGTATATAATGA-3′ and 5′-TTATTCTTGCGATAATGGCATA-3′. The resulting ≈1.8-kbp PCR product was then ligated into vector pCR T7/NT TOPO, which added oligonucleotides encoding an N-terminal extension that contains a hexahistidine sequence and a recognition epitope for the anti-Xpress antibody. The resulting protein product was designated rSsoPK2, which stands for recombinant S. solfataricus protein kinase 2. The resulting plasmid was used to transform Escherichia coli strain TOPO 10 F′. The transformed cells were cultured overnight on Luria-Bertani media containing 0.1 mg of ampicillin/ml and the plasmid isolated therefrom. DNA sequence analysis of the cloned DNA was performed to verify the fidelity of PCR amplification.
Site-directed mutagenesis of the cloned ORF for sso2387 was performed by using Promega's Gene Editor in vitro site-directed mutagenesis system according to the manufacturer's instructions. DNA encoding a truncated form of rSsoPK2, spanning residues 314 to 583, was amplified by PCR as described above by using 20 ng of the plasmid containing sso2387 as a template. The sequence of the forward primer was 5′-ATGGATCTGAGAATGAGTGTC-3′, while the reverse primer was identical to that described above.
Recombinant expression of rSsoPK2.
Recombinant expression of rSsoPK2, as well as mutagenically altered or truncated forms thereof, was carried out by using E. coli strain BL21(DE3)LysS. Briefly, E. coli isolates were transformed with the expression plasmids described above, and expression of recombinant protein was induced by using standard procedures (58). Cells were harvested by centrifugation and stored at −20°C until needed. The cell pellet from a 200-ml culture was thawed, resuspended in 4 ml of 50 mM MOPS (morpholinepropanesulfonic acid) (pH 7.0) containing 0.1 mg of lysozyme/ml, 5 g of RNase A/ml, 2 U of DNase I/ml, and 1 mM phenylmethylsulfonyl fluoride, and incubated on ice for 10 min. The cells were then lysed by sonication with three bursts of 30 s each at 30% power of a Heat Systems-Ultrasonics Model W140 Sonifier Cell Disruptor equipped with a microprobe. The lysate was centrifuged for 15 min at 3,000 × g at a temperature of 4°C, and the supernatant liquid was discarded.
The pellet was resuspended in 50 mM MOPS (pH 7.0) containing 5 M urea by passing the mixture several times through a pipette, and then it was centrifuged for 15 min at 3,000 × g at 4°C. The supernatant liquid was purified by metal chelate chromatography by following established procedures (58), with the exception that 5 M urea was included in all solutions.
Assay of protein kinase activity.
Protein kinase activity was routinely assayed in solution by the filter paper method (12). Briefly, rSsoPK2 (5- to 35-μg portions of the urea-solubilized pellet or 0.05- to 0.10-μg portions of metal chelate-purified protein) was incubated at 37°C in 100 μl of 50 mM MOPS (pH 7.0) containing 50 μM ATP (300 mCi of [γ-32P]ATP/ml), 5 mM MnCl2, 2 mM dithiothreitol, 5 M urea, and a phosphoacceptor substrate such as casein at a concentration of 1.0 mg/ml unless otherwise indicated. Inhibitors such as tamoxifen and trifluoperazine that do not readily dissolve in water were delivered as 10-fold-concentrated solutions in ethanol (ethanol alone had no effect on enzyme activity). Reactions were initiated by the addition of ATP. Following incubation at 37°C for periods of 15 to 60 min, reactions were terminated by spotting a 30-μl portion of the reaction mixture onto a 2- by 2-cm square of Whatman 3MM paper and immediately immersing the paper in a solution of 10% (wt/vol) trichloroacetic acid containing 4% (wt/vol) sodium pyrophosphate. After gently stirring for 20 min, filter papers were transferred to a solution of 5% (wt/vol) trichloroacetic acid containing 2% (wt/vol) sodium pyrophosphate and stirred for an additional 20 min. After repeating the last step three times more, the filter paper squares were air dried and the quantity of [32P]phosphate bound with the proteins precipitated on the filter was determined by liquid scintillation counting in 1 ml of ScintiSafe Plus 50% (Fisher) by use of a Beckman model LS 6500 liquid scintillation counter.
Radiolabeling of rSsoPK2 by autophosphorylation using [γ-32P]ATP.
For radiolabeling by autophosphorylation with [γ-32P]ATP, rSsoPK2 (5 to 35 μg of either urea-solubilized pellet or metal chelate-purified protein) was incubated as described above for the assay of protein kinase activity, except that no exogenous phosphoacceptor substrate was present and the temperature was increased to 65°C.
Phosphoamino acid analysis.
Phosphoamino acid analysis was performed essentially as described by Kamps and Sefton (29). Radiolabeled proteins were isolated by SDS-PAGE and transferred to an Immobilon P membrane. The portion of the membrane containing the radiolabeled protein was incubated for 1 h in 6 N HCl at 95°C, and the supernatant fluid was concentrated by using a Speed Vac. The hydrolysate was then applied to a 10- by 10-cm silica gel thin-layer chromatographic plate, along with a mixture of phosphoserine, phosphothreonine, and phosphotyrosine standards. The plate was subjected to two-dimensional thin-layer electrophoresis. The pH of the buffer used for the first dimension was 1.9, and that for the second dimension was 3.5. Standards were visualized by ninhydrin staining, and radiolabeled species were located by electronic autoradiography.
Mass spectroscopy.
The section of a Coomassie brilliant blue-stained gel containing the protein of interest was excised by using a clean razor blade and chopped into pieces approximately 1 mm3 in size, and the pieces were placed in a 1.5-ml Eppendorf tube. One hundred microliters of 25 mM ammonium bicarbonate (pH 8.0) containing 50% (vol/vol) acetonitrile was added, and the mixture was agitated for 10 min by using a Vortex mixer. The supernatant liquid was removed, and the process was repeated three more times. Next, a sufficient amount of 25 mM ammonium bicarbonate (pH 8.0) containing 10 mM dithiothreitol was added to cover the gel fragments, and the mixture was incubated for 1 h at 56°C. The mixture was cooled to room temperature, and the supernatant liquid was removed and replaced by an equal volume of 25 mM ammonium bicarbonate (pH 8.0) containing 55 mM iodoacetamide. The mixture was kept at room temperature, protected from light, and occasionally agitated with a Vortex mixer. After 45 min, the supernatant liquid was removed, replaced with 100 μl of 25 mM ammonium bicarbonate (pH 8.0), and agitated for 10 min with a Vortex mixer. The supernatant liquid was removed and replaced with 100 μl of ammonium bicarbonate (pH 8.0) containing 50% (vol/vol) acetonitrile. Following continuous mixing for 10 min, the supernatant liquid was removed and replaced with 100 μl of 25 mM ammonium bicarbonate (pH 8.0), and then the entire process was repeated.
The gel fragments were dried for 30 min in a Speed Vac, and then an equivalent volume of 25 mM ammonium bicarbonate (pH 8.0) containing 0.1 mg of trypsin/ml was added. Following initial agitation for 5 min with a Vortex mixer, the mixture was incubated for 12 to 16 h at 37°C. Next, 2 volumes of distilled water were added, the mixture was agitated for 5 min with a Vortex mixer, and the supernatant liquid, which contained the tryptic peptides, was removed and transferred to a fresh Eppendorf tube. The gel slices were washed twice by adding 2 volumes of 50% (vol/vol) acetonitrile containing 5% (vol/vol) trifluoroacetic acid (TFA), agitating the mixture for 5 min on a Vortex mixer, and removing the supernatant liquid. The supernatant liquids were pooled, reduced in volume to ≈10 μl by using a Speed Vac, and then brought up to a volume of ≈25 μl by the addition of 50% (vol/vol) acetonitrile containing 0.1% (vol/vol) TFA and stored at −20°C.
For mass spectral analysis, 0.5-μl portions of the tryptic peptide mixture were mixed with 0.5 μl of a saturated solution of α-hydroxycinnamic acid in 0.1% (vol/vol) TFA containing 50% (vol/vol) acetonitrile. Mass spectral determination of peptide masses was performed with a Kompact Seq matrix-assisted laser desorption ionization-time of flight mass spectrometer equipped with a nitrogen UV laser from Kratos Analytical (Chestnut Ridge, N.Y.). Gel slices from regions containing no visible proteins were used as controls to identify peaks arising from autoproteolysis of trypsin and other nonspecific sources. Peptide mass profiles were matched to potential protein sources by using the web-based ProFound Software package available from Rockefeller University (http://www.proteometrics.com [75]) assuming that all peptides bore a charge of +1.
RESULTS
The members of the eukaryotic protein kinase family are characterized by a catalytic core domain roughly 280 amino acids in length containing 12 conserved sequence motifs, each ranging from 4 to 20 residues in length, referred to as subdomains I to V, VIa, VIb, and VII to XI (20, 72). These subdomains can be grouped into three structural units: an N-terminal nucleotide binding domain containing subdomains I to IV, a C-terminal phosphotransfer and protein substrate binding domain containing subdomains VIa to XI, and an intervening linker containing subdomain V. Only a few of the amino acid residues comprising these subdomains are universally conserved among the members of this extraordinarily large protein superfamily. Not surprisingly, the majority of these are found within those regions of the protein most directly involved in executing the core functions common to all members of the superfamily, i.e., binding of their shared nucleotide substrate (subdomains I to III), catalysis of phosphotransfer (subdomain VIb), or interaction with their metal ion cofactor, usually Mg2+ (subdomain VII) (20, 72).
Since subdomains I to III, VIa, and VII embodied the core functions universal to the eukaryotic protein kinase paradigm, they were selected as templates against which to search the recently released genome sequence of S. solfataricus P2 (57). Six ORFs potentially encoding plausible candidates for nucleotide-dependent phosphotransferases were identified (Table 1). The protein product of one of the ORFs listed in Table 1, sso2387 (Fig. 1), was selected for further study because (i) it was encoded by one of the first portions of the S. solfataricus P2 genome to be made publicly accessible and hence was one of the earliest of the six to be identified, and (ii) its calculated molecular mass, 66.5 kDa, closely matched the molecular mass of a polypeptide of undetermined sequence previously demonstrated to exhibit protein kinase activity (39).
TABLE 1.
Salient features of ORFs potentially encoding eukaryotic protein kinases from S. solfataricus
| ORF | Length (amino acids) | Sequencea for subdomain:
|
||||
|---|---|---|---|---|---|---|
| I | II | III | VIb | VII | ||
| Consensus | ogxG52 xogxv | oaoK72 xo | E91 xxoo | oohrD166ok+xNooo | oko+D184fgo+ | |
| sso0197 | 287 | IGiG103keslv | IiVK122fh | E141kksW | ItHgD227LsPyNVLI | pyLiD245wpqA |
| sso0433 | 223 | IkrG10 aesni | rikK33 sY | E52 akII | IaHgD125LtTnNLIL | IfIiD144FGLS |
| sso2291 | 554 | IaiG253gssyI | YAMK271ip | E297finL | YVHlD392VKPqNIYF | VKLGD426lGsA |
| sso2387 | 583 | MsiG324vtGag | esMK346iV | E355irLA | dpksD496afTiNLdA | nrIeE514aGLn |
| sso2605 | 486 | Lssa113sLGqV | VAIK131vn | E187afYL | YFHaD262phPgNIAV | LvLyD280FGMS |
| sso3182 | 537 | IGvG247gtsyi | YALK265ip | E281sskL | YVHcD378VKPqNVLF | VKLAD412LGsA |
| sso3207 | 669 | LGnG375gMGyV | YAMK393vM | E407vakM | YtHcD498IKpsNILF | pKLSD531lGssv |
Consensus sequence as described by Hanks and Hunter (20). Symbols in the consensus sequence: uppercase letter, universally conserved amino acid residue; lowercase letter, highly conserved amino acid residue; o, position conserving nonpolar residue; +, position conserving small residue with near neutral polarity. Residue position within a prototypical catalytic domain was indicated by using the sequence of the Cα subunit of murine cAMP-dependent protein kinase. Shown below the consensus sequence for each subdomain are postulated homologous sequences from the deduced proteins encoded by the indicated ORFs. Residues that match the consensus are in uppercase letters; all others are in lowercase letters.
FIG. 1.
The predicted protein product of ORF sso2387 contains sequence features characteristic of eukaryotic protein kinases. Shown is the DNA-derived amino acid sequence of SsoPK2, the predicted protein product of ORF sso2387 from the genome of S. solfataricus (57) (GenBank accession no. AE006641). The positions of plausible candidates for the conserved sequence motifs, called subdomains, characteristic of members of the eukaryotic superfamily of protein kinases are indicated, with key residues highlighted in bold type (see Table 1, footnote a). The first residue of the truncated catalytic domain construct utilized in these studies is indicated by an asterisk underneath. The regions corresponding to the putative phosphopeptides identified by mass spectrometry (see Table 3) are underlined.
ORF sso2387 was cloned into vector pCR T7/NT TOPO, and its protein product was expressed as an N-terminally His-tagged fusion protein in E. coli. The expressed fusion protein, rSsoPK2, was recovered in inclusion bodies under all conditions during which recombinant gene expression was induced. The inclusion bodies could be dispersed in buffers containing 5 M urea and subsequently purified to near-electrophoretic homogeneity (Fig. 2, left side) by metal chelate chromatography. The removal of urea resulted in drastic losses of enzyme activity (see below) that could not be mitigated by the addition of nonionic detergents such as Triton X-100 or octyl glucoside. Consequently, all experiments used 5 M urea. This behavior is somewhat analogous to that of the NAD-dependent glutamate dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandium, whose catalytic activity was stimulated severalfold by denaturants such as guanidine hydrochloride, acetonitrile, and tetrahydrofuran (33), as well as malic enzyme from S. solfataricus, whose activity was stimulated severalfold by water-miscible organic solvents in vitro (19).
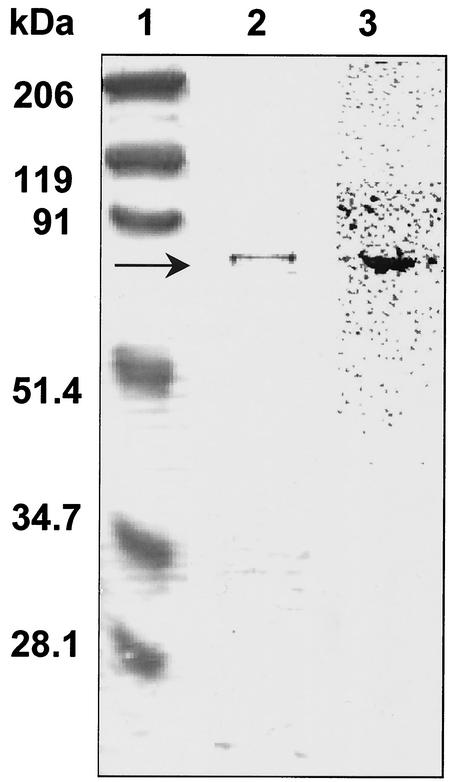
FIG. 2.
rSsoPK2 phosphorylates itself when incubated with [γ-32P]ATP in vitro. Metal chelate-purified rSsoPK2 (2 μg) was incubated for 60 min at 65°C with [γ-32P]ATP as described in Materials and Methods. The incubation mixture was then subjected to SDS-PAGE. Shown at left is the Coomassie blue-stained gel with the positions and relative molecular masses of the protein standards indicated at left. Shown on the right is an electronic autoradiogram of the same gel.
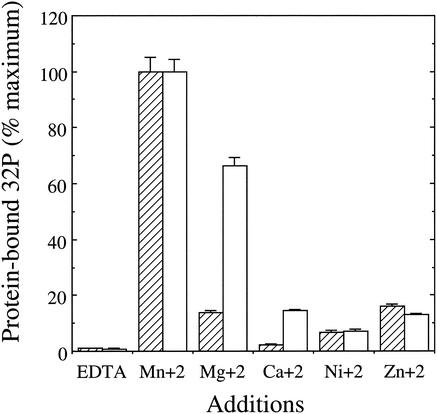
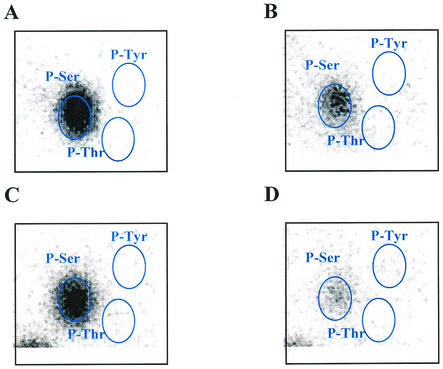
As no endogenous phosphoproteins from S. solfataricus or other thermophilic archaeons were available for use as test substrates, rSsoPK2 was challenged with a variety of mesophilic phosphoproteins, several of which could be phosphorylated by the membrane-associated protein kinase (39). As seen from the data in Table 2, rSsoPK2 catalyzed the phosphorylation of several exogenous proteins with [γ-32P]ATP as a phosphodonor substrate. However, it did not phosphorylate the tyrosine-rich polymers poly(Glu:Tyr) or poly(Glu4:Tyr). Mn2+ was the most effective divalent metal ion cofactor with regard to phosphotransfer to exogenous proteins (Fig. 3). Phosphoamino acid analysis indicated that each of the exogenous protein substrates tested was phosphorylated exclusively on serine residues (Fig. 4). Little or no phosphorylation took place when [γ-32P]GTP was substituted for [γ-32P]ATP (data not shown). These observations indicated that ORF sso2387 did not encode the membrane-associated protein kinase described in earlier studies, since the latter enzyme displayed a strong preference for phosphorylating threonine residues and was capable of utilizing a variety of purine nucleotides as phosphodonor substrates in vitro (39). Also, we recently determined that our initial estimate of the molecular mass of the membrane-associated protein kinase was high—a consequence of the presence of covalently bound carbohydrate (40).
TABLE 2.
Phosphorylation of exogenous proteins and peptides by rSsoPK2 and its putative catalytic domain in vitro
| Substrate | Avg activity ± SE (pmol of 32P/min · mg)a
|
|
|---|---|---|
| rSsoPK2 | Putative catalytic domain | |
| BSA | 578 ± 27 | 2,000 ± 40 |
| Casein | 372 ± 17 | 142 ± 7 |
| Mixed histones | 1,932 ± 216 | 145 ± 16 |
| RCM-lysozyme | 664 ± 22 | 132 ± 18 |
| Poly(Glu:Tyr) | NDb | ND |
| Poly(Glu4:Tyr) | ND | ND |
From triplicate determinations.
ND, not detectable (<1.0 pmol of 32P/min · mg).
FIG. 3.
Metal ion preference of rSsoPK2. The metal ion preference of rSsoPK2 for autophosphorylation (hatched bars) was assessed by incubating 35 μg of urea-solubilized pellet at 65°C for 60 min with 50 μM [γ-32P]ATP and the indicated metal ions, each at a concentration of 5 mM. The autophosphorylated protein was isolated by SDS-PAGE, and the quantity of [32P]phosphate incorporated was measured by electronic autoradiography. For determination of the metal ion preference of rSsoPK2 for phosphorylation of an exogenous protein substrate (open bars), 0.06 μg of metal chelate-purified rSsoPK2 was incubated with 1 mg of BSA/ml as a phosphoacceptor substrate for 60 min at 37°C in the presence of 50 μM [γ-32P]ATP and the indicated metal ions, each at a concentration of 5 mM. For further details, see Materials and Methods. Shown are the averages ± standard errors of duplicate determinations.
FIG. 4.
rSsoPK2 phosphorylates serine residues in vitro. Metal chelate-purified rSsoPK2 was incubated with [γ-32P]ATP with either no added proteins (A), casein (B), BSA (C), or mixed histones (D). The polypeptides within each reaction mixture were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane, and the section of the membrane containing the phosphoprotein of interest was incubated with 6 N HCl for 1 h at 95°C. The phosphoamino acids within the acid hydrolysate were then separated by two-dimensional thin-layer electrophoresis. Shown are electronic autoradiograms of the thin-layer chromatographic plates, with the positions of the following phosphoamino acid standards, which were visualized by staining with ninhydrin (circled): P-Ser, phosphoserine; P-Thr, phosphothreonine; and P-Tyr, phosphotyrosine. The quantities of rSsoPK2 utilized were 5 μg in the absence of exogenous protein substrates and 0.05 μg in their presence. For further details, see Materials and Methods.
It was next asked whether the predicted catalytic domain of rSsoPK2 was in fact the source of the protein kinase activity detected. As predicted, mutagenic alteration of Asp496, which corresponds to the catalytically essential aspartate found in the catalytic loop of eukaryotic protein kinases (Table 1), produced an inactive protein product. Perhaps more significantly, a truncated form of rSsoPK2 consisting only of the putative catalytic domain (Fig. 1), residues 314 to 583, phosphorylated exogenous proteins at rates comparable to that of rSsoPK2, although some differences in relative substrate preferences were observed (Table 2). As was the case with full-length rSsoPK2, the truncated form of the enzyme required the presence of molar concentrations of urea to maintain it in an active state after being solubilized from inclusion bodies.
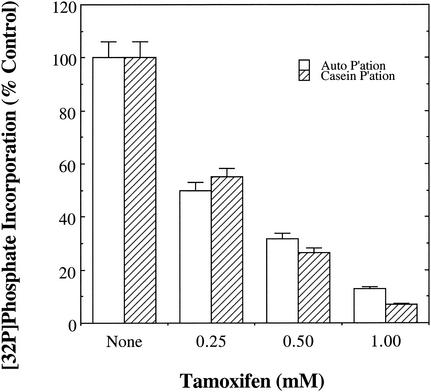
Compounds known to inhibit several well-characterized members of the eukaryotic protein kinase superfamily were tested for their ability to block the catalytic activity of rSsoPK2. Staurosporine, a broad-specificity inhibitor of eukaryotic protein kinases that targets the binding site for the nucleotide substrate (43), had no effect on rSsoPK2 when present at concentrations in considerable excess over those at which it acts on its established targets (Table 3). Several other compounds that display various degrees of selectivity for the ATP binding sites of protein kinases (i.e., genistein [3], ML-9 [47], and H-7 [47]) as well as PKI-peptide (9, 55), which targets the protein substrate binding site of the cAMP-dependent protein kinase, also failed to inhibit the enzyme (Table 3). However, tamoxifen inhibited both the autophosphorylation of rSsoPK2 and the phosphorylation of an exogenous protein substrate, casein, with a 50% inhibitory concentration of roughly 250 μM (Fig. 5). This result was somewhat surprising, as tamoxifen acts by antagonizing the activation of protein kinase C by phospholipids or phorbol esters (46, 71) and of calmodulin-dependent protein kinases (71) and phosphodiesterases (36) by calmodulin.
TABLE 3.
General properties of several known inhibitors of eukaryotic protein kinases and the highest concentrations tested against rSsoPK2a
| Inhibitor | Known targets | Mode of action | IC50 or Ki | Maximum concn |
|---|---|---|---|---|
| Staurosporine | Broad specificity | ATP binding site | 1 nM-3 μM | 20 μM |
| Genistein | PTKs | ATP binding site | 1-30 μM | 2 mM |
| ML-9 | cAPK, PKC, MLCK | ATP binding site | 4-54 μM | 80 μM |
| H-7 | Broad specificity | ATP binding site | 3-780 μM | 2 mM |
| PKI-peptide | cAPK-specific | Peptide binding site | 2.3 nM | 0.9 μM |
| Tamoxifen | PKC, CaM kinases | Antagonizes activators | 2-100 μM | 2.5 mM |
| Trifluoperazine | CaM kinases | Antagonizes CaM | ≈30 μM | 0.5 mM |
Abbreviations: cAPK, cAMP-dependent protein kinase; PKC, protein kinase C; PTK, protein-tyrosine kinase; CaM kinase, calmodulin-dependent protein kinases; MLCK, myosin light chain kinase; IC50, 50% inhibitory concentration.
FIG. 5.
rSsoPK2 is inhibited by tamoxifen. Metal chelate-purified rSsoPK2 was assayed for its ability to phosphorylate itself (open bars) or an exogenous protein substrate, casein (hatched bars), under standard conditions as described in Materials and Methods, with the exception that tamoxifen was included at the indicated concentrations. Shown are the averages ± standard errors (from duplicate assays) reported as the percentage of [32P]phosphate transferred to rSsoPK2 or casein relative to control incubations from which tamoxifen had been omitted. The quantities of rSsoPK2 present in the assays for autophosphorylation and casein phosphorylation were 2.5 and 2.1 μg, respectively.
To assess the specificity with which tamoxifen acted on rSsoPK2, another hydrophobic compound known to antagonize the ability of calmodulin to activate its target enzymes, trifluoperazine (51), was tested. At concentrations at which tamoxifen inhibited the activity of rSsoPK2 by 50% or more, 250 to 500 μM, trifluoperazine had no discernible effect on the phosphorylation of the exogenous substrate protein casein (Table 3). Polylysine, but not acidic glutamate-rich polymers or other polybasic compounds such as polyarginine, spermine, or spermidine, stimulated the activity of rSsoPK2 toward itself as well as exogenous protein substrates roughly twofold (data not shown).
rSsoPK2 underwent autophosphorylation when incubated with [γ-32P]ATP (Fig. 2, right side). As was the case for the phosphorylation of exogenous substrates, autophosphorylation took place on serine residues (Fig. 4). However, autophosphorylation occurred only at temperatures of 60°C or more. Mn2+ was the preferred cofactor for autophosphorylation (Fig. 3), but Mg2+ also proved moderately effective. It is unclear whether the greater efficacy of Mg2+ with regard to autophosphorylation reflected differences in the mechanisms by which self-phosphorylation and the phosphorylation of exogenous proteins proceed or whether it was an effect of the higher temperature at which the former process was assayed, 65 versus 37°C, on the conformation of the protein. Assessment of the phosphorylation of exogenous proteins at 65°C was precluded by the lack of a thermostable protein or peptide substrate. The truncated catalytic domain of rSsoPK2 also phosphorylated itself on serine residues, indicating that both the necessary catalytic machinery and the site(s) (or at least a portion of the sites) at which it becomes autophosphorylated were contained within this domain.
In an effort to map the site(s) of autophosphorylation, rSsoPK2 was incubated with ATP, isolated by SDS-PAGE, and digested with trypsin, and the resulting tryptic peptides were analyzed by matrix-assisted laser desorption-time of flight mass spectrometry. Three peptides whose masses corresponded to those of predicted tryptic peptides containing one or more 80-atomic-mass-unit phosphoryl groups were detected (Table 4). These phosphopeptides originated from two distinct regions of the protein spanning residues 266 to 285 and 524 to 553 (Fig. 1). The first region lies outside the N-terminal boundary of the catalytic domain, while the second corresponds to the predicted activation or T loop that resides between subdomains VII and VIII of prototypical eukaryotic protein kinases (reviewed in references 1, 25, and 27). Intriguingly, the activation loop is a frequent target of phosphorylation in many established members of the eukaryotic kinase family, either via autophosphorylation or the intervention of an exogenous protein kinase.
TABLE 4.
Identification of possible autophosphorylation sites in rSsoPK2 by mass spectrometry
| Peptidea | Residues | Mass (Da)
|
Phosphoryl groups (calculated) | ||
|---|---|---|---|---|---|
| Observed | Predicted | Difference | |||
| VAATFRREVSASGSSVVIRR | 266-285 | 2,308 | 2,148 | 160 | 2 |
| LLASSREIIQNEESYKLVK | 535-553 | 2,300 | 2,200 | 80 | 1 |
| REMERRALFLKLLASSR | 523-540 | 2,236 | 2,076 | 160 | 2 |
Potential phosphoacceptor serine residues are underlined.
In many instances, phosphorylation of the activation loop is necessary to render a protein kinase competent to phosphorylate exogenous substrates (reviewed in references 1, 25, and 27). However, preincubation with ATP at a temperature permissive for autophosphorylation (65°C) showed no significant effect when the activity of rSsoPK2 toward exogenous protein substrates was subsequently assayed at 37°C (data not shown). Mutagenic alteration of one of the serine residues in the putative activation loop of SsoPK2, Ser548, to a nonphosphorylatable alanine residue resulted in the complete loss of autophosphorylation (Table 5). Despite the loss of the ability to autophosphorylate, a variant of rSsoPK2 in which Ser548 had been altered to alanine was unimpaired in its ability to phosphorylate casein. Forms of rSsoPK2 in which the serine residues immediately N or C terminal to Ser548, i.e., Ser538, Ser539, Ser554, or Ser560, were mutagenically altered to serine residues all retained the ability to autophosphorylate as well as to phosphorylate casein at rates comparable to that of the unmodified enzyme (Table 5).
TABLE 5.
Effects of mutagenic alteration of serine residues in the putative activation loop on the activity of rSsoPK2a
| Mutational alteration(s) | Mutagenic oligonucleotide | Casein phosphorylation | Autophosphorylation |
|---|---|---|---|
| Ser538Ala, Ser539Ala | 5′-CTAAAGTTGTTAGCGGCTGCCAGAGAGATA-3′ | + | + |
| Ser548Ala | 5′-CAAAATGAGGAGGCTTATAAGCTTGTGAAG-3′ | + | − |
| Ser554Ala | 5′-GAGTTATAAGCTTGTGAAGGCCTATATAATAAAATACAGC-3′ | + | + |
| Ser560Ala | 5′-GTGAAGAGCTATATAATAAAATACGCCTTAAAACCCGAAG-3′ | + | + |
+, phosphorylation observed; −, phosphorylation not observed.
DISCUSSION
The genome of the extreme acidothermophilic archaeon S. solfataricus contained at least six ORFs whose predicted products display sequence features faintly reminiscent of the eukaryotic protein kinase paradigm. The lack of discernible representatives of the other well-characterized protein kinase families, i.e., the histidine kinases, HPr kinases, isocitrate dehydrogenase kinase/phosphatases, myosin heavy-chain/elongation factor 2 kinases (57), suggests that the eukaryotic protein kinase paradigm constitutes a predominant, and perhaps the sole, source of protein-serine/threonine/tyrosine kinase activity in this archaeon. In order to ascertain whether any of these hypothetical protein kinases possessed the catalytic potential inferred from sequence comparisons, the recombinant protein product of ORF sso2387, rSsoPK2, was expressed in E. coli. rSsoPK2 phosphorylated itself, as well as a variety of exogenous proteins, on serine residues in vitro—firmly establishing that it possessed phosphotransferase activity. A truncated form of rSsoPK2 encompassing only the presumed catalytic domain proved to be as effective a protein kinase as rSsoPK2 itself, while alteration of an aspartate residue predicted from sequence comparisons to be essential for protein kinase activity produced an inactive protein product. These observations corroborated the perceived structural and functional relationship between this archaeal enzyme and prototypical eukaryotic protein kinases.
As is typical of many eukaryotic protein kinases, rSsoPK2 can undergo self- or autophosphorylation. Mass peptide profiling, coupled with phosphoamino acid analysis, mapped at least a portion of the sites of autophosphorylation to two areas, one closely preceding the predicted N-terminal boundary of the catalytic domain and a second spanning the region between predicted subdomains VII and VIII. The latter region corresponds to the activation loop of prototypical eukaryotic protein kinases, whose phosphorylation via either an autocatalytic process or, in other cases, by an exogenous protein kinase, is oftentimes necessary for realization of full catalytic efficiency (reviewed in references 1, 25, and 27). Many bacterial members of the eukaryotic protein kinase superfamily also catalyze their self-phosphorylation (reviewed in reference 30). However, in no instance has either the identity of the site(s) of phosphorylation or the effect of autophosphorylation, if any, upon activity been reported.
The rate at which rSsoPK2 phosphorylated the most efficacious protein substrate tested, mixed histones, was 1.9 nmol/min · mg at 37°C—a temperature dictated by the necessity of using eukaryotic phosphoproteins as potential substrates. At the normal growth temperature of S. solfataricus, 75 ± 10°C (mean ± standard error), this projected to a reaction rate of roughly 30.4 nmol/min · mg or, for an ≈67-kDa protein, 2.0 min−1 assuming simple Arrhenius behavior. This figure falls at the lower end of the 1,000-fold range of kcat values, 4.3 to 3,100 min−1, reported to date for established members of the eukaryotic protein kinase superfamily (14). It should be noted that rSsoPK2 was roughly 1,000-fold more efficient at phosphorylating exogenous proteins in vitro than were members of the extended eukaryotic protein kinase superfamily that phosphorylate aminoglycoside antibiotics in vivo (13). Nor does SsoPK2 contain motif III, a conserved sequence element found in antibiotic kinases but not eukaryotic protein kinases (56).
For several reasons, we believe that this comparison underestimates, perhaps to a very large degree, the full catalytic potential of rSsoPK2 as a protein kinase. First, we are comparing a specific activity that was measured by using an arbitrary concentration of a completely nonphysiologic eukaryotic phosphoprotein to kcat values that represent theoretical maximum velocities extrapolated from measurements performed by using physiological phosphoprotein substrates or synthetic peptides of optimized sequence. Second, enzymes from thermophilic organisms oftentimes exhibit biphasic temperature behavior, transitioning to a more catalytically efficient conformation as the temperature approaches that of their natural environment (reviewed in reference 74). Examples include α-amylase from Pyrococcus furiosus (34), the P-type ATPase from Methanococcus jannaschii (44), an iron hydrogenase from Thermotoga maritima (28), and the pyruvate dehydrogenases from S. solfataricus (74) and P. furiosus (5). Several enzymes from mesophilic organisms also have been demonstrated to undergo similar thermal activation and cold inactivation, including ribulose-1,5-bisphosphate carboxylase-oxygenase from tobacco (10), tyrosine aminotransferase from chick liver (60), and sialic acid synthase from E. coli (42), Lastly, our measurements cannot account for the influence of second messengers or other ligands, covalent modification, or heterologous protein-protein interactions that so commonly activate protein kinases of all types in vitro and in vivo.
The ability to uncouple the phosphorylation of exogenous protein substrates from the autophosphorylation of rSsoPK2 with either variations in temperature or mutagenic alteration of the enzyme further suggests that the rates of histone phosphorylation reported herein may significantly underestimate the true catalytic potential of the enzyme. While phosphorylation of exogenous substrates proceeded at appreciable rates up to the limits imposed by the thermal stability of the eukaryotic proteins utilized, autophosphorylation was detected only at temperatures of 60°C or higher. Similarly, while substitution of Ser548 abolished autophosphorylation altogether, phosphotransfer to exogenous proteins proceeded unimpaired. This pattern of behavior, along with the broadening of metal ion specificity with increased temperature, suggests that rSsoPK2 underwent a conformational transition between 37 and 60°C.
Since autophosphorylation of eukaryotic protein kinases is associated with their activation, it is possible that the putative high-temperature conformer would be much more active toward exogenous protein substrates than was indicated by steady-state assays performed at lower temperatures. The failure of preincubation with ATP at 65°C to stimulate activity toward exogenous proteins at 37°C was not surprising. Detailed studies of the effects of phosphorylation upon the thermal stability of proteins indicate that the transition temperature generally is shifted less than 10°C (6, 54, 70). Hence, it is unlikely that the ability of autophosphorylation to promote adoption of the putative activated conformation by rSsoPK2 was sufficient to overcome the countervailing effects of the >30°C difference in the temperatures at which the respective measurements on protein conformation were performed.
The conclusion that proteins constitute the physiological target of SsoPK2's phosphotransferase activity still must be viewed with caution, however. Covalent modification by autophosphorylation is not an exclusive property of protein kinases; for example, nucleoside diphosphate kinase (45) and the multifunctional enzyme CAD, which contains carbamoyl phosphatase, aspartate transcarbamoylase, and dihydrooratase (61), both catalyze self-phosphorylation events in vitro. Moreover, the eukaryotic protein kinase superfamily itself has recently been determined to include kinases that target nonprotein substrates (23). It also should be noted that the deduced amino acid sequence of SsoPK2 displayed significant deviations from the eukaryotic protein kinase consensus. For example, SsoPK2 does not contain a basic residue between the conserved aspartate and asparagine residues in subdomain VIb.
In most eukaryotic protein kinases, the presence of a lysine residue of subdomain VIb, i.e., Asp-Xaa-Lys-Xaa-Xaa-Asn, is diagnostic of a protein-serine/threonine kinase, while the presence of an arginine, i.e., Asp-Xaa-Arg-Xaa-Xaa-Asn or Asp-Xaa-Xaa-Xaa-Arg-Asn, is indicative of a protein-tyrosine kinase (21). However, the lack of an arginine or lysine in subdomain VIb does not preclude the possibility that SsoPK2 is a protein kinase. To date, at least three protein kinases that lack either lysine or arginine in subdomain VIb, Rio1p (4) and PID261/BUD32 (15) from yeast and Pkn6 from the bacterium Myxococcus xanthus (76), have been characterized. It also should be noted that only three of the potential protein kinase ORFs in S. solfataricus contain a lysine or arginine in subdomain VIb, i.e., sso2291, sso3182, and sso3207, while all of the potential protein kinase ORFs identified in other archaeal genomes also lack this feature (31, 38, 59).
Potentially more-significant deviations are evident in that portion of SsoPK2 corresponding to the C-terminal half of the predicted catalytic/protein substrate binding lobe. Specifically, the distance between subdomain VIb, the catalytic loop, and the carboxyl terminus of SsoPK2 is ≈25 to 30 residues shorter than is the corresponding region in the eukaryotic protein kinase consensus. The subdomain assignments presented in Fig. 1, which align Pro-Glu564 of SsoPK2 with the conserved Pro-Glu sequence of subdomain VIII, do so at the expense of completely omitting subdomains X and XI. On the other hand, if one assumes that the sole arginine residue present in the final C-terminal 43 amino acids of SsoPK2, Arg575, corresponds to the conserved arginine of subdomain XI in eukaryotic protein kinases, there exists neither sufficient space to accommodate subdomains VIII, IX, and X nor obvious sequence matches thereto. Thus, while the alignment presented in Fig. 1 may ultimately prove flawed in some respects, it seems clear that the fold in this region of SsoPK2 must deviate significantly from that of prototypical eukaryotic protein kinases. As was the case with the conserved basic residue in subdomain VIb, apparent C-terminal truncations and other deviations are a common feature of many potential archaeal protein kinases, suggesting that they belong to subgroups that had diverged from the line of development from which the majority of the eukaryotic protein kinases found in eucaryal and bacterial organisms emerged (38). A definitive determination of the physiologic function of SsoPK2 and other potential archaeal protein kinases must therefore await the identification of their natural substrates.
Acknowledgments
This work was supported by grant number MCB 0077484 from the National Science Foundation.
We thank Monique Coy for assistance in characterizing the effects of polycations on protein kinase activity.
REFERENCES
- 1.Adams, J. A. 2001. Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 101:2271-2290. [DOI] [PubMed] [Google Scholar]
- 2.Aizawa, S.-I., C. S. Harwood, and R. J. Kadner. 2000. Signaling components in bacterial locomotion and sensory perception. J. Bacteriol. 182:1459-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiyama, T., J. Ishida, S. Nakagawa, H. Ogawara, S.-I. Watanabe, N. Itoh, M. Shibuya, and Y. Fukami. 1987. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J. Biol. Chem. 262:5592-5595. [PubMed] [Google Scholar]
- 4.Angermayr, M., A. Roidl, and W. Bandlow. 2002. Yeast Rio1p is the founding member of a novel subfamily of protein serine kinases involved in the control of cell cycle progression. Mol. Microbiol. 44:309-324. [DOI] [PubMed] [Google Scholar]
- 5.Blamey, J. M., and M. W. W. Adams. 1993. Purification and characterization of pyruvate ferrodoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta 1161:19-27. [DOI] [PubMed] [Google Scholar]
- 6.Blanes-Mira, C., C. Ibanez, G. Fernandez-Ballester, R. Planells-Cases, E. Perez-Paya, and A. Ferrer-Montiel. 2001. Thermal stabilization of the catalytic domain of botulinum neurotoxin E by phosphorylation of a single tyrosine residue. Biochemistry 40:2234-2242. [DOI] [PubMed] [Google Scholar]
- 7.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and simple method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, H.-C., B. E. Kemp, R. B. Pearson, A. J. Smith, L. Misconi, S. M. Van Patten, and D. A. Walsh. 1986. A potent synthetic peptide inhibitor of the cAMP-dependent protein kinase. J. Biol. Chem. 261:989-992. [PubMed] [Google Scholar]
- 10.Chollet, R., and L. L. Anderson. 1977. Conformational changes associated with the reversible cold inactivation of ribulose-1,5-bisphosphate carboxylase oxygenase. Biochim. Biophys. Acta 482:228-240. [DOI] [PubMed] [Google Scholar]
- 11.Condo, I., D. Ruggero, R. Reinhardt, and P. Londel. 1998. A novel aminopeptidase associated with the 60 kDa chaperonin in the thermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 29:775-785. [DOI] [PubMed] [Google Scholar]
- 12.Corbin, J. D., and E. M. Reimann. 1971. Assay of cAMP-dependent protein kinases. Methods Enzymol. 38:287-290. [DOI] [PubMed] [Google Scholar]
- 13.Daigle, D. M., G. A. McKay, P. R. Thompson, and G. D. Wright. 1999. Aminoglycoside antibiotic phosphotransferases are also serine protein kinases. Chem. Biol. 6:11-18. [DOI] [PubMed] [Google Scholar]
- 14.Enke, D. A., P. Kaldis, and M. J. Solomon. 2000. Kinetic analysis of the cyclin-dependent kinase-activating kinase (Cak1p) from budding yeast. J. Biol. Chem. 275:33267-33271. [DOI] [PubMed] [Google Scholar]
- 15.Facchin, S., S. Sarno, O. Marin, R. Lopreiato, G. Sartori, and L. A. Pinna. 2002. Acidophilic character of yeast PID261/BUD32, a putative ancestor of eukaryotic protein kinases. Biochem. Biophys. Res. Commun. 296:1366-1371. [DOI] [PubMed] [Google Scholar]
- 16.Fairbanks, G., T. L. Steck, and D. F. H. Wallach. 1971. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry 10:2606-2617. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, E. H. 1999. Cell signaling by protein tyrosine phosphorylation. Adv. Enzyme Regul. 39:359-369. [DOI] [PubMed] [Google Scholar]
- 18.Graves, J. D., and E. G. Krebs. 1999. Protein phosphorylation and signal transduction. Pharmacol. Ther. 82:111-121. [DOI] [PubMed] [Google Scholar]
- 19.Guagliardi, A., G. Manco, M. Rossi, and S. Bartolucci. 1989. Stability and activity of a thermostable malic enzyme in denturants and water-miscible organic solvents. Eur. J. Biochem. 183:25-30. [DOI] [PubMed] [Google Scholar]
- 20.Hanks, S. K., and T. Hunter. 1995. The eukaroytic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9:576-596. [PubMed] [Google Scholar]
- 21.Hanks, S. K., and R. A. Lindberg. 1991. Use of degenerate oligonucleotide probes to identify clones that encode protein kinases. Methods Enzymol. 200:525-532. [DOI] [PubMed] [Google Scholar]
- 22.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 23.Hon, W.-C., G. A. McKay, P. R. Thompson, R. M. Sweet, D. S. C. Yang, G. D. Wright, and A. M. Berghius. 1997. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell 89:887-895. [DOI] [PubMed] [Google Scholar]
- 24.Hunter, T. 1995. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80:225-236. [DOI] [PubMed] [Google Scholar]
- 25.Huse, M., and J. Kuriyan. 2002. The conformational plasticity of protein kinases. Cell 109:275-282. [DOI] [PubMed] [Google Scholar]
- 26.Jeon, S.-J., S. Fuliwara, M. Takagi, T. Tanaka, and T. Imanaka. 2002. Tk-PTP, protein-tyrosine/serine phosphatase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1: enzymatic characteristics and identification of its substrate proteins. Biochem. Biophys. Res. Commun. 295:508-514. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, L. N., M. E. M. Noble, and D. J. Owen. 1996. Active and inactive protein kinases: structural basis for regulation. Cell 85:149-158. [DOI] [PubMed] [Google Scholar]
- 28.Juszczak, A., S. Aono, and M. W. W. Adams. 1991. The extremely thermophilic eubacterium Thermotoga maritima contains a novel iron-hydrogenase whose cellular activity is dependent upon tungsten. J. Biol. Chem. 266:13834-13841. [PubMed] [Google Scholar]
- 29.Kamps, M. P., and B. M. Sefton. 1989. Acid and base hydrolysis of phosphoproteins bound to immobilon facilitates analysis of phosphoamino acids in gel-fractionated proteins. Anal. Biochem. 176:22-27. [DOI] [PubMed] [Google Scholar]
- 30.Kennelly, P. J. 2002. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol. Lett. 206:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Kennelly, P. J. 2003. Archaeal protein kinases and protein phosphatases—insights from genomics and biochemistry. Biochem. J. 370:373-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennelly, P. J., K. A. Oxenrider, J. Leng, J. S. Cantwell, and N. Zhao. 1993. Identification of a serine/threonine-specific protein phosphatase from the Archaebacterium Sulfolobus solfataricus. J. Biol. Chem. 268:6505-6510. [PubMed] [Google Scholar]
- 33.Kujo, C., and T. Oshima. 1998. Enzymological characterization of the hyperthermostable NAD-dependent glutamate dehydrogenase from the archaeon Pyrobaculum islandicum and effects of denaturants and organic solvents. Appl. Environ. Microbiol. 64:2152-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laderman, K. A., K. Asada, T. Uemori, H. Mukai, Y. Taguchi, I. Kato, and C. Anfinsen. 1993. α-Amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. Cloning and sequencing of the gene and expression in Escherichia coli. J. Biol. Chem. 268:24402-24407. [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Lam, H.-Y. P. 1984. Tamoxifen is a calmodulin antagonist in the activation of cAMP phosphodiesterase. Biochem. Biophys. Res. Commun. 118:27-32. [DOI] [PubMed] [Google Scholar]
- 37.Leng, J., A. J. Cameron, S. Buckel, and P. J. Kennelly. 1995. Isolation and cloning of a protein-serine/threonine phosphatase from an archaeon. J. Bacteriol. 177:6510-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leonard, C. J., L. Aravind, and E. V. Koonin. 1998. Novel families of putative protein kinases in Bacteria and Archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 8:1038-1047. [DOI] [PubMed] [Google Scholar]
- 39.Lower, B. H., K. M. Bischoff, and P. J. Kennelly. 2000. The archaeon Sulfolobus solfataricus contains a membrane-associated protein kinase activity that preferentially phosphorylates threonine residues. J. Bacteriol. 182:3452-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lower, B. H., and P. J. Kennelly. 2002. The membrane-associated protein-serine/threonine kinase from Sulfolobus solfataricus is a glycoprotein. J. Bacteriol. 184:2614-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mai, B., G. Frey, R. V. Swanson, E. J. Mathur, and K. O. Stetter. 1998. Molecular cloning and functional expression of a protein-serine/threonine phosphatase from the hyperthermophilic archaeon Pyrodictium abyssi TAG11. J. Bacteriol. 180:4030-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merker, R. I., and F. A. Troy. 1990. Biosynthesis of the polysialic acid capsule in Escherichia coli K1. Cold inactivation of sialic acid synthase regulates capsule expression below 20 degrees C. Glycobiology 1:93-100. [DOI] [PubMed] [Google Scholar]
- 43.Meyer, T., U. Regenass, D. Fabbro, E. Alteri, J. Rosel, M. Muller, G. Caravatti, and A. Matter. 1989. A derivative of staurosporine (CGP 41 251) shows selectivity for protein kinase C inhibition and in vitro anti-proliferative as well as in vivo anti-tumor activity. Int. J. Cancer 43:851-856. [DOI] [PubMed] [Google Scholar]
- 44.Morsomme, P., M. Chami, S. Marco, J. Nader, K. A. Ketchum, A. Goffeau, and J.-L. Rigaud. 2002. Characterization of a hyperthermophilic P-type ATPase from Methanococcus jannaschii expressed in yeast. J. Biol. Chem. 277:29608-29616. [DOI] [PubMed] [Google Scholar]
- 45.Munoz-Dorado, J., N. Almaula, S. Inouye, and M. Inouye. 1993. Autophosphorylation of nucleoside diphosphate kinase from Myxococcus xanthus. J. Bacteriol. 175:1176-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Brian, C. A., R. M. Liskamp, D. H. Solomon, and I. B. Weinstein. 1985. Inhibition of protein kinase C by tamoxifen. Cancer Res. 45:2462-2465. [PubMed] [Google Scholar]
- 47.Ono-Saito, N., I. Niki, and H. Hidaka. 1999. H-Series protein kinase inhibitors and potential clinical applications. Pharmacol. Ther. 82:123-131. [DOI] [PubMed] [Google Scholar]
- 48.Osorio, G., and C. A. Jerez. 1996. Adaptive response of the archaeon Sulfolobus acidocaldarius BC65 to phosphate starvation. Microbiology 142:1531-1536. [DOI] [PubMed] [Google Scholar]
- 49.Oxenrider, K. A., M. E. Rasche, M. V. Thorsteinsson, and P. J. Kennelly. 1993. An okadaic acid-sensitive protein phosphatase from the archaeon Methanosarcina thermophila TM-1. FEBS Lett. 331:291-295. [DOI] [PubMed] [Google Scholar]
- 50.Ponting, C. P., L. Aravind, J. Schultz, P. Bork, and E. V. Koonin. 1999. Eukaryotic signaling domain homologues in Archaea and Bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 289:729-745. [DOI] [PubMed] [Google Scholar]
- 51.Prozialeck, W. C., and B. Weiss. 1982. Inhibition of calmodulin by phenothiazines and related drugs: structure-activity relationships. J. Pharmacol. Exp. Ther. 222:509-516. [PubMed] [Google Scholar]
- 52.Rudolph, J., and D. Oesterhelt. 1995. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarium. EMBO J. 14:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudolph, J., N. Tolliday, C. Schmitt, S. C. Schuster, and D. Oesterhelt. 1999. Phosphorylation in halobacterial signal transduction. EMBO J. 14:4249-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saitoh, T., and J.-P. Changeaux. 1981. Change in the state of phosphorylation of acetylcholine receptor during maturation of the electromotor synapse in Torpedo marmorata electric organ. Proc. Natl. Acad. Sci. USA 78:4430-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott, J. D., E. H. Fischer, K. Takio, J. G. Demaille, and E. G. Krebs. 1985. Amino acid sequence of the heat-stable inhibitor of the cAMP-dependent protein kinase from rabbit skeletal muscle. Proc. Natl. Acad. Sci. USA 82:4379-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. K. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. R. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi, L., K. M. Bischoff, and P. J. Kennelly. 1999. The icfG gene cluster of Synechocystis sp. strain PCC 6803 encodes an Rsb/Spo-like protein kinase, protein phosphatase, and two phosphoproteins. J. Bacteriol. 181:4761-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi, L., M. Potts, and P. J. Kennelly. 1998. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms. A family portrait. FEMS Microbiol. Rev. 22:229-253. [DOI] [PubMed] [Google Scholar]
- 60.Shioji, K., H. Imai, I. Ueda, Y. Tanigawa, and M. Shimoyama. 1978. Tyrosine aminotransferase from chick liver. Heat activation and cold inactivation of the enzyme. Biochim. Biophys. Acta 522:96-103. [DOI] [PubMed] [Google Scholar]
- 61.Sigoillot, F. D., D. R. Evans, and H. I. Guy. 2002. Autophosphorylation of the mammalian multifunctional protein that inhibits de novo pyrimidine biosynthesis. J. Biol. Chem. 277:24809-24817. [DOI] [PubMed] [Google Scholar]
- 62.Skorko, R. 1984. Protein phosphorylation in the Archaebacterium Sulfolobus acidocaldarius. Eur. J. Biochem. 145:617-622. [DOI] [PubMed] [Google Scholar]
- 63.Skorko, R. 1989. Polyphosphate as a source of phosphoryl group in protein modification in the archaebacterium Sulfolobus acidocaldarius. Biochimie 71:1089-1093. [DOI] [PubMed] [Google Scholar]
- 64.Smith, R. F., and K. Y. King. 1995. Identification of a eukaryotic-like protein kinase gene in the Archaebacteria. Protein Sci. 4:126-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith, S. C., B. McCartney, P. J. Kennelly, and M. Potts. 1997. Protein-tyrosine phosphorylation in the Archaea. J. Bacteriol. 179:2418-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Solow, B., K. M. Bischoff, M. J. Zylka, and P. J. Kennelly. 1998. Archaeal phosphoproteins. Identification of a hexosephosphate mutase and the α-subunit of succinyl-CoA synthetase in the extreme acidothermophile Sulfolobus solfataricus. Protein Sci. 7:105-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Solow, B., J. C. Young, R. H. White, and P. J. Kennelly. 1997. Gene cloning and expression and characterization of a toxin-sensitive protein phosphatase from the methanogenic archaeon Methanosarcina thermophila TM-1. J. Bacteriol. 179:5072-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spudich, E. N., and J. L. Spudich. 1981. Photosensitive phosphoproteins in Halobacteria: regulatory coupling of transmembrane proton flux and protein dephosphorylation. J. Cell Biol. 91:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spudich, J. L., and W. Stoeckenius. 1980. Light-regulated retinal-dependent reversible phosphorylation of Halobacterium proteins. J. Biol. Chem. 255:5501-5503. [PubMed] [Google Scholar]
- 70.Stemmer, C., A. Schwander, G. Bauw, P. Fojan, and K. D. Grasser. 2002. Protein kinase CK2 differentially phosphorylates maize chromosomal high mobility group B (HMGB) proteins modulating their stability and DNA interactions. J. Biol. Chem. 277:1092-1098. [DOI] [PubMed] [Google Scholar]
- 71.Su, H.-D., G. J. Mazzei, W. R. Vogler, and J. F. Kuo. 1985. Effect of tamoxifen, a nonsteroidal antiestrogen, on phospholipid/calcium-dependent protein kinase and phosphorylation of its endogenous substrate proteins from the rat brain and ovary. Biochem. Pharmacol. 34:3649-3653. [DOI] [PubMed] [Google Scholar]
- 72.Taylor, S. S., D. R. Knighton, J. Zheng, J. M. Sowadski, C. S. Gibbs, and M. J. Zoller. 1993. A template for the protein kinase family. Trends Biochem. Sci. 18:84-89. [DOI] [PubMed] [Google Scholar]
- 73.Tonks, N. K., C. D. Diltz, and E. H. Fischer. 1988. Purification of the major protein-tyrosine-phosphatases of human placenta. J. Biol. Chem. 263:6722-6730. [PubMed] [Google Scholar]
- 74.Witzmann, S., and H. Bisswanger. 1998. The pyruvate dehydrogenase complex from thermophilic organisms: thermal stabilty and re-association from the enzyme components. Biochim. Biophys. Acta 1385:341-352. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, W., and B. T. Chait. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 72:2482-2489. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, W., M. Inouye, and S. Inouye. 1996. Reciprocal regulation of the differentiation of Myxococcus xanthus by Pkn5 and Pkn6, eukaryote-like Ser/Thr kinases. Mol. Microbiol. 20:435-447. [DOI] [PubMed] [Google Scholar]